Abstract
G-quadruplexes represent a versatile sensing platform for the construction of label-free molecular detection assays owing to their diverse structures that can be selectively recognized by G-quadruplex-specific luminescent probes. In this Survey and Summary, we highlight recent examples of the application of the label-free strategy for the development of G-quadruplex-based luminescent detection platforms with a view towards the potential application of tetraplex structures in the design of DNA logic gates.
INTRODUCTION
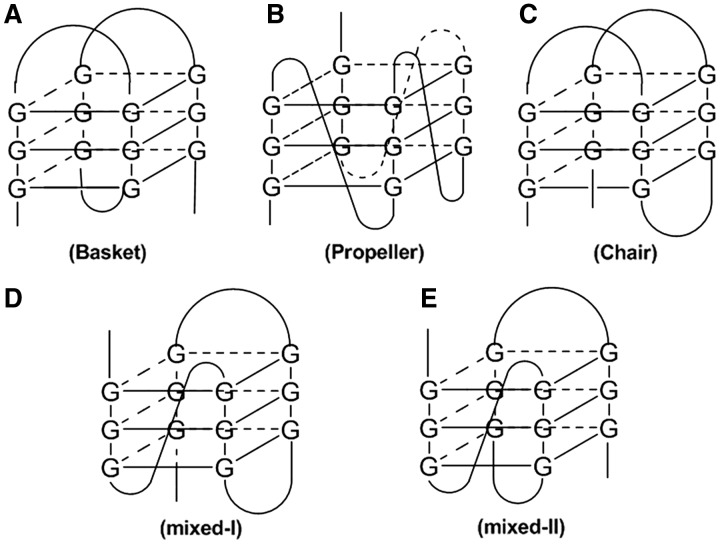
The G-quadruplex structure consists of planar stacks of guanine tetrads stabilized by eight Hoogsteen and Watson–Crick hydrogen bonds (N1–O6, N2–N7), with the O6 atoms orientated centrally into the ring to form an anionic bipyramidal cage that can coordinate to monovalent cations such as Na+ or K+ (Figure 1). The overall G-quadruplex structure may be formed from an intramolecular (a single strand folds on itself) or intermolecular (formed by two or more strands) arrangement of guanine-rich oligonucleotides. The π-stacked guanine tetrads are connected by intervening variable-length sequences that form loops that are situated on the exterior of the core. These may be classified as diagonal, lateral (also called edgewise) or propeller (also called double chain reversal) (1).
Figure 1.

Chemical structure of a guanine tetrad.
In contrast to the standard B-form of double-helical DNA, G-quadruplexes show a rich diversity in structural topologies that can be sensitive to the number and length of the guanine tracts, the lengths of the intervening loop regions and the character of the metal ion in solution (Figure 2) (2). For example, the orientation of the strands may be either parallel or anti-parallel, or both conformations (termed hybrid) may be present in some G-quadruplexes, such as the (3 + 1) G-quadruplex motif in human telomeric DNA (3). Furthermore, it has been proposed that up to 26 possible topologies are available for a three-loop G-quadruplex (4). An oligonucleotide in solution may therefore exist as one or a mixture of several different quadruplex forms that may possibly be in dynamic equilibrium with each other. Consequently, the presence and precise function of G-quadruplexes in vivo is still an active area of debate. The fascinating variety of folding topologies, molecular interfaces and novel interaction surfaces available to DNA/RNA G-quadruplexes has been recently reviewed by Collie and Parkinson (5).
Figure 2.
Schematic representation of G-quadruplex topologies.
The extensive structural polymorphism of G-quadruplexes has rendered them as attractive signal-transducing elements for the development of DNA-based probes. DNA oligonucleotides are versatile components for the construction of sensing platforms owing to their low cost, ease of synthesis, high solubility, biocompatibility and stability in aqueous solution and biological media (6–9). Early studies in the field of DNA-based sensing typically used labelled oligonucleotides that are covalently conjugated with donor/acceptor or fluorophore/quencher pairs (10–12). However, fluorescent labelling can be relatively expensive and time-consuming, and the covalent attachment of the fluorophore may influence the binding affinity or selectivity of the functional oligonucleotide, thus potentially interfering with the operation of the assay (9). The ‘label-free’ strategy has emerged as a simple and cost-effective alternative to the use of fluorescently labelled oligonucleotides in DNA-based sensing, whereby luminescent probes are not covalently attached to the nucleic acid backbone but instead interact non-covalently with DNA through a number of binding modes such as intercalation, groove binding, end stacking or electrostatic interactions (13). In this context, we define the term label-free to include ‘natural’ oligonucleotides that are not modified with fluorescent nucleobases or covalent fluorophores.
The label-free approach relies on the specific interaction between oligonucleotides and selective DNA probes. In recent years, immense efforts have been invested into the development of specific probes for detecting and distinguishing G-quadruplexes from other DNA conformations likely to be present in the cellular environment, including the predominant double helix. G-quadruplex-selective ligands tend to contain planar aromatic or heteroaromatic systems that can interact with the terminal faces of the G-quadruplex through π–π stacking interactions (14). Further selectivity for a particular sequence may be achieved by targeting the more structurally heterogeneous groove and loop regions that are distinctive for each G-quadruplex topology (15).
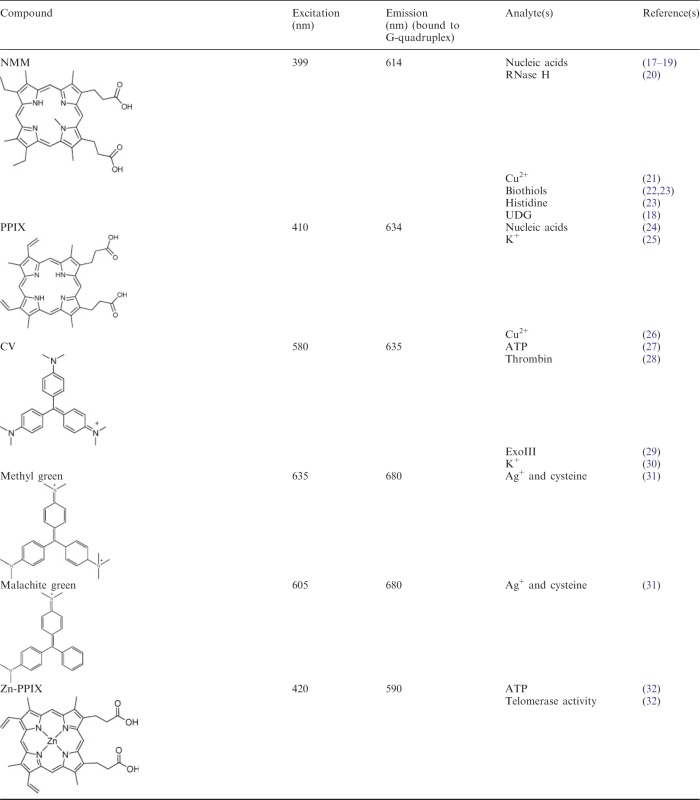
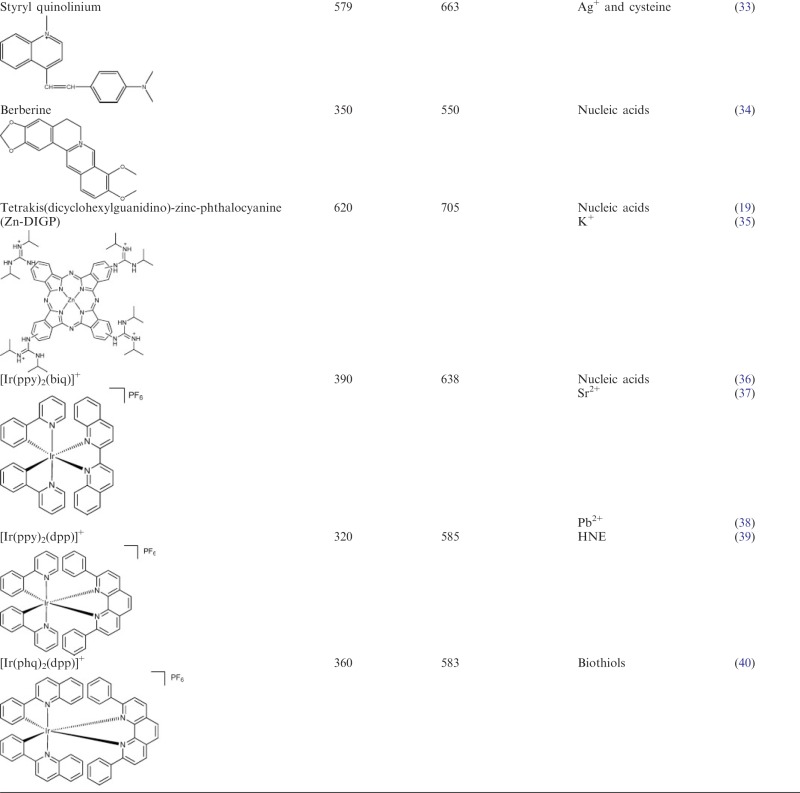
Both organic dyes and inorganic metal complexes have found use for the development of label-free G-quadruplex-based detection platforms. The most popular classes of fluorescent G-quadruplex dyes include those derivatives based on carbocyanine, porphyrin, ethidium, carbazole and triphenylmethane, whereas phosphorescent G-quadruplex-selective metal complexes based on platinum(II), ruthenium(II) and iridium(III) centres have been reported (16). A summary of the luminescent G-quadruplex probes used for the construction of label-free DNA assays described in this review are presented in Table 1.
Table 1.
Luminescent G-quadruplex-selective probes utilized for label-free DNA-based detection platforms and logic gates
 |
 |
Luminescence offers a versatile combination of simplicity, low cost and high sensitivity that compares favourably with colorimetric or electrochemical detection methods. Furthermore, portable spectrofluorometers can be used for the in-field monitoring of analytes, which cannot readily be achieved for instrumental techniques that require extensive sample preparation and expensive instrumentation. Recent summary articles by Wang and co-workers (41) and Uttamchandani and co-workers (42) describe general luminescent and colorimetric G-quadruplex-based approaches for the detection of analytes. Our review highlights recent examples of the ‘label-free’ strategy for the construction of simple and cost-efficient G-quadruplex-based detection platforms with a view towards potential application of tetraplex structures in the design of label-free DNA logic gates.
CASE STUDIES
Detection of DNA
DNA and RNA play pivotal roles in controlling the function and behaviour of cells through the central processes of transcription and translation, and mutations or malfunction in the genetic code can represent potential biomarkers for human disease. This has stimulated the development of analytical tools for the specific detection of nucleic acid sequences for potential application in genetic profiling and disease diagnosis (43). In 1996, Tyagi and Kramer (10) developed the ‘molecular beacon’ strategy for the detection of specific DNA sequences in homogenous solution. The classical molecular beacon is a doubly labelled hairpin oligonucleotide whose fluorescence is internally quenched in the absence of the target. However, binding of a complementary target sequence ‘opens up’ the molecular beacon and separates the fluorophore from the quenching, thereby restoring fluorescence. Other molecular beacons based on the covalent conjugation of oligonucleotides to a signalling component such as organic dyes, quantum dots or gold nanoparticles have also been reported (44–48).
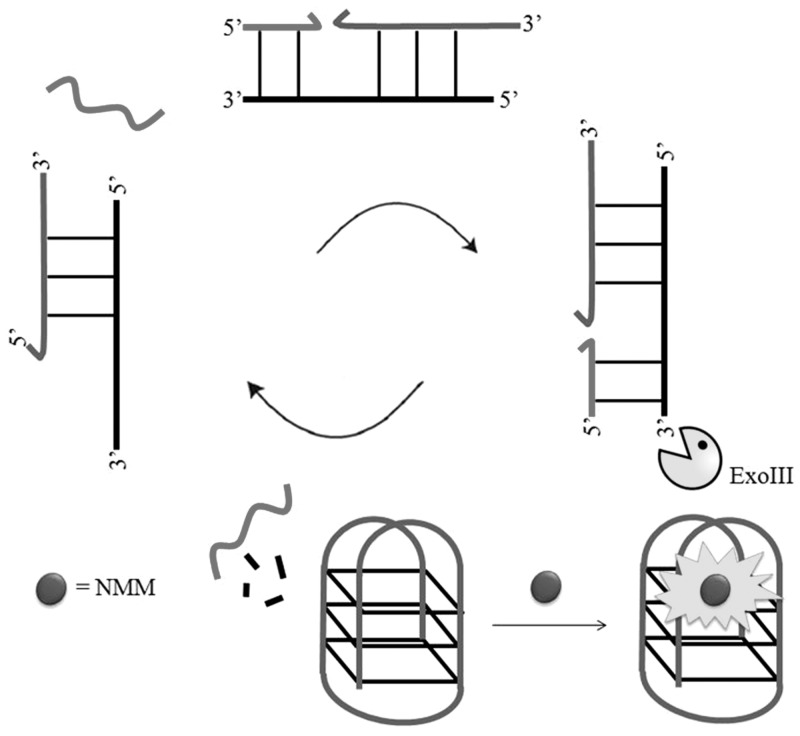
The catalytic activity of DNA-cleaving enzymes has been harnessed to amplify luminescence signals arising from target recognition, thereby potentially enhancing the sensitivity of the assay. For example, Plaxco and co-workers (49) have developed a simple and rapid exonuclease III (ExoIII)-aided target recycling method for amplified DNA detection using labelled oligonucleotides with astounding attomolar sensitivity. In the context of label-free sensing, Qu and co-workers (17) have used ExoIII to generate an amplified DNA detection system based on assisted strand-cleavage cycles and ligand-responsive quadruplex formation. A duplex DNA probe was designed with one strand as a G-quadruplex-forming oligomer and the other as a complementary sequence to the target DNA (Figure 3). This duplex probe was specifically designed with 3′-protuding termini at both ends to prevent ExoIII digestion in the absence of analyte. The addition and hybridization of the target DNA with the duplex probe forms a blunt terminus that allows ExoIII digestion of the complementary sequence from the 3′-terminus, thereby releasing both the target DNA and the G-quadruplex-forming oligomer. The free G-quadruplex-forming oligomer folds into a quadruplex structure that is detected by the anionic porphyrin N-methyl mesoporphyrin IX (NMM) with a switch-on fluorescence response. The released target DNA is free to bind to another 5′-protruding terminus of duplex probe to trigger a new hydrolysis reaction, which leads to significant amplification of the signal. This method exhibited sub-picomolar sensitivity for the DNA target, which compared favourably with other amplification methods using labelled oligonucleotides.
Figure 3.
Enzyme-assisted G-quadruplex-based detection platform for DNA.
Ren and co-workers (18) have developed a G-quadruplex-based strand displacement assay for DNA detection using NMM as a luminescent probe. The duplex DNA probe consists of a probe strand (P2: 5′-CTCGATCGCACT2A2GA2T3C-3′) partially hybridized to a G-quadruplex-forming oligomer (P1: 5′-G3T4G3A3T2CT2A2GTGCG3T4G3-3′) (Figure 4). In the absence of the target, the system is only weakly fluorescent owing to the weak interaction between the DNA probe and NMM. The addition of the target strand (P3: 5′-GA3T2CT2A2GTGCGATCGAG-3′) forms a more stable P2–P3 duplex, which displaces P1, and the resulting G-quadruplex formed by the free P1 is detected by NMM with a switch-on fluorescence response.
Figure 4.
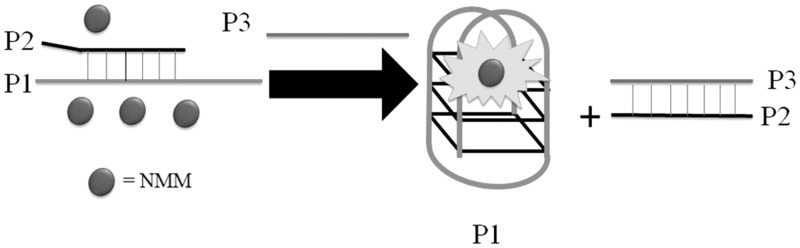
Luminescent switch-on detection of DNA based on the analyte-induced strand dissociation and subsequent G-quadruplex formation.
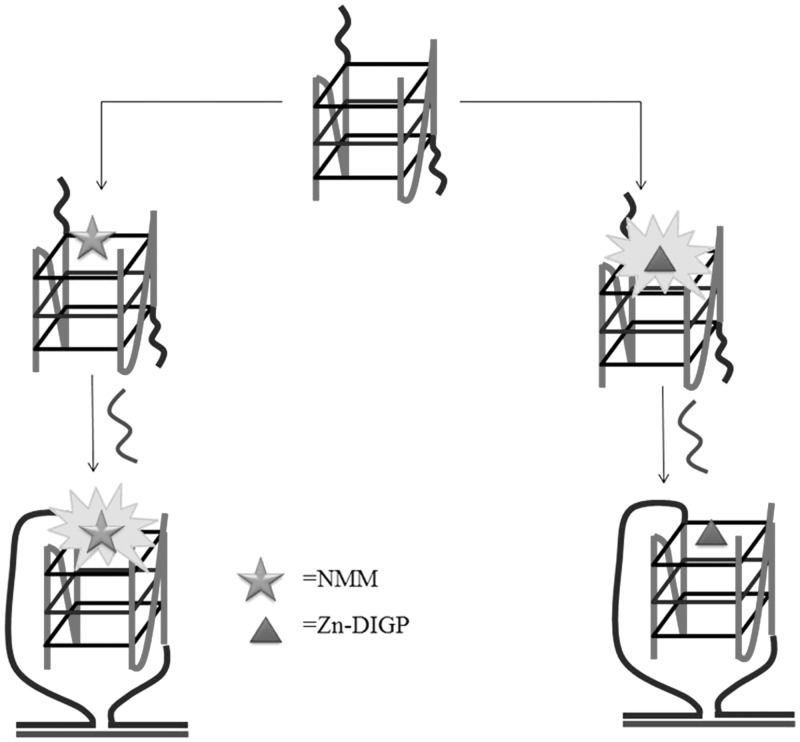
Wang and co-workers (19) used NMM and the cationic tetrakis(dicyclohexylguanidino)-zinc-phthalocyanine (Zn-DIGP) as G-quadruplex-selective probes to construct a G-quadruplex-based DNA detection assay (Figure 5). The authors designed a bimolecular c-myc G-quadruplex comprised from two fragment oligomers (5′-GCAGACACATC2AGT5GAG3TG4-3′ and 5′-AG3TG4A2T4CGATAGC2AG2ACA2-3′) that each contains flanking segments complementary to the target DNA sequence, derived from the hepatitis B viral gene (5′-T2GTC2TG2CTATCGCTG2ATGTGTCTGC-3′). The addition of complementary DNA to this solution further stabilizes the G-quadruplex structure while introducing additional conformational constraints into the complex, thereby changing the local environment and fluorescence properties of the dye molecules. Interestingly, NMM displayed enhanced fluorescence on target binding, whereas reduced fluorescence was observed for Zn-DIGP. Both the switch-on and switch-off modes of detection exhibited nanomolar sensitivity for DNA and could discriminate mismatched from perfectly matched target DNAs. Furthermore, the authors demonstrated the application of this assay for the detection of DNA in diluted biological fluids such as human serum or urine.
Figure 5.
G-quadruplex-based sensing platform for single-stranded DNA with switch-on (left) and switch-off (right) detection modes.
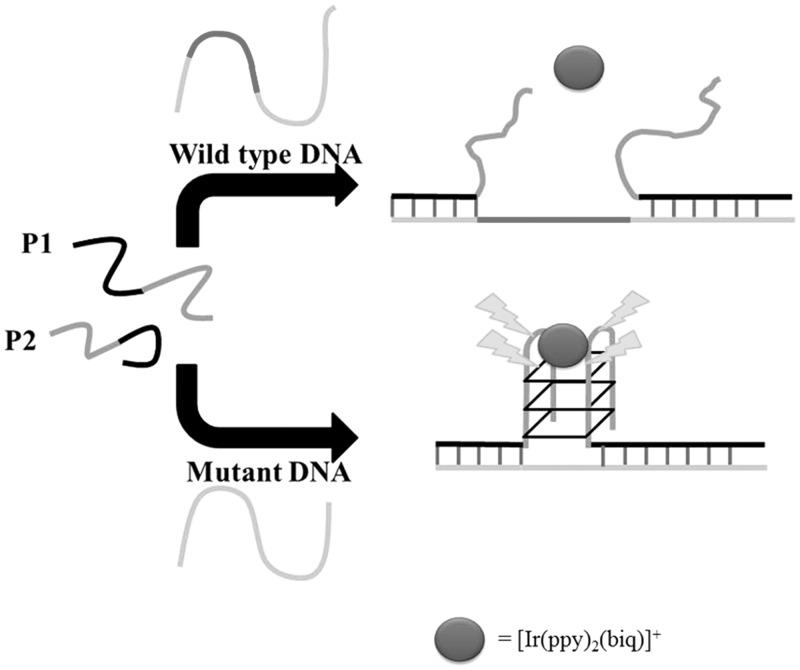
Our group has used the G-quadruplex-selective luminescent iridium(III) complex [Ir(ppy)2(biq)]+ (where ppy = 2-phenylpyridine and biq = 2,2′-biquinoline) as a signal transducer to develop a G-quadruplex-based detection platform for gene deletion (Figure 6) (36). We designed a split G-quadruplex comprising two short oligonucleotides, P1 (5′-ATGACTATCT3A2TG3TAG3-3′) and P2 (5′-G3T2G3CGTAG2A4TGAG-3′), that both contain complementary regions recognizing the DNA sequences flanking the deletion site of the chemokine receptor gene CCR5 (5′-CTCAT4C2ATACAT2A3GATAGTCAT-3′). The oligonucleotides also contain pendant guanine-rich overhangs that can form a split G-quadruplex when brought into close proximity. The CCR5 protein has been reported to be a co-receptor for macrophage-tropic Human immunodeficiency virus type 1 (HIV-1) strains and can be exploited by HIV-1 to gain entry into CD4+ T cells. However, the mutant allele CCR5-Δ32 does not produce a functional protein and thus confers resistance against HIV infection. In this assay, the longer wild-type CCR5 DNA sequence (5′-CTCAT4C2ATACAGTCAGTATCA2T2CTG2A2GA2T3C2AGACAT2A3GATAGTCAT-3′) is able to hybridize with P1 and P2, but the large spatial separation of the guanine-rich overhangs prevents effective split G-quadruplex formation. This results in a reduced luminescence signal of the iridium(III) complex in response to the single and double-stranded regions of the DNA. However, hybridization with the shorter mutant DNA sequence brings the two G-quadruplex-forming sequences into close proximity, promoting the formation of the split G-quadruplex and thereby producing a significant switch-on luminescence enhancement. Importantly, the assay was able to effectively discriminate between deletion mutants differing by only a single base, and the simple design of the system potentially allows specificity for a target sequence of any length by judicious modification of the split G-quadruplex oligonucleotides.
Figure 6.
Split G-quadruplex-based luminescent switch-on detection strategy for the detection of gene deletion. [Reproduced from (36) by permission of The Royal Society of Chemistry].
Similar split G-quadruplex-based strategies were subsequently reported by other groups for the selective detection of DNA length and sequence. Wang and co-workers (24) used protoporphyrin IX (PPIX) and a 3:1 split G-quadruplex as a ‘caliper’ to measure the length of DNA. The fluorescence of PPIX was found to be gradually and linearly reduced, as the DNA base length was increased. Consequently, calibration of PPIX fluorescence could allow the caliper to be used to measure the length of an unknown DNA sequence. This assay may be a convenient tool in recombination studies to determine whether the foreign DNA has been inserted into the cleavage site. Recently, Chen and co-workers (34) used the G-quadruplex-selective probe berberine for the construction of a fluorescent assay for the DNA fragment from the transgene cauliflower mosaic virus 35S promoter (5′-ATTGTGCGTCATCCCTTACGTCAGTGGAG-3′). Hybridization of the target DNA with two oligonucleotides derived from the human telomeric repeat, (5′-CTCCACTGACGTAATTAGGGTTAGGG-3′) and (5′-TTAGGGTTAGGGGGATGACGCACAAT-3′), leads to the formation of a split G-quadruplex that is strongly bound by berberine, thereby resulting in an enhanced fluorescence response. This assay had a detection limit of 2.0 nM.
Aptamer-based detection
Nucleic acid aptamers are available for a wide variety of different targets such as organic dyes, nucleotides, biological cofactors, amino acids, peptides and enzymes (50). Some aptamers adopt a G-quadruplex structure on binding to their cognate ligands. This conformational transition can be effectively monitored by the use of G-quadruplex selective luminescent molecules that are able to transduce the analyte binding event into a luminescent response.
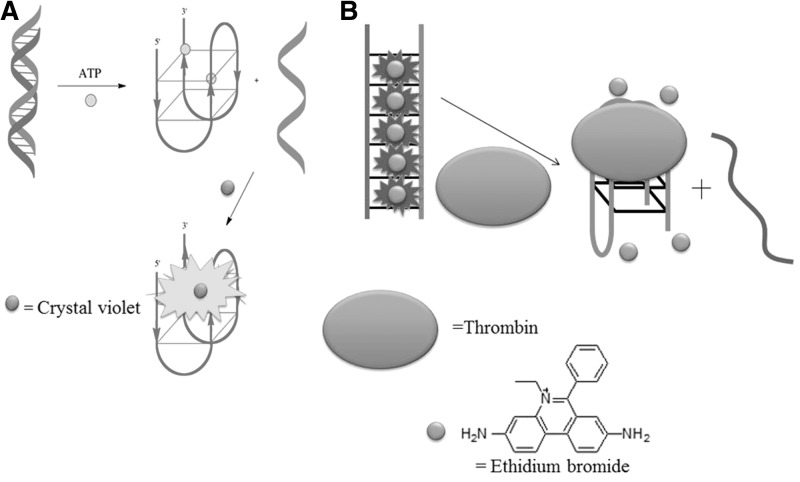
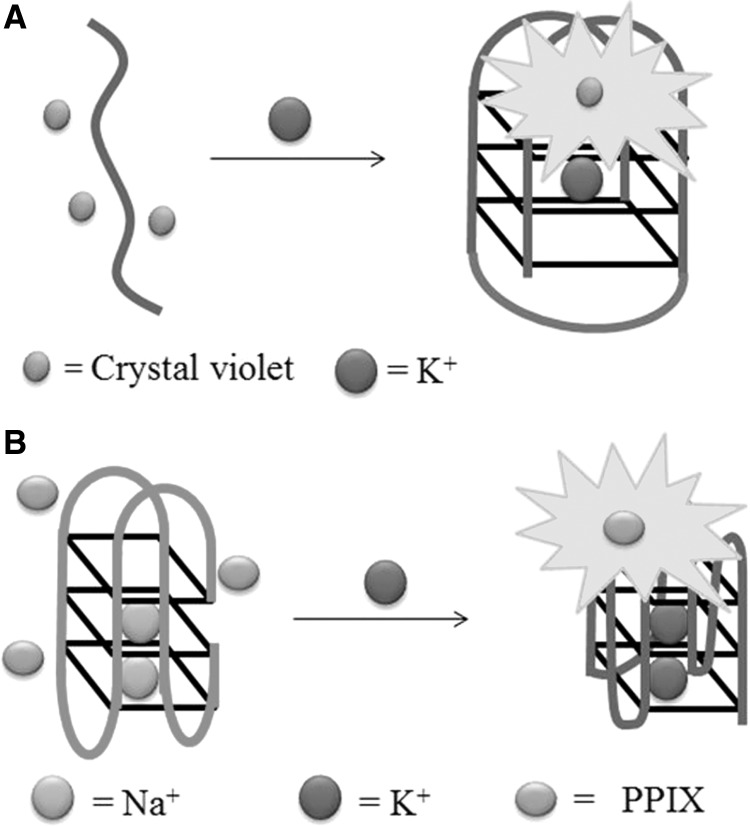
Our group envisaged that a switch-on aptamer-based detection method for adenosine triphosphate (ATP) could be developed using a duplex-to-quadruplex conversion strategy (27). The ATP aptamer is known to fold into a G-quadruplex on binding to ATP. We designed a duplex DNA probe consisting of the ATP aptamer (5′-A2C2TG5AGTAT2GCG2AG2A2G2T-3′) hybridized with its complementary sequence (5′-AC2T2C2TC2GCA2TACTC5AG2T2-3′). In the absence of the target DNA, the fluorescence of the G-quadruplex-selective crystal violet (CV) is weak. However, the addition of ATP promotes the dissociation of the duplex and subsequent formation of the G-quadruplex motif, which is detected by CV with a switch-on luminescence response (Figure 7A). We further demonstrated the selectivity of the assay for ATP over other nucleotide analogues and chemical species likely to be present in biological samples and applied this methodology to the detection of ATP in HeLa cell extracts. A similar approach was used by Willner and co-workers (32) who used zinc protoporphyrin-IX (Zn-PPIX) in conjunction with the free aptamer for the detection of ATP.
Figure 7.
G-quadruplex-based aptamer-based detection assay for (A) ATP and (B) thrombin.
The Dong group has developed a switch-off detection platform for thrombin by using the thrombin aptamer (5′-G2T2G2TGTG2T2G2-3′) in conjunction with ethidium bromide (EB) as a duplex probe (51). Initially, the thrombin aptamer is hybridized with a complementary DNA sequence to form a duplex structure, thus promoting intercalation and fluorescence emission of EB (Figure 7B). The addition of thrombin induces the dissociation of the thrombin aptamer due to the formation of the thrombin-aptamer G-quadruplex ensemble, resulting in a reduced fluorescence signal of EB owing to its weak interaction with G-quadruplex. A switch-on label-free detection platform for thrombin was devised by Jin and co-workers (28) using a single strand-to-quadruplex approach. This assay using CV as a G-quadruplex-selective dye achieved a detection limit of 8 pM for the detection of thrombin with selectivity over IgG and lysozyme.
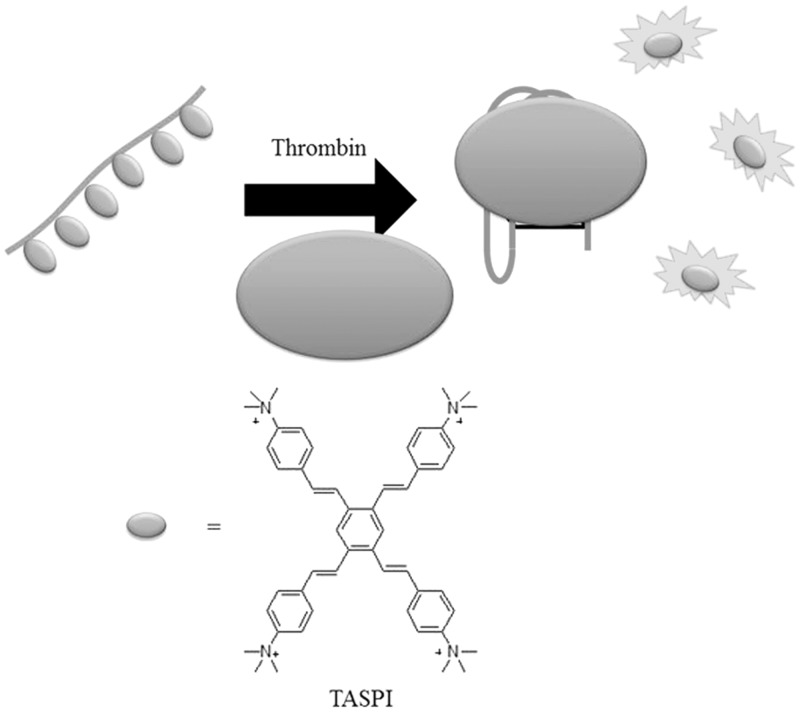
The Yan group has recently used the pyrazine derivative TASPI (E,E,E,E,-2,3,5,6-tetrakis[4-(trimethyl-amino)styryl]-pyrazinyl iodide) as a luminescent probe for the aptamer-based detection of thrombin (52). In the absence of thrombin, the emission of TASPI is quenched by its interaction with the aptamer owing to aggregation-induced quenching. However, the formation of the thrombin-aptamer G-quadruplex ensemble releases TASPI from the DNA and fluorescence signal of the dye is recovered (Figure 8). This method exhibited a detection limit of 50 nM and was selective for thrombin over trypsin, bovine serum albumin and tyrosinase.
Figure 8.
Switch-on detection of thrombin using TASPI.
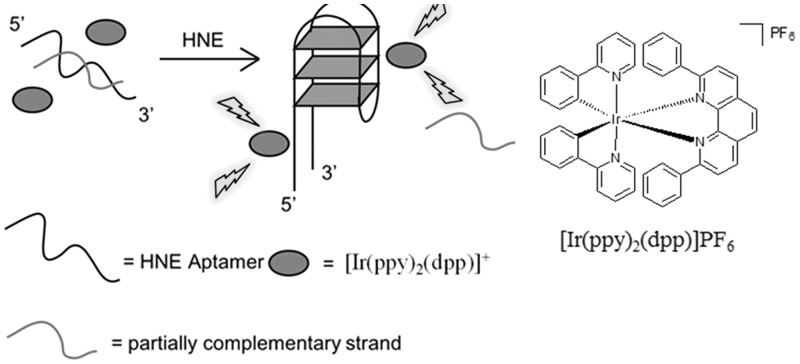
Recently, our research group has fabricated a switch-on aptamer-based detection method for human neutrophil elastase (HNE) using a duplex-to-quadruplex conversion strategy. A G-quadruplex-selective iridium(III) complex [Ir(ppy)2(dpp)]+ (where ppy = 2-phenylpyridine and dpp = 2,9-diphenyl-1,10-phenanthroline) was used as the signal transducer of the assay (Figure 9) (39). In this assay, the HNE aptamer (5′-TAGCGATACTGCGTG3T2G4CG2TAG3C2AGCAGTCTCGT-3′) is initially hybridized with a partially complementary strand (5′-G2C3TAC3GC4A2C3AC-3′), and the weak binding of the iridium(III) complex to the DNA duplex results in a weak luminescence signal. However, the addition of HNE induces the dissociation of the duplex structure and promotes the formation of the HNE protein-aptamer G-quadruplex ensemble. The strong interaction of the iridium(III) complex with the G-quadruplex motif results in a significant luminescence enhancement. This assay could detect as low as 0.04 nM of HNE with a linear dynamic range of in the range of 0.04–6 nM.
Figure 9.
G-quadruplex-based aptamer-based detection assay for HNE using a luminescent iridium(III) complex [Ir(ppy)2(dpp)]+. [Reproduced from (39) by permission of The Royal Society of Chemistry].
Detection of metal ions
Metal ions are prevalent in biological systems and are centrally involved in essential life processes, such as energy metabolism and storage, signal transduction and nucleic acids processing. In recent years, oligonucleotides have been found represent a versatile sensing platform for the sensing of various metal ions (54). Besides K+ and Na+ ions, divalent metal cations such as Ca2+ (55), Sr2+ (56) or Pb2+ (57) ions have also been reported to stabilize the G-quadruplex. Furthermore, Ono and co-workers (58,59) have pioneered the application of the thymine–Hg2+–thymine and cytosine–Ag+–cytosine interactions to develop fluorescent molecular beacons for detection of these cations. We highlight recent examples of label-free strategies that have been used to harness the metal ion-binding properties of nucleic acids for the construction of G-quadruplex-based luminescent detection platforms.
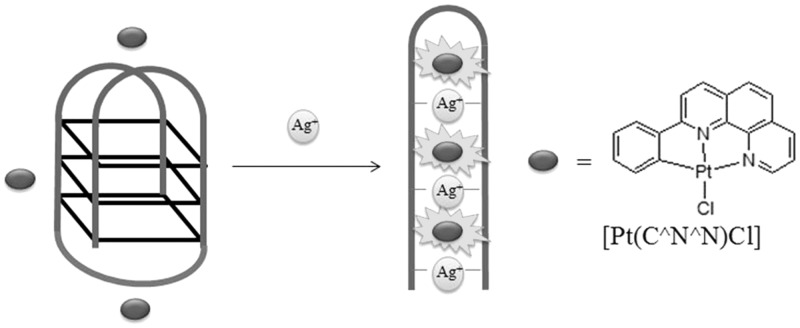
Our group has developed a label-free G-quadruplex-based assay for Ag+ ions based on the Ag+-induced quadruplex-to-duplex transition of a specially designed guanine and cytosine-rich oligonucleotide (Figure 10) (53). Instead of using a G-quadruplex-selective ligand, we chose to use a platinum(II) metallointercalator to achieve a switch-on luminescence in response to Ag+ ions. In the absence of Ag+, the weak interaction between the platinum(II) complex and the G-quadruplex motif resulted in a low luminescence background. The addition of Ag+ ions converts the G-quadruplex into a duplex conformation via the formation of cytosine–Ag+–cytosine mismatched base pairing, resulting in the intercalation of the platinum(II) metallointercalator and thus recovering its luminescence signal.
Figure 10.
G-quadruplex-to-duplex transition for the detection of Ag+ ions using a platinum(II) metallointercalator.
Kong and co-workers (30) have used CV to study the effect of K+ ions on the topology of the human telomeric G-quadruplex sequence [5′-(G3T2A)3G3-3′]. The addition of K+ ions was found to induce the formation of the G-quadruplex motif, thereby resulting in a switch-on fluorescence response (Figure 11A). Interestingly, the use of other G-quadruplex-forming sequences in conjunction with CV generated switch-on detection modes for the K+ ion (60). Wang, Dong and co-workers (25) have developed a variation of this strategy using ion-responsive oligonucleotide PS2.M (5′-GTG3TAG3CG3T2G2-3′) and the PPIX as a parallel G-quadruplex fluorescent probe. In the presence of Na+ ions, PS2.M exists in an anti-parallel quadruplex topology that interacts weakly with PPIX (Figure 11B). Upon the addition of K+, the anti-parallel G-quadruplex converts into a parallel conformation and interacts strongly with PPIX to create a significant luminescence enhancement (Figure 11B). A similar strategy was reported by Wang and co-workers (35) using the c-myc G-quadruplex sequence (5′-TGAG3TG4AG3TG4A2-3′) and Zn-DIGP. This assay exhibited a 3500-fold selectivity for K+ ions over Na+ ions.
Figure 11.
G-quadruplex-based switch-on detection assays for K+ ions using G-quadruplex-selective luminescent probes.
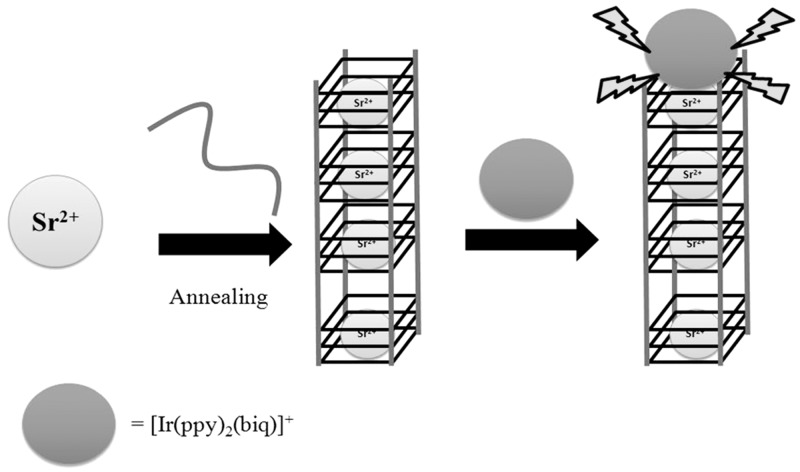
Besides potassium, Pb2+- (38,61,62) or Sr2+- (37,63) mediated G-quadruplex structures have also been used for the label-free luminescent detection of these metal ions. Our group has used the G-quadruplex-forming sequence T2 (5′-G4T2G4T2G4T2G4-3′) and a phosphorescent iridium(III) complex [Ir(ppy)2(biq)]+ to construct a DNA-based detection platform for Sr2+ (37). In the absence of Sr2+ ions, the oligonucleotide T2 exists in a random coil conformation that interacts only weakly with the iridium(III) complex, resulting in a low luminescent signal. However, the addition of Sr2+ ions induces the formation of the G-quadruplex motif, thereby enhancing the luminescence of the iridium(III) complex (Figure 12). Qu and co-workers (63) have reported a similar strategy using single-walled carbon nanotubes (SWNTs) and the luminescent dye thiazole orange (TO) to develop a G-quadruplex-based for Sr2+ ion detection. In the absence of analyte, the human telomeric DNA sequence (HTS: 5′-AG3T2AG3T2AG3T2AG3-3′) is strongly adsorbed onto the SWNT surface, and the fluorescence of TO is low. The addition of Sr2+ ions triggers the dissociation of HTS from the SWNTs, and the resulting G-quadruplex structure is strongly bound by TO with a switch-on fluorescence response. Both assays were able to selectively detect nanomolar Sr2+ ions. Recently, our research group have reported a similar strategy for sub-nanomolar Pb2+ detection by using the oligonucleotide PS2.M and a luminescent iridium(III) complex [Ir(ppy)2(biq)]+ (38). On binding to Pb2+ ions, the single-stranded PS2.M is induced into a G-quadruplex conformation, which greatly enhances the luminescence emission of the iridium(III) complex. Furthermore, the application of the assay for the detection of nanomolar Pb2+ ions in spiked river water samples was demonstrated.
Figure 12.
Schematic representation of the G-quadruplex-selective probe for the detection of Sr2+ ions. [Reproduced from (37) by permission of The Royal Society of Chemistry].
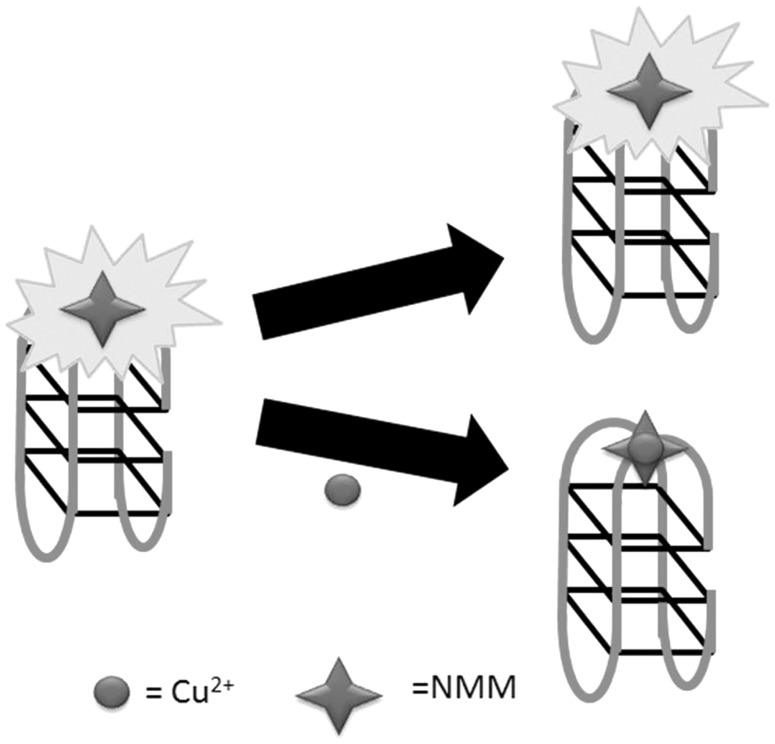
In 2010, Wang, Wang and co-workers (21) fabricated a switch-off fluorescent assay for the detection of Cu2+ ions using the G-quadruplex probe NMM as a signal transducer (Figure 13). In the absence of Cu2+ ions, the G-quadruplex formed from 24GT (5′-G3T4G3T4G3T4G3-3′) is strongly bound by NMM with a strong fluorescence response. However, the fluorescence of the system was gradually quenched by the continuous addition of Cu2+ ions. The assay could detect as low as 83 nM of Cu2+ with linear detection range from 0.083 to 10 mM. Later on, the same group reported a similar strategy for the switch-off detection of Cu2+ ions using the PS5.M oligonucleotide (5′-GTG3TCAT2GTG3TG3TGTG2-3′) and the G-quadruplex probe PPIX (26).
Figure 13.
Schematic representation of the G-quadruplex-selective probe for the switch-off detection of Cu2+ ions.
Detection of biothiols
Cysteine (Cys), homocysteine (Hcy) and glutathione (GSH) are biological thiols (biothiols) that play important roles in maintaining cellular functions. GSH is the most abundant thiol in animal cells and is responsible for regulating redox homeostasis within the cell, whereas the cysteine/cystine redox ratio largely determines the redox state of human plasma. Aberrations in the levels of biothiols have been implicated in the development of various diseases such as psoriasis, cardiovascular disease and cancers (64).
Ren, Qu and co-workers (22) designed a label-free detection fluorescent assay for the detection of nanomolar biothiols in aqueous solution and human plasma using a silver metallization-engineered conformational switch (Figure 14). In the absence of biothiols, silver metallization effectively prevents the oligonucleotide [5′-G3(T4G3)3-3′] from forming a G-quadruplex structure, resulting in the weak fluorescence of NMM. However, biothiols are able to sequester silver via the strong Ag+–S interaction and release the DNA from the silver surface. The free oligonucleotide is able to fold into a G-quadruplex and interact strongly with NMM, thus producing a significant luminescence enhancement. The principle of DNA/biothiol competition for Ag+ ions was also used by the group of Qiu and Chen (33) and by the group of Kong (31) for the dual detection of Ag+ and cysteine using the G-quadruplex-selective probes styryl quinolinium and triphenylmethane dye (methyl green or malachite green), respectively. Qiu, Chen and co-workers further demonstrated the application of this assay for the detection of cysteine in human urine.
Figure 14.
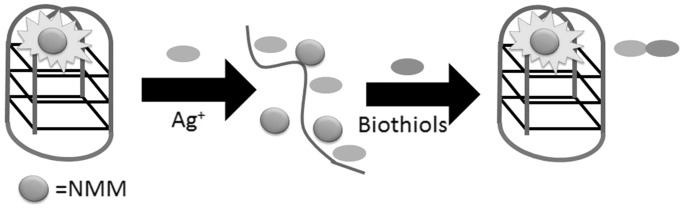
Schematic illustration of the G-quadruplex-based assay for the detection of biothiols based on the selective complexation between silver ion and biothiols.
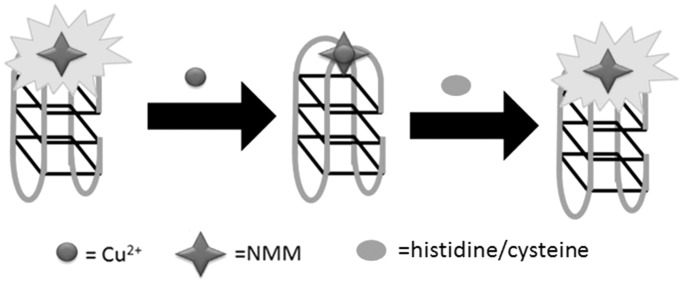
Liu, Wang and co-workers (23) designed a label-free, G-quadruplex-based approach for the simultaneous detection of histidine and cysteine by using the fluorescence quencher Cu2+ (Figure 15). In the presence of G-quadruplex formed from 24GT (5′-G3T4G3T4 G3T4G3-3′), the fluorescence intensity of NMM was dramatically enhanced. However, addition of Cu2+ effectively quenched the fluorescence of the NMM/G-quadruplex ensemble owing to the chelation of Cu2+ ions by NMM as well as the Cu2+-induced unfolding of the G–quadruplex motif. The fluorescence of the system could be recovered by the addition of histidine or cysteine owing to the strong binding affinity of Cu2+ to the imidazole group of histidine or the thiol group of cysteine. In this strategy, a high selectivity is conferred by the use of the cysteine-masking agent N-ethylmaleimide (NEM), which helps to discriminate histidine from cysteine. This assay could detect as low as 3 nM of histidine and 5 nM of cysteine in aqueous solution.
Figure 15.
Schematic illustration of the G-quadruplex-based assay for the detection of histidine/cysteine.
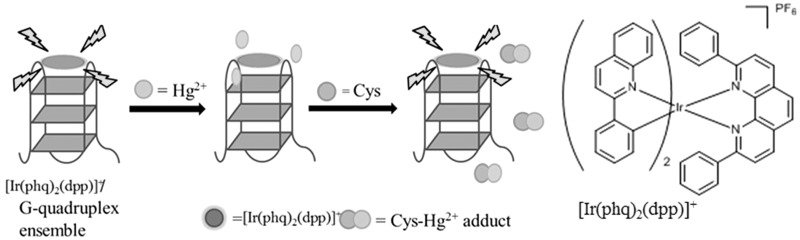
Recently, our group used a G-quadruplex-selective luminescent iridium(III) complex and mercury(II) ions for the selective detection of biothiols in aqueous solution (Figure 16) (40). The iridium(III) complex [Ir(phq)2(dpp)]+ (where phq = 2-phenylquinoline and dpp = 2,9-diphenyl-1,10-phenanthroline) is highly emissive in the presence of the PS2.M G-quadruplex. The addition of Hg2+, a well-known fluorescence quencher, results in significant quenching of the luminescence signal of the metal complex, presumably due to various electron-transfer mechanisms. However, biothiols can effectively sequester Hg2+ ions by the formation of the strong Hg(II)–S bond, resulting in the restoration of luminescence signal of the complex–G-quadruplex ensemble. Significantly, the assay exhibited a tunable range for the detection of biothiols, which could be controlled by varying the concentration of Hg2+ ions.
Figure 16.
Schematic illustration of the label-free G-quadruplex-based assay for the detection of Cys using [Ir(phq)2(dpp)]+ as signal transducing element. [Reproduced from (40) by permission of The Royal Society of Chemistry].
Detection of enzyme activity
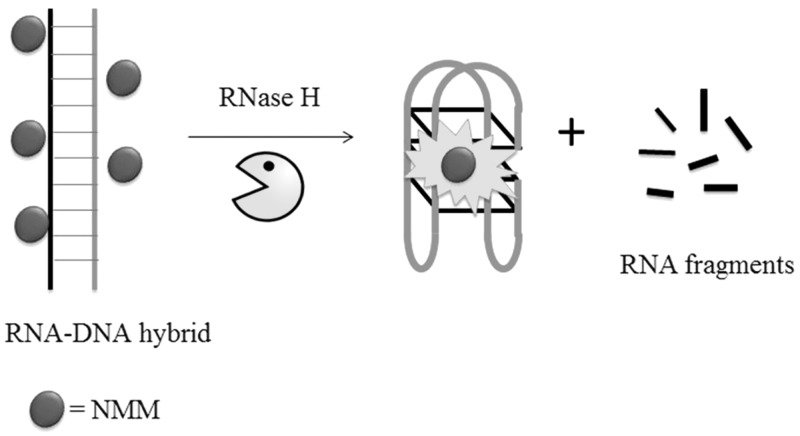
In label-free G-quadruplex-based enzyme assays, the design of the oligonucleotide substrate is highly flexible and depends primarily on the activity of the targeted enzyme. Ren and co-workers (20) have designed a DNA–RNA hybrid duplex motif for the label-free detection of RNase H activity. RNase H is a ribonuclease that specifically degrades the RNA strand of an RNA–DNA duplex (66). Importantly, the RNase H activity of HIV-1 reverse transcriptase is required for retroviral DNA synthesis, and thus wide efforts have been invested into the discovery of inhibitors against this activity (67). The DNA–RNA hybrid consists of a short G-rich oligonucleotide hybridized to a complementary RNA sequence, and the G-quadruplex-specific fluorescent dye NMM is added to the sample mixture as a signal transducing unit (Figure 17). In the absence of RNase H, the fluorescence signal of NMM is low owing to the weak interaction of the dye with the intact hybrid duplex. The addition of RNase H results in the digestion of the RNA strand, and subsequently releasing the guanine-rich DNA oligonucleotide that could fold into an intramolecular G-quadruplex. The NMM dye thus interacts strongly with the nascent G-quadruplex, producing a significant ‘switch-on’ luminescence response. The authors further demonstrated the potential application of this assay for screening RNase H inhibitors.
Figure 17.
Schematic diagram of the label-free luminescent detection assay for RNase H activity using a duplex-to-quadruplex approach.
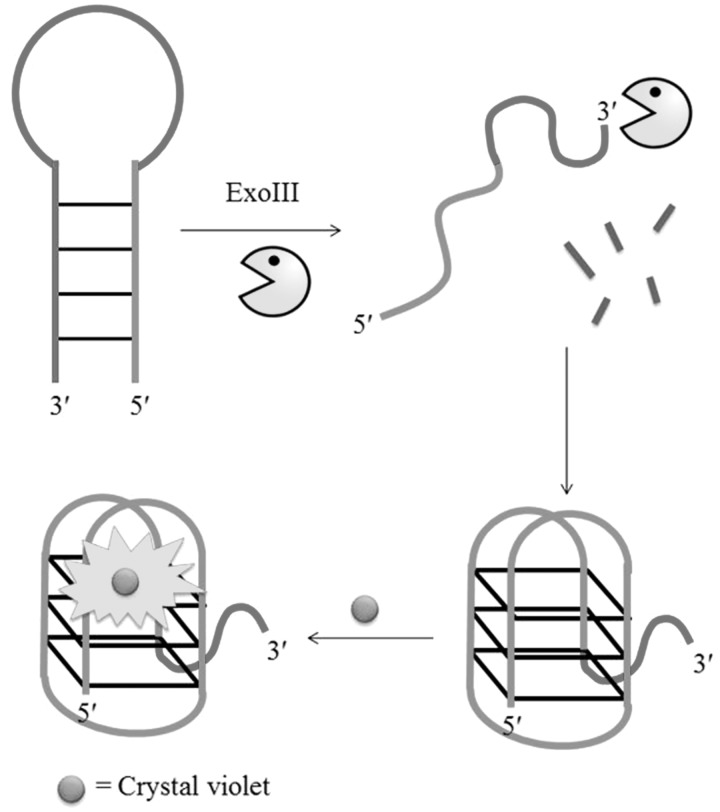
Our research group has reported a G-quadruplex-based strategy for the detection of 3′ → 5′ exonucleases (29), which are enzymes that play important roles in a variety of key cellular and physiological processes such as DNA proofreading (68). In this study, we designed an unlabelled DNA hairpin sequence [5′-AG3(T2AG3)3CAGA2G2AT2A(C3TA2)3C3T-3′] consisting of a 22-bp G-quadruplex-forming sequence at the 5′-terminus, a 11-bp loop region, and a 22-bp complementary C-rich sequence at the 3′-terminus. Initially, the oligonucleotide is hybridized in a stem-loop arrangement, and the fluorescence of the G-quadruplex-selective dye CV is weak (Figure 18). The prokaryotic 3′ → 5′ ExoIII is able to digest DNA specifically from the 3′-terminus in the duplex stem region but is halted at the loop region, as it is unable to accept single-stranded DNA as substrate. The free G-quadruplex sequence at the 5′-terminus is released and folds into G-quadruplex sequence, which is detected by CV with a strong fluorescence enhancement. The assay was readily amenable to a high-throughput format, and the exonucleolytic activity of the enzyme could be monitored in real-time. Furthermore, the system was selective for ExoIII over other nucleases such as the 5′ → 3′ exonuclease T7, the single-strand-specific 3′ → 5′ exonuclease I and the non-specific DNase I.
Figure 18.
Schematic representation of the label-free luminescent detection assay for 3′ → 5′ exonuclease activity.
Some enzymes are able to recognize mismatched or modified DNA bases and excise them from DNA for subsequent DNA repair processes. Ren and co-workers (18) have devised a duplex-to-G-quadruplex strategy for the label-free detection of uracil DNA-glycosylase (UDG) activity. A G-quadruplex-forming DNA sequence is initially hybridized with a uracil-containing oligonucleotide to form a duplex structure (Figure 19). In the absence of the target enzyme, the fluorescence of the quadruplex-specific dye NMM is weak. The addition of UDG results in the selective cleavage of uracil bases, resulting in the release of the G-quadruplex-forming sequence. The nascent G-quadruplex structure is strongly bound by NMM, leading to a switch-on fluorescence response.
Figure 19.
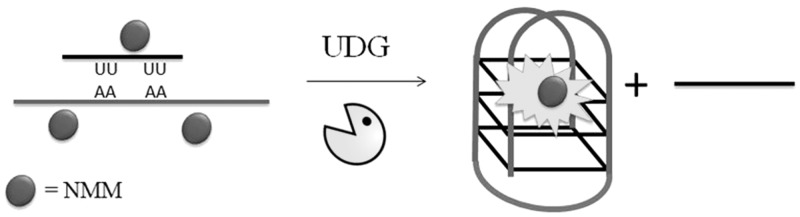
Schematic diagram of the luminescent label-free detection assay for UDG activity using a duplex-to-quadruplex transition.
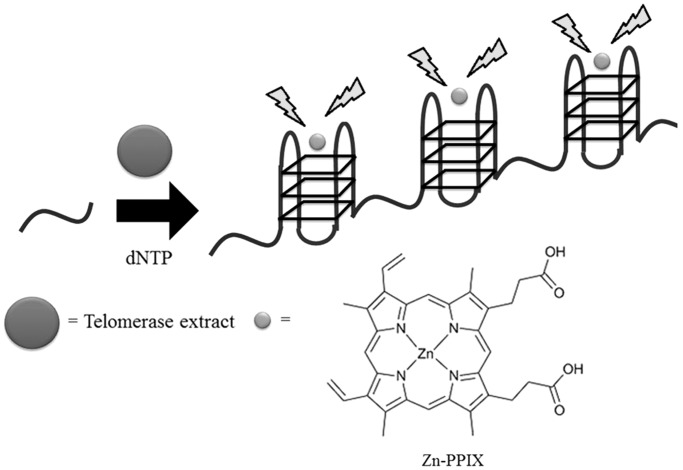
Enzymes with elongation activities are able to initiate the extension of a DNA or RNA strand by the stepwise incorporation of nucleotides to the substrate. The real-time polymerase chain reaction is one basic method for monitoring polymerase activity. Intricate label-free assays have also been devised to selectively detect various enzymes with elongation activities. For example, Willner and co-workers (32) have designed a label-free luminescent assay for monitoring the activity of telomerase. The authors used a synthetic oligomer containing the telomeric TTAGGG sequence as a primer for telomerization (Figure 20). This DNA is incubated with cell extracts containing telomerase and dNTPs. The telomerization of the template sequence by telomerase produces a longer DNA strand-containing tandem TTAGGG repeats. This longer sequence is able to fold into a G-quadruplex and is recognized by the G-quadruplex-selective dye Zn-PPIX with a switch-on luminescence response. However, cell extracts lacking telomerase activity would not be able to promote elongation of the short primer sequence, and therefore the resulting fluorescence emission of the system would be low.
Figure 20.
Schematic diagram for the label-free luminescent detection of telomerase activity.
LABEL-FREE TETRAPLEX DNA-BASED LOGIC GATES
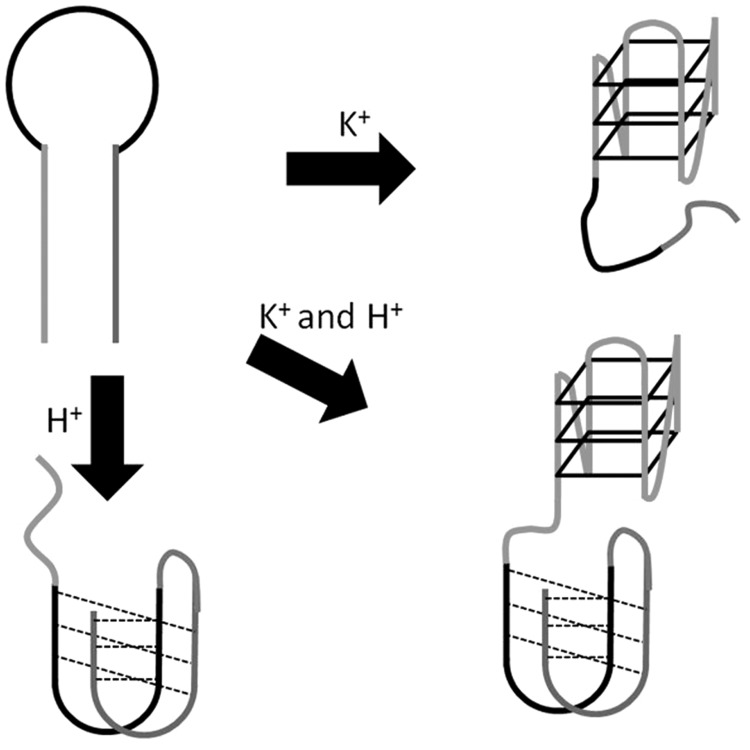
Molecular computation, a term that includes a number of distinct ‘bottom-up’ approaches towards the design of chemical and/or biological computers, has risen as a fascinating potential alternative to silicon-based computing. This has stimulated the development of molecular logic gates, such as ‘OR’, ‘AND’ and ‘NOT’, that parallel the Boolean functions that are fundamental basis for modern digital information integration. As the seminal work of Adleman (69), who pioneered the idea of using DNA to solve the famous ‘traveling salesman’ problem in the mid-1990s, the design and construction of DNA systems that can perform Boolean logic operations in response to chemical and/or physical inputs has attracted immense interest (65,70). In recent decades, a variety of DNA structures and functional oligonucleotides have been incorporated into the design of basic Boolean logic devices. In particular, both G-quadruplex and i-motif tetraplexes have received tremendous attention owing to their rich polymorphic nature and adaptive responses towards various stimuli (71). The i-motif is a four-stranded DNA structure formed from cytosine-rich oligonucleotides under acidic condition and characterized by intercalative C–C+ hemiprotonated base pairing (72,73).
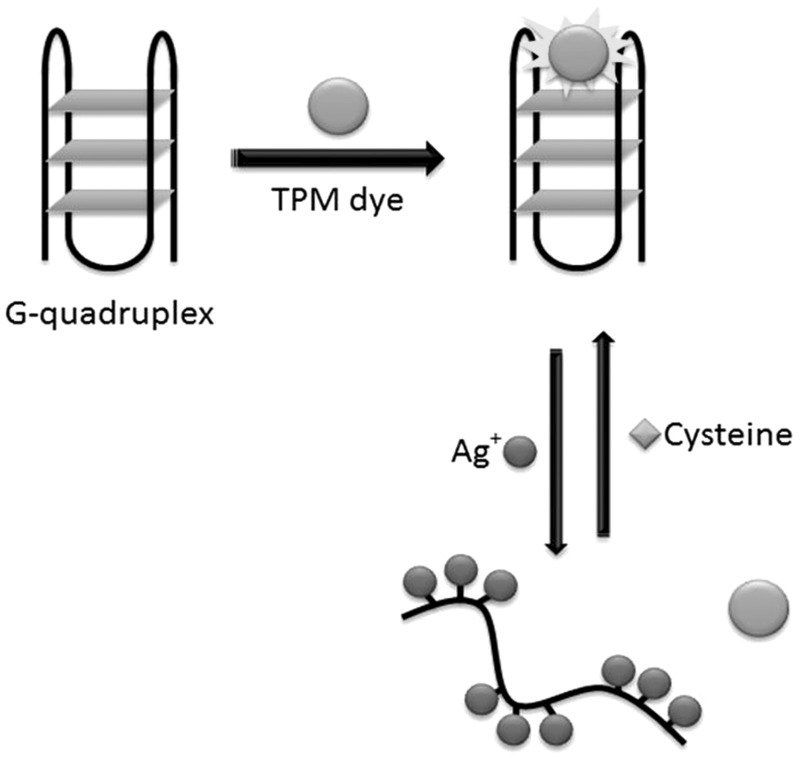
Kong and co-workers (31) have presented label-free variation of the IMPLICATION Ag+/cysteine logic gate using a G-quadruplex-selective fluorescent triphenylmethane dye (Figure 21). This logic device is ‘turned on’ with a strong fluorescence signal in the absence of any inputs (i1 = 0, i2 = 0). The addition of Ag+ (i1 = 1, i2 = 0) disrupts the G-quadruplex structure and weakens the binding of the fluorescent dye, thereby producing an ‘off’ output. The subsequent addition of cysteine (i1 = 1, i2 = 1) sequesters Ag+ ions and restores the fluorescence of the assay.
Figure 21.
Illustration of a label-free IMPLICATION logic gate using Ag+ and cysteine as inputs and a fluorescent triphenylmethane dye. The fluorescence output of the triphenylmethane dye is dependent on the integrity of the G-quadruplex structure.
In 2010, Ren and co-workers (74) devised a logic platform capable of multiple logic operations using the conjugated polymer PFP (poly [(9,9-bis(6′-N,N,N-trimethylammonium)hexyl)fluorenylene phenylene dibromide] and human telomeric DNA [(AG3(T2AG3)3/(C3TA2)3C3T)]. In this system, the cationic conjugated polymer acts as a donor, and the intercalating dye Genefinder functions as an acceptor (Figure 22). At neutral pH, the probe exists in the duplex form and allows intercalation by the organic dye, whereas the positively charged conjugated polymer associates closely with the negatively charged duplex DNA through electrostatic interactions. This brings PFP and the intercalating dye into close proximity, and the resulting Fluorescence resonance energy transfer (FRET) interaction amplifies the fluorescence of the intercalating dye while quenching the emission from PFP. At low pH, the cytosine-rich strand dissociates from the DNA duplex to form a stable i-motif. This prevents intercalation by the organic dye and reduces the FRET response. Taking advantage of the distinct fluorescence properties of the PFP/Genefinder/DNA system at different pH, the authors demonstrated that INH/NINH logic gates could be implemented by using duplex DNA and H+ ions as inputs (pH 4.0). Similarly, a XNOR logic gate could also be constructed using hydrogen chloride and sodium hydroxide as inputs. Finally, the authors demonstrated the construction of a NOR logic gate with the physiologically useful parameters of temperature and pH as inputs. A similar logic system was devised by Huang, Huang and co-workers using the conjugated polymer PFEP poly{[9,9-bis(6′-(N,N,N-diethylmethylammonium)hexyl)-2,7-fluorenylene ethynylene]-alt-co-[2,5-bis(3′-(N,N,N-diethylmethylammonium)-10-oxapropyl)-1,4-phenylene] tetraiodide} and two intercalating dyes (TO and EB) (75).
Figure 22.
Schematic representation of label-free logic gate system based on the conformational switch of telomeric DNA and the FRET signal from a conjugated polymer to the DNA-binding dye.
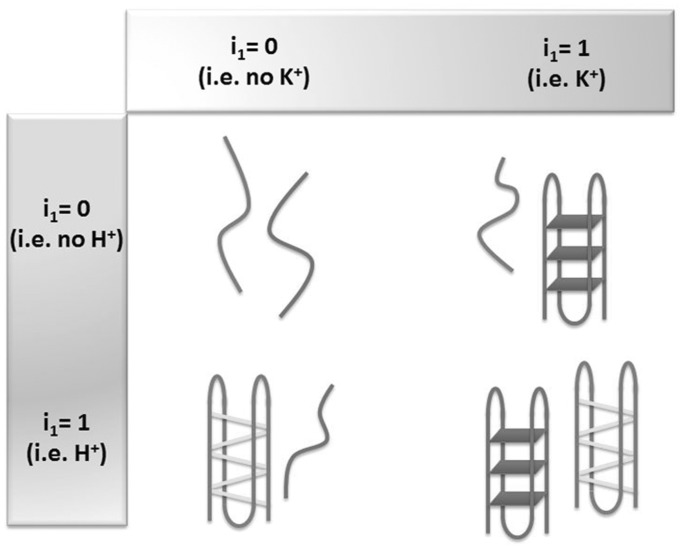
Our group has developed a label-free ‘‘OR’’ logic gate for H+ and K+ ions (76). The system comprises a cytosine-rich oligonucleotide [5′-(AC3T)4-3′] and a guanine-rich oligonucleotide [5′-A(G3T2A)3G3CAGA2G2ATA2-3′] in conjunction with the triphenylmethane dye CV, which selectively interacts with tetraplex DNA over duplex or single-stranded DNA. In the absence of H+ and K+ ions (i1 = 0, i2 = 0), the guanine- and cytosine-rich oligonucleotides exist as random coil structures that interact weakly with CV, resulting in an ‘off’ state (Figure 23). In the presence K+ and/or H+ ions, the guanine-rich and cytosine-rich DNA can be induced into a G-quadruplex or i-motif structure, respectively. Owing to the strong binding of CV to either tetraplex structure, the fluorescence of the system is turned to an ‘on’ state in the presence of either input.
Figure 23.
Schematic representation of the structural interconversion of the guanine- and cytosine-rich sequences into G-quadruplex or i-motif structures induced H+ or K+ ions, respectively.
In 2011, Wang, Dong and co-workers (77) constructed a versatile logic device with NOT, NOR and AND functions using DNA/Ag fluorescent nanoclusters with K+ and H+ as inputs (Figure 24). The authors designed an oligonucleotide HP26 (5′-GGGTTAGGGTCCCCCCACCCTTACCC-3′) that acts as a stabilizer for the synthesis of fluorescent Ag nanoclusters. In the absence of input, HP26 adopts a hairpin conformation that facilitates the templated synthesis of yellow (λem = 570 nm) and red (λem = 646 nm) fluorescent emitters. However, the addition of K+ or H+ ions induces the formation of G-quadruplex or i-motif structures, respectively. This alters the degree of encapsulation of the Ag nanoclusters and thereby changes the fluorescent properties of the system. Furthermore, the system was able to perform multiple logic operations via multichannel fluorescence output, and different logic gates could be constructed by simply altering the specific sequence of the DNA stabilizer.
Figure 24.
Schematic representation of the structural interconversion of the guanine- and cytosine-rich hairpin sequence (HP26) into G-quadruplex or i-motif structure induced by K+ or H+ ions, respectively. The DNA acts as a stabilizer for the templated synthesis of Ag nanoclusters.
CONCLUSION
The versatility of the G-quadruplex motif has vastly expanded the number of potential applications of G-quadruplexes in fields as diverse as therapeutic and diagnostic tools (78,79) and anti-cancer drug screening platforms (80–82). Compared with organic chemosensors or protein antibodies, DNA oligonucleotides offer the distinct advantages of low cost, high thermostability and solubility, ease of production and modification and a rich structural polymorphism that can be sensitive to the presence of particular analytes. In particular, we have focused on the ‘label-free’ strategy that uses G-quadruplex DNA in conjunction with G-quadruplex-selective probes for sensing. The label-free approach has recently gained popularity owing to its simplicity and economy, as it does not require costly and time-consuming labelling and immobilization steps (83,84).
In this Survey and Summary, we have described a large variety of G-quadruplex DNA designs and G-quadruplex-selective probes that have been used for the construction of luminescent sensing platforms for DNA, metal ions, enzyme detection and biothiols. Furthermore, we have discussed the use of the G-quadruplex and the related i-motif tetraplex structure for the design of luminescent DNA logic gates. The unique structural features exhibited by the diverse array of G-quadruplex topologies offers the potential for highly specific interactions between the luminophore and the functional oligonucleotide that may avoid non-specific interference from single- or double-stranded nucleic acid species likely to be present in the biological environment. However, such a system may still be potentially affected by the large amounts of telomeric DNA present in eukaryotic biological samples, which could sequester the luminescent probe and interfere with the signal of the assay. Nevertheless, several of the label-free G-quadruplex-based luminescent assays described in this review have been successfully applied for analyte detection in biological media such as cell extracts and urine.
Towards the future, the selectivity of the label-free DNA-based detection platforms again appears to be a key issue that could be addressed and improved. In many cases, the selectivity of the label-free assay may be limited by the relatively modest selectivity of the G-quadruplex ligand for the G-quadruplex motif over other conformations such as duplex or single-stranded DNA, thereby somewhat negating the potent advantages of sensitivity and selectivity offered by nucleic acid aptamers or other functional oligonucleotides. Another drawback of the label-free approach is the lack of true multiplex capability that is possible with labelled oligonucleotides, though some progress has been made to mimic this using the multichannel fluorescence properties of silver nanoclusters as reported by Wang, Dong and co-workers (77) for the construction of a label-free logic device.
An ideal label-free DNA-based sensing system should contain a luminescent reporter molecule that displays a selective ‘switch-on’ emission response on binding to G-quadruplex DNA, which is in turn formed from the highly specific interaction between a precursor oligonucleotide with its cognate analyte. As many of the luminescent G-quadruplex probes used for label-free sensing described in this Survey and Summary have only been recently discovered or only recently been applied for G-quadruplex detection, we envision that more avid and selective G-quadruplex-binding ligands will continue to be developed in the future. Based on the multitude of examples of label-free luminescent G-quadruplex-based detection assays and logic gates reported recently, we anticipate that this field would continue to thrive and mature in the years to come.
FUNDING
Funding for open access charge: Hong Kong Baptist University [FRG2/11-12/009]; Centre for Cancer and Inflammation Research, School of Chinese Medicine (CCIR-SCM, HKBU); the Health and Medical Research Fund [HMRF/11101212]; the Research Grants Council [HKBU/201811 and HKBU/204612]; the French National Research Agency/Research Grants Council Joint Research Scheme [A-HKBU201/12]; the Science and Technology Development Fund, Macao SAR [001/2012/A] and the University of Macau [MYRG091(Y1-L2)-ICMS12-LCH and MYRG121(Y1-L2)-ICMS12-LCH].
Conflict of interest statement. None declared.
REFERENCES
- 1.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 5.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willner I, Shlyahovsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 8.Li D, Song S, Fan C. Target-responsive structural switching for nucleic acid-based sensors. Acc. Chem. Res. 2010;43:631–641. doi: 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- 9.Wang RE, Zhang Y, Cai J, Cai W, Gao T. Aptamer-based fluorescent biosensors. Curr. Med. Chem. 2011;18:4175–4184. doi: 10.2174/092986711797189637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 11.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA·RNA hybridization in living cells. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Dong S, Wang E. Homogeneous analysis: label-free and substrate-free aptasensors. Chem. Asian J. 2010;5:1262–1272. doi: 10.1002/asia.200900660. [DOI] [PubMed] [Google Scholar]
- 14.Monchaud D, Teulade-Fichou M-P. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 15.Haider SM, Neidle S, Parkinson GN. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011;93:1239–1251. doi: 10.1016/j.biochi.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Ma D-L, Chan DS-H, Yang H, He H-Z, Leung C-H. Luminescent G-quadruplex probes. Curr. Pharm. Des. 2012;18:2058–2075. doi: 10.2174/138161212799958314. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Wu L, Ren J, Qu X. A label-free fluorescent turn-on enzymatic amplification assay for DNA detection using ligand-responsive G-quadruplex formation. Chem. Commun. 2011;47:5461–5463. doi: 10.1039/c1cc11396h. [DOI] [PubMed] [Google Scholar]
- 18.Hu D, Huang Z, Pu F, Ren J, Qu X. A label-free, quadruplex-based functional molecular beacon (LFG4-MB) for fluorescence turn-on detection of DNA and nuclease. Chem. Eur. J. 2011;17:1635–1641. doi: 10.1002/chem.201001331. [DOI] [PubMed] [Google Scholar]
- 19.Ren J, Qin H, Wang J, Luedtke NW, Wang E, Wang J. Label-free detection of nucleic acids by turn-on and turn-off G-quadruplex-mediated fluorescence. Anal. Bioanal. Chem. 2011;399:2763–2770. doi: 10.1007/s00216-011-4669-0. [DOI] [PubMed] [Google Scholar]
- 20.Hu D, Pu F, Huang Z, Ren J, Qu X. A quadruplex-based, label-free, and real-time fluorescence assay for RNase H activity and inhibition. Chem. Eur. J. 2010;16:2605–2610. doi: 10.1002/chem.200902166. [DOI] [PubMed] [Google Scholar]
- 21.Qin H, Ren J, Wang J, Wang E. G-quadruplex facilitated turn-off fluorescent chemosensor for selective detection of cupric ion. Chem. Commun. 2010;46:7385–7387. doi: 10.1039/c0cc01695k. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Lin Y, Zhao C, Ren J, Qu X. Silver metallization engineered conformational switch of G-quadruplex for fluorescence turn-on detection of biothiols. Chem. Commun. 2012;48:11428–11430. doi: 10.1039/c2cc36690h. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Liu J, Fang Y, Qin Y, Xu S, Liu Y, Wang E. G-quadruplex-based ultrasensitive and selective detection of histidine and cysteine. Biosens. Bioelectron. 2013;41:563–568. doi: 10.1016/j.bios.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Zhang L, Wang E. Measurement of the base number of DNA using a special calliper made of a split G-quadruplex. Chem. Commun. 2012;48:11990–11992. doi: 10.1039/c2cc36693b. [DOI] [PubMed] [Google Scholar]
- 25.Li T, Wang E, Dong S. Parallel G-quadruplex-specific fluorescent probe for monitoring DNA structural changes and label-free detection of potassium ion. Anal. Chem. 2010;82:7576–7580. doi: 10.1021/ac1019446. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Zhu J, Ai J, Zhou Z, Jia X, Wang E. Label-free G-quadruplex-specific fluorescent probe for sensitive detection of copper(II) ion. Biosens. Bioelectron. 2013;39:268–273. doi: 10.1016/j.bios.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 27.He H-Z, Pui-Yan Ma V, Leung K-H, Shiu-Hin Chan D, Yang H, Cheng Z, Leung C-H, Ma D-L. A label-free G-quadruplex-based switch-on fluorescence assay for the selective detection of ATP. Analyst. 2012;137:1538–1540. doi: 10.1039/c2an15999f. [DOI] [PubMed] [Google Scholar]
- 28.Jin Y, Bai J, Li H. Label-free protein recognition using aptamer-based fluorescence assay. Analyst. 2010;135:1731–1735. doi: 10.1039/c0an00014k. [DOI] [PubMed] [Google Scholar]
- 29.Leung C-H, Chan DS-H, Man BY-W, Wang C-J, Lam W, Cheng Y-C, Fong W-F, Hsiao W-LW, Ma D-L. Simple and convenient G-quadruplex-based turn-on fluorescence assay for 3′ → 5′ exonuclease activity. Anal. Chem. 2010;83:463–466. doi: 10.1021/ac1025896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong D-M, Guo J-H, Yang W, Ma Y-E, Shen H-X. Crystal violet–G-quadruplex complexes as fluorescent sensors for homogeneous detection of potassium ion. Biosens. Bioelectron. 2009;25:88–93. doi: 10.1016/j.bios.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Guo J-H, Kong D-M, Shen H-X. Design of a fluorescent DNA IMPLICATION logic gate and detection of Ag+ and cysteine with triphenylmethane dye/G-quadruplex complexes. Biosens. Bioelectron. 2010;26:327–332. doi: 10.1016/j.bios.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Sharon E, Freeman R, Liu X, Willner I. Fluorescence detection of DNA, adenosine-5′-triphosphate (ATP), and telomerase activity by zinc(II)-protoporphyrin IX/G-quadruplex labels. Anal. Chem. 2012;84:4789–4797. doi: 10.1021/ac300348v. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y-J, Ma N, Li Y-J, Lin Z-Y, Qiu B, Chen G-N, Wong K-Y. Styryl quinolinium/G-quadruplex complex for dual-channel fluorescent sensing of Ag+ and cysteine. Sens. Actuators B Chem. 2012;173:295–299. [Google Scholar]
- 34.Qiu B, Zhang YS, Lin YB, Lu YJ, Lin ZY, Wong KY, Chen GN. A novel fluorescent biosensor for detection of target DNA fragment from the transgene cauliflower mosaic virus 35S promoter. Biosens. Bioelectron. 2013;41:168–171. doi: 10.1016/j.bios.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Qin H, Ren J, Wang J, Luedtke NW, Wang E. G-quadruplex-modulated fluorescence detection of potassium in the presence of a 3500-fold excess of sodium ions. Anal. Chem. 2010;82:8356–8360. doi: 10.1021/ac101894b. [DOI] [PubMed] [Google Scholar]
- 36.He H-Z, Chan DS-H, Leung C-H, Ma D-L. A highly selective G-quadruplex-based luminescent switch-on probe for the detection of gene deletion. Chem. Commun. 2012;48:9462–9464. doi: 10.1039/c2cc32253f. [DOI] [PubMed] [Google Scholar]
- 37.Leung K-H, Ma VP-Y, He H-Z, Chan DS-H, Yang H, Leung C-H, Ma D-L. A highly selective G-quadruplex-based luminescent switch-on probe for the detection of nanomolar strontium(ii) ions in sea water. RSC Adv. 2012;2:8273–8276. [Google Scholar]
- 38.He H-Z, Leung K-H, Yang H, Shiu-Hin Chan D, Leung C-H, Zhou J, Bourdoncle A, Mergny J-L, Ma D-L. Label-free detection of sub-nanomolar lead(II) ions in aqueous solution using a metal-based luminescent switch-on probe. Biosens. Bioelectron. 2013;41:871–874. doi: 10.1016/j.bios.2012.08.060. [DOI] [PubMed] [Google Scholar]
- 39.Leung K-H, He H-Z, Ma VP-Y, Yang H, Chan DS-H, Leung C-H, Ma D-L. A G-quadruplex-selective luminescent switch-on probe for the detection of sub-nanomolar human neutrophil elastase. RSC Adv. 2013;3:1656–1659. [Google Scholar]
- 40.Leung K-H, He H-Z, Ma VP-Y, Chan DS-H, Leung C-H, Ma D-L. A luminescent G-quadruplex switch-on probe for the highly selective and tunable detection of cysteine and glutathione. Chem. Commun. 2013;49:771–773. doi: 10.1039/c2cc37710a. [DOI] [PubMed] [Google Scholar]
- 41.Lv L, Guo Z, Wang J, Wang E. G-quadruplex as signal transducer for biorecognition events. Curr. Pharm. Des. 2012;18:2076–2095. doi: 10.2174/138161212799958459. [DOI] [PubMed] [Google Scholar]
- 42.Neo JL, Kamaladasan K, Uttamchandani M. G-quadruplex based probes for visual detection and sensing. Curr. Pharm. Des. 2012;18:2048–2057. doi: 10.2174/138161212799958341. [DOI] [PubMed] [Google Scholar]
- 43.Qin WJ, Yim OS, Lai PS, Yung L-YL. Dimeric gold nanoparticle assembly for detection and discrimination of single nucleotide mutation in Duchenne muscular dystrophy. Biosens. Bioelectron. 2010;25:2021–2025. doi: 10.1016/j.bios.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 44.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 45.Levy M, Cater SF, Ellington AD. Quantum-dot aptamer beacons for the detection of proteins. ChemBioChem. 2005;6:2163–2166. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 46.Beni V, Zewdu T, Joda H, Katakis I, O’Sullivan C. Gold nanoparticle fluorescent molecular beacon for low-resolution DQ2 gene HLA typing. Anal. Bioanal. Chem. 2012;402:1001–1009. doi: 10.1007/s00216-011-5493-2. [DOI] [PubMed] [Google Scholar]
- 47.Martí AA, Jockusch S, Li Z, Ju J, Turro NJ. Molecular beacons with intrinsically fluorescent nucleotides. Nucleic Acids Res. 2006;34:e50. doi: 10.1093/nar/gkl134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimoto K, Shimizu H, Inouye M. Unambiguous detection of target dnas by excimer-monomer switching molecular beacons. J. Org. Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]
- 49.Zuo X, Xia F, Xiao Y, Plaxco KW. Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J. Am. Chem. Soc. 2010;132:1816–1818. doi: 10.1021/ja909551b. [DOI] [PubMed] [Google Scholar]
- 50.Famulok M, Mayer G, Blind M. Nucleic acid aptamersfrom selection in vitro to applications in vivo. Acc. Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 51.Li B, Wei H, Dong S. Sensitive detection of protein by an aptamer-based label-free fluorescing molecular switch. Chem. Commun. 2007:73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 52.Yan S, Huang R, Zhou Y, Zhang M, Deng M, Wang X, Weng X, Zhou X. Aptamer-based turn-on fluorescent four-branched quaternary ammonium pyrazine probe for selective thrombin detection. Chem. Commun. 2011;47:1273–1275. doi: 10.1039/c0cc02792h. [DOI] [PubMed] [Google Scholar]
- 53.Man BY-W, Chan DS-H, Yang H, Ang S-W, Yang F, Yan S-C, Ho C-M, Wu P, Che C-M, Leung C-H, et al. A selective G-quadruplex-based luminescent switch-on probe for the detection of nanomolar silver(i) ions in aqueous solution. Chem. Commun. 2010;46:8534–8536. doi: 10.1039/c0cc01201g. [DOI] [PubMed] [Google Scholar]
- 54.Ma D-L, Chan DS-H, Man BY-W, Leung C-H. Oligonucleotide-based luminescent detection of metal ions. Chem. Asian J. 2011;6:986–1003. doi: 10.1002/asia.201000870. [DOI] [PubMed] [Google Scholar]
- 55.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen FM. Strontium(2+) facilitates intermolecular G-quadruplex formation of telomeric sequences. Biochemistry. 1992;31:3769–3776. doi: 10.1021/bi00130a006. [DOI] [PubMed] [Google Scholar]
- 57.Kotch FW, Fettinger JC, Davis JT. A lead-filled G-quadruplex: insight into the G-Quartet's selectivity for Pb2+ over K+ Org. Lett. 2000;2:3277–3280. doi: 10.1021/ol0065120. [DOI] [PubMed] [Google Scholar]
- 58.Ono A, Togashi H. Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew. Chem. Int. Ed. Engl. 2004;43:4300–4302. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]
- 59.Ono A, Cao S, Togashi H, Tashiro M, Fujimoto T, Machinami T, Oda S, Miyake Y, Okamoto I, Tanaka Y. Specific interactions between silver(i) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 2008:4825–4827. doi: 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- 60.Kong D-M, Ma Y-E, Guo J-H, Yang W, Shen H-X. Fluorescent sensor for monitoring structural changes of G-quadruplexes and detection of potassium ion. Anal. Chem. 2009;81:2678–2684. doi: 10.1021/ac802558f. [DOI] [PubMed] [Google Scholar]
- 61.Guo L, Nie D, Qiu C, Zheng Q, Wu H, Ye P, Hao Y, Fu F, Chen G. A G-quadruplex based label-free fluorescent biosensor for lead ion. Biosens. Bioelectron. 2012;35:123–127. doi: 10.1016/j.bios.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 62.Li T, Dong S, Wang E. A lead(II)-driven DNA molecular device for turn-on fluorescence detection of lead(II) ion with high selectivity and sensitivity. J. Am. Chem. Soc. 2010;132:13156–13157. doi: 10.1021/ja105849m. [DOI] [PubMed] [Google Scholar]
- 63.Qu K, Zhao C, Ren J, Qu X. Human telomeric G-quadruplex formation and highly selective fluorescence detection of toxic strontium ions. Mol. Biosyst. 2012;8:779–782. doi: 10.1039/c2mb05446a. [DOI] [PubMed] [Google Scholar]
- 64.Townsend DM, Tew KD, Tapiero H. The importance of glutathione in human disease. Biomed. Pharmacother. 2003;57:145–155. doi: 10.1016/s0753-3322(03)00043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okamoto A, Tanaka K, Saito I. DNA logic gates. J. Am. Chem. Soc. 2004;126:9458–9463. doi: 10.1021/ja047628k. [DOI] [PubMed] [Google Scholar]
- 66.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klarmann GJ, Hawkins ME, Le GSFJ. Uncovering the complexities of retroviral ribonuclease H reveals its potential as a therapeutic target. AIDS Rev. 2002;4:183–194. [PubMed] [Google Scholar]
- 68.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 69.Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 70.Stojanovic MN, Mitchell TE, Stefanovic D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 71.Alberti P, Bourdoncle A, Sacca B, Lacroix L, Mergny J-L. DNA nanomachines and nanostructures involving quadruplexes. Org. Biomol. Chem. 2006;4:3383–3391. doi: 10.1039/b605739j. [DOI] [PubMed] [Google Scholar]
- 72.Choi J, Kim S, Tachikawa T, Fujitsuka M, Majima T. pH-induced intramolecular folding dynamics of i-Motif DNA. J. Am. Chem. Soc. 2011;133:16146–16153. doi: 10.1021/ja2061984. [DOI] [PubMed] [Google Scholar]
- 73.Jin KS, Shin SR, Ahn B, Rho Y, Kim SJ, Ree M. pH-dependent structures of an i-motif DNA in solution. J. Phys. Chem. A. 2009;113:1852–1856. doi: 10.1021/jp808186z. [DOI] [PubMed] [Google Scholar]
- 74.Pu F, Wang C, Hu D, Huang Z, Ren J, Wang S, Qu X. Logic gates and pH sensing devices based on a supramolecular telomere DNA/conjugated polymer system. Mol. Biosyst. 2010;6:1928–1932. doi: 10.1039/c004215c. [DOI] [PubMed] [Google Scholar]
- 75.Li J, Huang Y-Q, Qin W-S, Liu X-F, Huang W. An optical-logic system based on cationic conjugated polymer/DNA/intercalating dyes assembly for label-free detection of conformational conversion of DNA i-motif structure. Polym. Chem. 2011;2:1341–1346. [Google Scholar]
- 76.Ma D-L, Kwan MH-T, Chan DS-H, Lee P, Yang H, Ma VP-Y, Bai L-P, Jiang Z-H, Leung C-H. Crystal violet as a fluorescent switch-on probe for i-motif: label-free DNA-based logic gate. Analyst. 2011;136:2692–2696. doi: 10.1039/c1an15091j. [DOI] [PubMed] [Google Scholar]
- 77.Li T, Zhang L, Ai J, Dong S, Wang E. Ion-tuned DNA/Ag fluorescent nanoclusters as versatile logic device. ACS Nano. 2011;5:6334–6338. doi: 10.1021/nn201407h. [DOI] [PubMed] [Google Scholar]
- 78.Gatto B, Palumbo M, Sissi C. Nucleic acid aptamers based on the G-quadruplex structure: therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 79.Tucker WO, Shum KT, Tanner JA. G-quadruplex DNA aptamers and their ligands: structure, function and application. Curr. Pharm. Des. 2012;18:2014–2026. doi: 10.2174/138161212799958477. [DOI] [PubMed] [Google Scholar]
- 80.Han H, Hurley LH. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 81.Mergny J-L, Helene C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 82.Kerwin SM. G-quadruplex DNA as a target for drug design. Curr. Pharm. Des. 2000;6:441–471. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 83.Ma D-L, He H-Z, Leung K-H, Zhong H-J, Chan DS-H, Leung C-H. Label-free luminescent DNA oligonucleotide-based probes. Chem. Soc. Rev. 2013;42:3427–3440. doi: 10.1039/c2cs35472a. [DOI] [PubMed] [Google Scholar]
- 84.Ma D-L, Ma VP-Y, Chan DS-H, Leung C-H. An overview of bioactive luminescent transition metal complexes as therapeutic agents. Angew. Chem. Int. Ed. 2013 doi: 10.1002/anie.201208414. doi:10.1002/anie.201208414. [DOI] [PubMed] [Google Scholar]