Abstract
Asp443 and Glu478 are essential active site residues in the RNase H domain of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). We have investigated the effects of substituting Asn for Asp443 or Gln for Glu478 on the fidelity of DNA-dependent DNA synthesis of phylogenetically diverse HIV-1 RTs. In M13mp2 lacZα-based forward mutation assays, HIV-1 group M (BH10) and group O RTs bearing substitutions D443N, E478Q, V75I/D443N or V75I/E478Q showed 2.0- to 6.6-fold increased accuracy in comparison with the corresponding wild-type enzymes. This was a consequence of their lower base substitution error rates. One-nucleotide deletions and insertions represented between 30 and 68% of all errors identified in the mutational spectra of RNase H-deficient HIV-1 group O RTs. In comparison with the wild-type RT, these enzymes showed higher frameshift error rates and higher dissociation rate constants (koff) for DNA/DNA template–primers. The effects on frameshift fidelity were similar to those reported for mutation E89G and suggest that in HIV-1 group O RT, RNase H inactivation could affect template/primer slippage. Our results support a role for the RNase H domain during plus-strand DNA polymerization and suggest that mutations affecting RNase H function could also contribute to retrovirus variability during the later steps of reverse transcription.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) sequence diversity constitutes a major hurdle for the design of effective and broadly applicable HIV vaccines and plays a major role in antiviral drug resistance. Major factors contributing to the genetic variability of HIV-1 are its high replication rate and the low fidelity of the viral reverse transcriptase (RT). The RT catalyses the conversion of the viral genomic single-stranded RNA into double-stranded proviral DNA, which is integrated into the genome of the host cell (1,2). During reverse transcription, the viral genomic RNA is used as template to synthesise a complementary DNA strand (minus-strand DNA). While this process takes place, the viral RNA is degraded. DNA-dependent DNA polymerization initiated from a short polypurine tract (PPT) resistant to endonucleolytic cleavage leads to the formation of the double-stranded proviral DNA.
The HIV-1 RT is a heterodimeric enzyme with subunits of 66 and 51 kDa, termed as p66 and p51, respectively. Both polypeptide chains share the same amino acid sequence, but amino acids 441 to 560 of the C-terminal region of p66 are missing in p51 (3). The large subunit contains separated DNA polymerase and RNase H domains at its N- and C-terminal regions, respectively. Three highly conserved acidic residues (Asp110, Asp185 and Asp186) form the catalytic triad in the DNA polymerase active site. On the other hand, the RNase H catalytic site contains four conserved carboxylates (Asp443, Glu478, Asp498 and Asp549) that form two metal-binding pockets required for the nucleotidyl transfer reaction involved in RNA degradation during reverse transcription (4,5). Mutational studies have demonstrated that the replacement of Asp443, Glu478 or Asp498 into their corresponding amides (Asn or Gln) renders RTs devoid of RNase H activity (6,7). The DNA polymerase and the RNase H domains are separated by 15–20 nucleotides. The RNase H cleavage specificity is controlled by primer grip residues (8) that help to position the DNA primer strand near the endonuclease active site (9). Amino acids of the RNase H domain contributing to the RNase H primer grip include the Thr473-Asn474-Gln475-Lys476 quartet, Tyr501 and Ile505 (9).
Mutational studies have shown that the fidelity of HIV-1 RT is mainly determined by dNTP-binding residues or amino acids located in their vicinity [for reviews, see refs. (10,11)]. However, other interactions affecting residues of the DNA polymerase domain with the template (e.g. Glu89) or the primer strand (e.g. Met230), or with the minor groove-binding track (e.g. Gly262 and Trp266) may also contribute to the accuracy of the RT. In some cases, amino acid substitutions at those positions could affect slippage of the template or primer strands and result in altered frameshift error rates, as observed with mutants E89G, G262A or W266A (12-14). In addition, other mutants such as M230I and M230L were prone to introduce tandem repeat deletions, particularly in the presence of biased dNTP concentrations (15).
The contribution of the RNase H domain to DNA polymerase fidelity has not been explored in detail. One round of replication experiments carried out ex vivo showed that the primer grip mutation Y501W produces a 4-fold increase in mutant frequency compared with the wild-type HIV-1 RT, although another mutation in the same region (I505A) had no effect on the mutation rate (16). Tyr501 is highly conserved in retroviruses. The equivalent residue in the murine leukaemia virus (MLV) RT is Tyr586. Substituting Phe for Tyr586 in such an RT produces a 5-fold increase in the retroviral mutation rate in a single replication cycle (17). Sequence analysis of progeny virus revealed that mutations induced by Y586F were frequently observed at adenine–thymine tracts. Modest reductions in accuracy were also observed with MLV vectors carrying other RNase H primer grip mutations such as S557A, A558V and Q559L (18).
It is widely accepted that RNase H activity may affect mutation and recombination through its coordinated action with the RT DNA polymerase during minus-strand DNA synthesis and plus-strand DNA transfer [see reviews in refs. (19,20)]. However, there is little information about its contribution to DNA-dependent DNA synthesis. In this study, we have used the HIV-1 group O RT as a model because of its biotechnological interest. This enzyme shares ∼80% sequence identity with prototypic HIV-1 group M subtype B RTs, frequently used as a reference in biochemical studies. However, the wild-type HIV-1 group O (O_WT) polymerase is resistant to non-nucleoside RT inhibitors (21) and shows increased thermal stability and fidelity of DNA-dependent DNA synthesis (22). In previous studies, we demonstrated that substituting Ile for Val75 in HIV-1 group O and group M RTs increased their fidelity around 2-fold (22,23), as determined with an M13mp2 lacZα-based forward mutation assay (24). Overall error rates of 1.4 × 10−4, 5.5 × 10−5 and 2.9 × 10−5 were obtained for wild-type BH10 (BH10_WT), O_WT and mutant O_V75I RTs, respectively (22). Now, we have used the forward mutation assay to study the effects of RNase H-inactivating mutations on the fidelity of RTs with different levels of accuracy. Our data show that RNase H mutations increase fidelity by lowering the base substitution error rates of those polymerases. The analysis of mutational spectra and the subsequent biochemical studies suggest that RNase H function may also have an influence on frameshift fidelity during DNA-dependent DNA synthesis, although this effect appears to be restricted to the HIV-1 group O RTs.
MATERIALS AND METHODS
Mutagenesis, expression and purification of recombinant RTs
Site-directed mutagenesis was carried out with the QuikChange site-directed mutagenesis kit (Stratagene) by following the manufacturer’s instructions. The templates used were derivatives of the plasmid p66RTB (25,26) containing the wild-type RT-coding sequences of the HIV-1BH10 (BH10) strain (26), an HIV-1ESP49 group O isolate (21) or a previously described derivative of this clone with the V75I substitution (22). Mutant HIV-1 group O RTs bearing the substitution D443N were obtained with the mutagenic primers 5′-GAAACCTATTATGTAAATGGAGCAGCTA-3′ and 5′-TAGCTGCTCCATTTACATAATAGGTTTC-3′. The amino acid change E478Q was introduced in the BH10 RT by using the mutagenic primers 5′-AAATCAGAAAACTCAGTTACAAGCAA-3′ and 5′-TTGCTTGTAACTGAGTTTTCTGATTT-3′. After mutagenesis, the entire RT-coding regions were sequenced and, if correct, used for RT expression and purification. O_WT RT and mutant derivatives V75I, V75I/E478Q and E478Q were obtained as previously described (22). All RTs were purified as p66/p51 heterodimers with a His6 tag at the C-terminal end of the 66-kDa subunit. For this purpose, the RT p66 subunit (encoded within the corresponding p66RTB derivative) was co-expressed with HIV-1 protease in Escherichia coli XL1 Blue, and the obtained heterodimers were then purified by ionic exchange followed by immobilized metal affinity chromatography on Ni2+-nitriloacetic acid agarose (25). Enzymes were quantified by active site titration before biochemical studies (27).
Nucleotide incorporation and RNase H activity assays
Kinetic parameters for nucleotide incorporation were determined by using 5′-32P-labelled 25PGA and D38, as primer and template, respectively (Figure 1). The 25PGA primer was previously labelled at its 5′ terminus with [γ-32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (New England Biolabs), and then annealed to D38 in 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl and 4 mM MgCl2. RTs were pre-incubated with the template–primer during 10 min at 37°C in 6 µl of 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. Reactions were initiated by adding 6 µl of preincubation buffer containing dTTP at concentrations ranging from 20 nM to 40 μM, and incubated at 37°C. To determine the rate of product formation (28), aliquots of 4 µl were removed after 15 s, quenched with sample loading buffer (10 mM EDTA in 90% formamide containing 3 mg/ml xylene cyanol FF and 3 mg/ml bromophenol blue), analysed by denaturing polyacrylamide gel electrophoresis and quantified with a BAS 1500 scannner (Fuji) using the program TINA version 2.09 (Raytest Isotopenmessgerate Gmbh, Staubenhardt, Germany). The concentration of template–primer in these assays was 30 nM, while the active RT concentration was ∼6–10 nM. The kcat and the apparent Km values were determined as previously described (29,30). Mispair extension assays were carried out under the conditions described above but using a DNA duplex containing the primer 25PIN that differed from 25PGA in having A instead of T at the 3′ end (Figure 1). Aliquots were removed 40 s after adding the dTTP, and assays were performed with nucleotide concentrations ranging from 7.5 to 300 μM. The RNase H activity of WT and mutant RTs was determined with the 31T-RNA/21P complex as previously described (22).
Figure 1.
Nucleotide sequences of DNA/DNA complexes used in the assays.
Primer extension assays in the presence of three dNTPs
The template–primer M54/3TRP (Figure 1) was used to determine the extent of misincorporation in the absence of one dNTP (23). The concentration of the template–primer in the reaction was 30 nM, and the active enzyme concentration was 12 nM. Assays were carried out for 0–60 min at 37°C in 10 µl of 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. Reactions were initiated by the addition of 10 µl of different combinations of dNTPs at a concentration of 1 mM each (*: dATP, dCTP, dGTP, dTTP); (-G: dATP, dCTP, dTTP); (-C: dATP, dGTP, dTTP); (-A: dCTP, dGTP, dTTP); (-T: dATP, dCTP, dGTP). At the end of the incubation period (5, 30 and 60 min), the reactions were stopped by adding 4 µl of sample loading buffer. Reactions products were analysed by denaturing polyacrylamide gel electrophoresis and phosphorimaging.
M13mp2 lacZα-based fidelity assays
The fidelity of DNA-dependent DNA synthesis was determined using the forward mutation assay (24), in the previously described conditions (31). Briefly, a gapped M13mp2 DNA substrate was obtained by deleting 407 nucleotides of one of the strands, including the lacZα-complementation target sequence. Gap-filling reactions were carried out at 37°C during 30 min in 10 μl of 25 mM Tris-HCl (pH 8.0) buffer containing 100 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 250 μM of each dNTP, 5 μg/ml of gapped duplex DNA and 100 nM RT. Polymerization products were electroporated into E. coli MC1061 host cells, and after a brief recovery period (10 min), transformants were plated on an E. coli CSH50 lawn in M9 plates containing 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside and isopropyl β-d-1-thiogalactopyranoside (24). Mutant plaques (light blue or colourless) were picked, and the phage double-stranded DNA replicative form was isolated. Phenotypes of mutant plaques were confirmed by nucleotide sequencing using primer 5′-GCTTGCTGCAACTCTCTCAG-3′ (Macrogen Inc., Seoul, South Korea).
Error frequencies were calculated as the ratio of confirmed mutant plaques to the total number of plaques screened. Specific error rates were derived by multiplying the overall error frequency with the percentage of all mutations represented by the particular class of mutations (e.g. base substitutions). This number is divided by 0.6 (the average probability of an error being expressed in the M13mp2 assay) (32), and by the total number of sites where this class of mutations can be detected (i.e. 125 for base substitutions, 148 for frameshifts and 79 and 69 for mutations in runs and non-runs, respectively).
Processivity assays
Qualitative measurements of the processivity of WT and mutant RTs were obtained with single-stranded M13mp2 DNA template, primed with a 32P-5′-end-labelled 15-mer (Lac110: 5′-GCGATTAAGTTGGGT-3′) complementary to positions 105–119 of the lacZα coding sequence (33). Processivity reactions were carried out at 37°C in 20 μl of 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 30 nM template–primer, 50 µM of each dNTP, 50 nM RT and 5 µg/µl salmon sperm DNA trap. Enzymes were pre-incubated with template–primer for 10 min before adding the dNTPs (either in the presence or absence of trap). Following nucleotide addition, aliquots were removed after 15 min, mixed with an equal volume of sample loading buffer, and analysed by electrophoresis on a denaturing 12% polyacrylamide gel containing 8 M urea and phosphorimaging. The effectiveness of the trap was previously determined by pre-incubating the template–primer with the DNA trap before adding the RT.
Determination of dissociation equilibrium constants (Kd) for RT and DNA/DNA duplexes
Template–primers used in these experiments (D38/25PGA and D38/25PIN) are shown in Figure 1. Both DNA/DNA duplexes are identical except for the nucleotide located at the 3′ end of the primer that originates a mismatch in the case of D38/25PIN. For the determination of the Kd of WT and mutant RTs and the D38/25PGA duplex, enzymes were pre-incubated with increasing concentrations of the 5′-32P-labelled 25/38-mer (1–30 nM) for 10 min at 37°C in 10 µl of a 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. Reactions were initiated by adding 10 µl of 1 mM dTTP (dissolved in the pre-incubation buffer). Aliquots of 4 μl were then removed at 10, 20, 30 and 40 s, quenched with sample loading buffer and analysed by denaturing polyacrylamide gel electrophoresis and phosphorimaging, as described above. In these experiments, the active RT concentration was around 3 nM. The burst amplitudes (RT bound to template–primer at time 0) were plotted as a function of the template–primer concentration, and the data were fitted to a quadratic equation to obtain the equilibrium dissociation constant for RT binding to template–primer (34).
The Kds for the mismatched DNA/DNA complex were determined by using an equilibrium competition assay (35). In these assays, RTs were preincubated with equimolar amounts of radioactively labelled and unlabelled template–primers, before the nucleotide incorporation reaction. Briefly, RTs (3 nM) were first preincubated at 37°C for 10 min, with a mixture of D38/[32P]25PGA and unlabelled D38/25PGA (each one at 7 nM concentration) in 10 μl of 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. Then, the elongation reaction was initiated by adding 10 μl of 1 mM dTTP (dissolved in the pre-incubation buffer). Aliquots of 4 μl were removed at 10, 20, 30 and 40 s, quenched with sample loading buffer and analysed by denaturing polyacrylamide gel electrophoresis. The fraction of extended labelled primer at time 0 (Exmatched) is determined by phosphorimaging. Then a similar experiment is performed by using as unlabelled challenge DNA the template–primer D38/25PIN, instead of unlabelled D38/25PGA. D38/25PIN contains an A:C mismatch at the 3′ end of the primer (Figure 1). The fraction of extended labelled primer in these experiments is designated as Exmismatched, and the Kd for the mismatched template–primer can be calculated from the expression:
Determination of dissociation rate constants (koff)
Dissociation rate constants for template–primers were determined with three different substrates (i.e. D38/25PGA, Lac46T/21P1 and 31T/21P) (Figure 1). Assays were carried out in 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. RTs (24 nM) were pre-incubated with the corresponding template–primer substrate (at 8 nM concentration) for 10 min at 37°C. Then, 2.5 μl of salmon sperm DNA trap (16.5 mg/ml) was added to 5 μl of the pre-incubated sample, and the incubation was continued for 0–60 s, before adding the corresponding dNTP. Elongation reactions carried out in the presence of 20 μM dATP (with Lac46T/21P1) or 100 μM dTTP (with 31T/21P and with D38/25PGA) were incubated for 7 s. The dNTPs were supplied in 2.5 μl of 25 mM Tris-HCl buffer pH 8.0, containing 100 mM KCl, 4 mM MgCl2 and 2 mM dithiothreitol. To determine whether the addition of only one nucleotide could affect the koff values, control experiments involving the addition of several nucleotides were carried out with primer 31T/21P. These assays were carried out under the same conditions, after adding a mixture of dTTP and dATP at concentrations of 200 μM and 500 μM, respectively, followed by a 1-min incubation at 37°C. Reactions were quenched with one volume of sample loading buffer and analysed by denaturing polyacrylamide gel electrophoresis and phosphorimaging. The data (amount of elongated primer) were fitted to the single exponential decay: y = exp(−koff × t), where koff is the dissociation rate constant and t is the time of incubation with the trap (in seconds) (36,37).
RESULTS
Enzyme activity
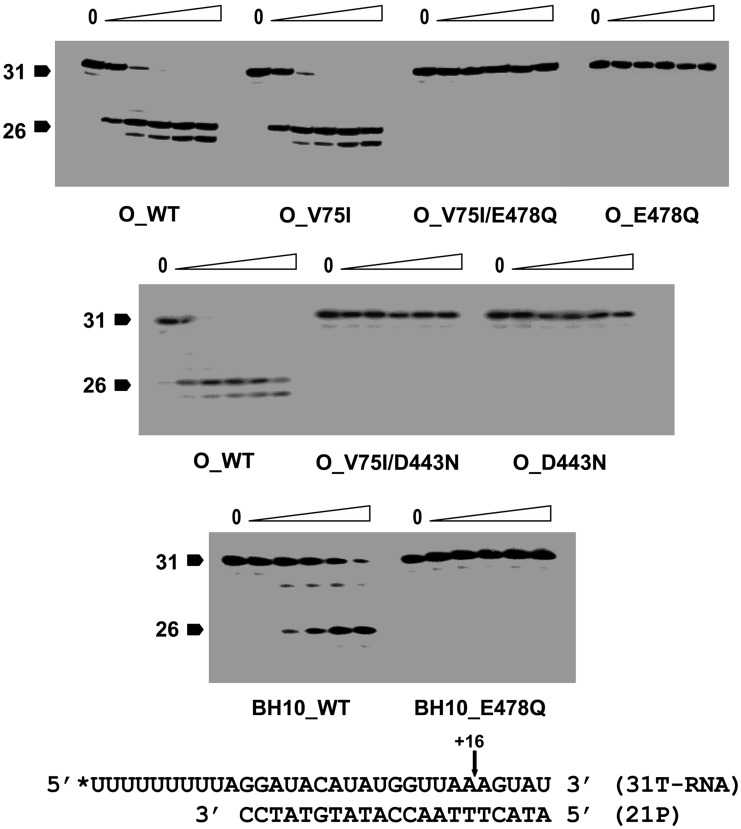
The active centre of the HIV-1 RT RNase H domain contains highly conserved acidic residues (Asp443, Glu478, Asp498 and Glu549) that coordinate two divalent cations required to hydrolyse the RNA template during reverse transcription. HIV-1 RTs with amino acid substitutions affecting residues of the RNase H catalytic centre (e.g. D443N or E478Q) were devoid of endonuclease activity, as determined with an RNA/DNA heteropolymeric substrate (Figure 2). The DNA polymerase activity of RNase H-deficient mutants was determined with a 38/25-mer heteropolymeric template–primer and dTTP, under steady-state conditions. As shown in Table 1, RNase H-inactivating mutations were not detrimental to nucleotide incorporation. All tested RTs showed similar or increased catalytic efficiencies (measured as the ratio kcat/Km) in comparison with the O_WT RT, and the mutant enzyme V75I (O_V75I), as well as with the BH10_WT RT. The substitution D443N produced a moderate increase in DNA polymerase activity that in the case of the double-mutant V75I/D443N was due in part to the higher kcat value obtained with this enzyme.
Figure 2.
RNase H activity of WT and mutant RTs. Cleavage of a [32P]RNA/DNA substrate containing a 31-nt template RNA and a 21-nt primer was carried out at 37°C in the presence of 100 nM RT (active enzyme concentration). Time points in the experiments were 0, 0.25, 1, 2, 4 and 8 min, respectively. Arrows indicate the position of the substrate and the cleavage product of 26 nucleotides.
Table 1.
Steady-state kinetic parameters for dTTP incorporation of WT and mutant RTsa
| RTs | kcat (min−1) | Km (µM) | kcat/Km (µM−1 min−1) |
|---|---|---|---|
| O_WT | 5.02 ± 0.62 | 0.136 ± 0.016 | 36.9 ± 6.3 |
| O_V75I | 6.43 ± 0.58 | 0.193 ± 0.041 | 33.3 ± 7.7 |
| O_D443N | 5.68 ± 0.06 | 0.104 ± 0.008 | 54.6 ± 4.3 |
| O_E478Q | 6.39 ± 0.32 | 0.143 ± 0.012 | 44.7 ± 4.4 |
| O_V75I/D443N | 11.46 ± 0.88 | 0.212 ± 0.052 | 54.1 ± 13.9 |
| O_V75I/E478Q | 5.04 ± 0.25 | 0.143 ± 0.020 | 35.2 ± 5.2 |
| BH10_WT | 5.92 ± 0.91 | 0.099 ± 0.020 | 59.8 ± 15.2 |
| BH10_E478Q | 6.88 ± 0.79 | 0.068 ± 0.009 | 101.2 ± 18.4 |
aD38/25PGA was used as the substrate. Data shown are the mean values ± standard deviation, obtained from at least four independent experiments.
Fidelity of DNA synthesis of RNase H-deficient RTs
Primer extension assays carried out with a 54/22-mer DNA/DNA complex and biased dNTP pools (i.e. mixtures of three nucleotides) showed that HIV-1 group O RNase H-deficient RTs, such as mutants D443N and V75I/E478Q, rendered shorter products than the WT enzyme (Supplementary Figure S1). The largest differences were detected in extension reactions carried out in the absence of dCTP or dATP. A limited analysis of the mispair extension fidelity of wild-type and RNase H-deficient RTs under steady-state conditions revealed that mutant enzymes were also three to four times less efficient in extending C:A mispairs (Supplementary Table S1). The results of those initial experiments suggested that RNase H inactivation could increase fidelity of DNA-dependent DNA synthesis, even though the RNase H domain is located away from the DNA polymerase catalytic site of the RT.
An M13mp2-based forward mutation assay was used to determine the fidelity of RTs devoid of RNase H activity. In these assays, RTs are used to synthesise a DNA complementary to the lacZα gene in reactions carried out in the presence of relatively high dNTP concentrations. Mutations generated during the polymerization reaction can be scored using an indicator strain, by the number of plaques with altered colour phenotype (pale blue or colourless). The mutant frequencies obtained with HIV-1 group O RTs lacking RNase H activity, such as mutants O_D443N, O_E478Q, O_V75I/D443N and O_V75I/E478Q, were ∼3–6.6 times lower than those obtained with the wild-type enzyme (Table 2). These effects were also observed in the BH10 RT, as the corresponding E478Q mutant showed 2-fold increased fidelity in comparison with the wild-type enzyme (Table 2). Mutations E478Q and D443N showed the largest increase in accuracy when introduced in the O_WT RT sequence context.
Table 2.
Accuracy of RT variants in M13mp2 lacZα forward mutation assays
| RTs | Total plaques | Mutant plaques | Mutant frequencya | Fidelity (fold increase) (relative to O_WT RT) |
|---|---|---|---|---|
| O_WT | ||||
| (exp. 1)b | 7579 | 63 | 0.00831 | |
| (exp. 2) | 3957 | 38 | 0.00960 | |
| O_V75Ib | 9894 | 47 | 0.00475 | 1.9 |
| O_D443N | 25697 | 51 | 0.00198 | 4.5 |
| O_E478Q | 40755 | 55 | 0.00135 | 6.6 |
| O_V75I/D443N | 27500 | 50 | 0.00182 | 4.9 |
| O_V75I/E478Q | 16854 | 49 | 0.00291 | 3.1 |
| BH10_WT | ||||
| (exp. 1) | 3792 | 43 | 0.01134 | 0.8 |
| (exp. 2) | 2192 | 29 | 0.01323 | 0.7 |
| BH10_E478Q | 18045 | 104 | 0.00576 | 1.5 (2.0)c |
aReported background frequencies in this assay (∼6 × 10−4) (24) are in most cases a consequence of M13mp2 DNA rearrangements that result in the loss of the lacZ gene. Phage DNA was obtained from all mutant plaques, and the sequence of the reporter gene was determined in all cases. No lacZ mutations were identified after sequencing phage DNA from >20 000 plaques obtained from two to three E. coli electroporations carried out with gapped M13mp2 DNA substrate (31). For each enzyme, mutant plaques were obtained after transfection with the products of four to six gap-filling reactions.
bReported values for O_WT and O_V75I RTs were taken from Álvarez et al. (22).
cFold-increase in fidelity relative to BH10_WT RT.
The mutational spectra of RNase H-deficient HIV-1 group O RTs show a relatively large proportion of one-nucleotide deletions that in several cases accumulated at specific hot spots (Supplementary Figure S2a–d). For example, the spectrum of O_D443N RT has one hot spot at position +123, and O_V75I/D443N has four major hot spots for one-nucleotide deletions at positions −61, +105, +141 and +149. Hotspots around positions +105/+106 and +149 were found in the spectra of O_V75I/D443N and O_V75I/E478Q RTs. Although position +149 could be considered as a common hot spot location for both mutant RTs, identified errors were one-nucleotide deletions in the case of O_V75I/D443N RT and base substitutions (mostly G→T mutations) in the case of O_V75I/E478Q RT. These results were rather different from those obtained with O_WT and mutant O_V75I RTs that showed a very low tendency to introduce insertions or deletions (22). A base substitution hot spot at position −36 was observed in the mutational spectra of O_D443N and O_E478Q RTs. This mutational hot spot was previously identified in the mutational analysis of O_WT and O_V75I RTs, but was absent in the RNase H-deficient polymerases carrying the V75I substitution. All RNase H-deficient RTs were shown to introduce large deletions, although they were more frequent in the case of mutant O_E478Q.
Unlike in the case of O_WT and O_V75I RTs, insertions and deletions represented 30–68% of all errors in the mutational spectra generated by the RNase H-deficient HIV-1 group O RTs (Table 3). The calculated error rates reveal that both D443N and E478Q increase the accuracy of the RT by reducing the base substitution error rate, although both mutant RTs also showed decreased frameshift fidelity. For example, the base substitution error rate of O_E478Q RT was estimated to be 10.9 times lower than the rate obtained with the O_WT enzyme. In contrast, the wild-type enzyme showed increased frameshift fidelity in comparison with mutant O_E478Q RT. Similar data were obtained with O_D443N RT, which in comparison with the WT RT introduced base substitutions errors with an 8.3-fold reduced frequency, but showed increased frameshift error rate. Most of the insertions and deletions introduced by RNase H-deficient RTs were caused by −1 frameshifts, rendering one-nucleotide deletions. Mutants O_D443N and O_V75I/D443N showed a higher tendency to introduce −1 frameshifts at heteropolymeric sequences (i.e. non-runs) in comparison with the RTs containing the E478Q substitution.
Table 3.
Summary of error rates for WT and mutant RTs, for various classes of mutations
| Mutation type | O_WT RTa |
O_V75I RTa |
O_D443N RT |
O_E478Q RT |
O_V75I/D443N RT |
O_V75I/E478Q RT |
BH10_WT RTa |
BH10_E478Q RT |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | |
| All classes | 49 | 1/17 166 | 47 | 1/29 936 | 51 | 1/71 673 | 57 | 1/101 644 | 54 | 1/72 403 | 49 | 1/48 916 | 49 | 1/7369 | 107 | 1/23 981 |
| Base substitutions | 49 | 1/9053 | 47 | 1/15 789 | 26 | 1/74 122 | 31 | 1/98 547 | 20 | 1/103 209 | 34 | 1/37 175 | 31 | 1/5760 | 95 | 1/14 246 |
| Transitions | 25 (51%) | 23 (48.9%) | 18 (69.2%) | 15 (48.4%) | 8 (40%) | 17 (50%) | 20 (64.5%) | 27 (28.4%) | ||||||||
| Transversions | 24 (49%) | 24 (51.1%) | 8 (30.8%) | 16 (51.6%) | 12 (60%) | 17 (50%) | 11 (35.5%) | 68 (71.6%) | ||||||||
| Frameshifts | 0 | <1/541 516 | 0 | <1/878 587 | 25 | 1/91 343 | 26 | 1/139 197 | 34 | 1/71 768 | 15 | 1/99 826 | 18 | 1/11 745 | 12 | 1/133 533 |
| Insertions | 0 | 0 | 4 (16%) | 1 (3.8%) | 1 (2.9%) | 1 (6.7%) | 3 (16.7%) | 11 (91.7%) | ||||||||
| Deletions | 0 | 0 | 21 (84%) | 25 (96.2%) | 33 (97.1%) | 14 (93.3%) | 15 (83.3%) | 1 (8.3%) | ||||||||
| At runsb | 0 | <1/280 371 | 0 | <1/498 947 | 8 | 1/152 173 | 9 | 1/214 574 | 1 | 1/1 340 801 | 12 | 1/66 553 | 18 | 1/6253 | 4 | 1/213 833 |
| At non-runs | 0 | <1/244 881 | 0 | <1/435 789 | 14 | 1/76 156 | 10 | 1/169 100 | 31 | 1/36 981 | 1 | 1/712 074 | 0 | <1/98 207 | 8 | 1/93 383 |
aReported values for O_WT, O_V75I and BH10 RTs were taken from Álvarez et al. (22).
bA run is considered when there is a row of three or more identical nucleotides. Insertions or deletions of two or more nucleotides are excluded from the run versus non-run analysis.
In the case of BH10 RT, the increased fidelity conferred by E478Q could be also attributed to its lower base substitution error rate, as compared with the BH10_WT RT (Table 3). However, in contrast with the well-known strong tendency to produce −1 frameshifts of WT HIV-1 group M subtype B RTs (10,11), this type of errors represented only 11.2% of all identified mutations in the spectrum of the BH10_E478Q RT (Table 3 and Supplementary Figure S2e). Mutational hot spots generated by BH10_E478Q RT, and involving base substitutions at positions −36, +81/+83 and +88/+90 were also observed in the mutational spectrum of BH10_WT RT (22). BH10_E478Q RT has a low tendency to produce large deletions. In our study, we could only identify one deletion after screening >100 plaques. Despite the low frequency of these events, RNase H-deficient HIV-1 group O RTs appear to be more prone to introduce deletions during DNA polymerization.
Template–primer binding and processivity
The increased number of errors due to frameshifts generated by RNase H-deficient HIV-1 group O RTs could be associated with an impaired interaction with the template–primer. Dissociation equilibrium constants (Kd) for O_WT and mutant RTs and DNA/DNA template–primers are given in Table 4. There were not significant differences in the Kd values obtained for O_WT RT and mutant enzymes lacking RNase H activity. Furthermore, the presence of a mismatched nucleotide at the 3′ end of the primer had no effect on template–primer binding affinity (Table 4).
Table 4.
Dissociation equilibrium constants for WT and mutant HIV-1 group O RTs and DNA/DNA template–primers
| RTs | Apparent Kd (nM) |
|
|---|---|---|
| Matched template–primer | Mismatched template–primer | |
| O_WT | 2.76 ± 0.29 | 2.74 ± 0.33 |
| O_D443N | 2.60 ± 0.19 | 2.94 ± 0.65 |
| O_E478Q | 2.92 ± 0.55 | ND |
| O_V75I/D443N | 2.41 ± 0.40 | ND |
| O_V75I/E478Q | 2.83 ± 0.59 | 3.27 ± 0.69 |
The Kd values were obtained with DNA/DNA template–primers D38/25PGA (containing a matched base pair at the 3′ end of the primer) and D38/25PIN (containing a mismatched 3′ terminus). Reported values are the averages ± standard deviations, obtained from at least three independent experiments.
ND, not determined.
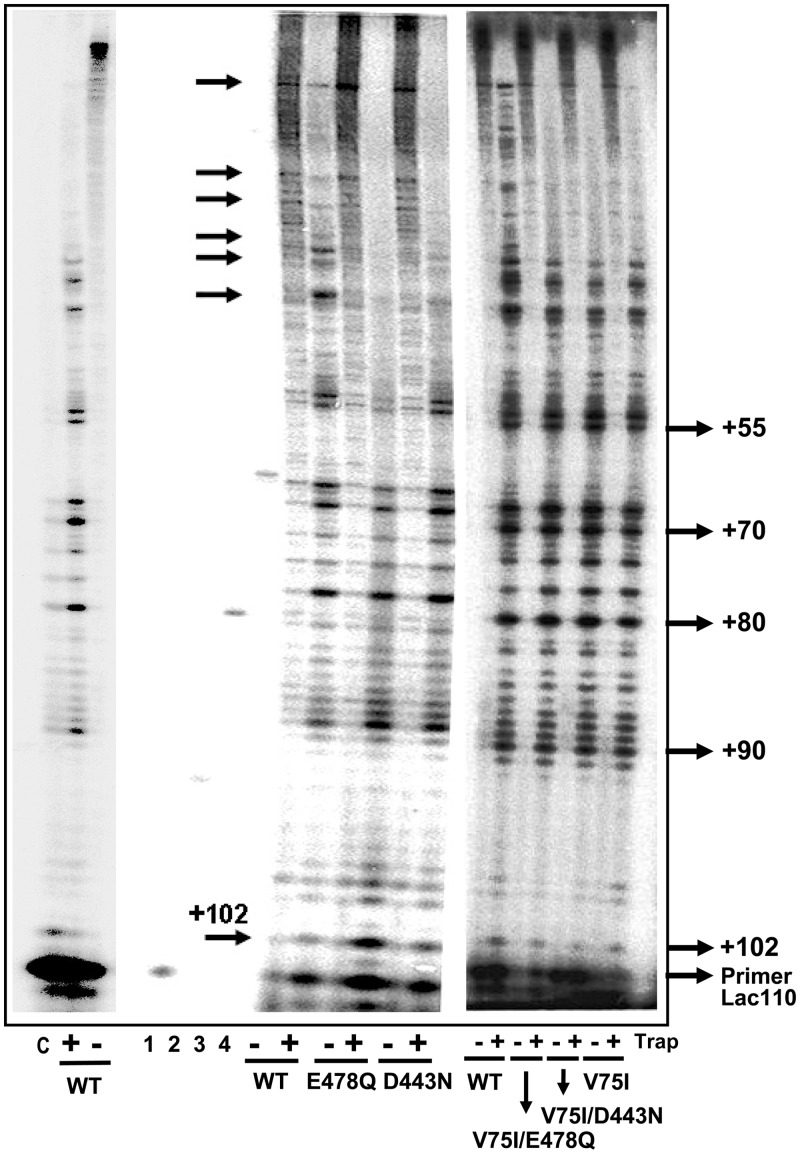
Earlier studies showed a correlation between the probability of termination of processive synthesis and frameshift fidelity (33,38). Therefore, we studied the processivity of RNase H-deficient mutants using single-stranded M13mp2 DNA as template and a primer mimicking the sequence of lacZα around position +110. As shown in Figure 3, all HIV-1 group O RTs were able to synthesise long DNA products in the absence of trap. However, the length of these products is decreased relative to the wild-type enzyme when reactions are carried out in the presence of DNA trap. Qualitative analysis of the processivity patterns reveals that O_WT RT produces longer extensions than the RNase H-deficient RTs. Termination sites appear to be similar for all enzymes, but prominent stops were observed at position +102 with mutants O_D443N and O_E478Q. Both RTs generated large deletions at this termination site (Figure 3). The three RTs having the mutation V75I (i.e. O_V75I, O_V75I/D443N and O_V75I/E478Q) showed intermediate processivity, while O_D443N RT and particularly O_E478Q were found to be the least processive polymerases. These results suggest that RNase H mutations may have an effect on fidelity by altering the dynamics of DNA/DNA template–primer binding.
Figure 3.
Processivity of wild-type and mutant HIV-1 group O RTs. Processivity assays were performed with M13mp2 single-stranded DNA as template and the 15-nt oligonucleotide Lac110 as primer. Reactions were carried out for 15 min at 37°C. Marker oligonucleotides of 15, 25, 38 and 54 nucleotides are shown in lanes 1–4, and correspond to primers Lac110, 25PGA, D38 and M54, respectively. C stands for control (obtained after pre-incubating the template–primer with the DNA trap before adding the RT). Numbers on the right indicate the location of specific stops in the lacZ sequence.
In this context, dissociation rate constants (koff) of RT and DNA/DNA complexes provide information on the kinetics of this equilibrium. We obtained koff values for all HIV-1 group O RTs using three different heteropolymeric template–primers to avoid any potential bias due to the local sequence context in the DNA duplex. As shown in Table 5, differences were relatively small but significant, at least in the case of template–primers 31T/21P and D38/25PGA. The mutant O_V75I/D443N showed the highest koff, followed by the RTs O_D443N, O_V75I/E478Q and O_E478Q, while the lowest koff values were obtained with the WT enzyme and the mutant O_V75I RT. Interestingly, these values correlate well with the calculated frameshift error rates that are highest for mutant O_V75I/D443N RT and lowest for O_WT and O_V75I RTs.
Table 5.
Dissociation rate constants (koff) for binary complexes of RT and DNA/DNA template–primers
| RTs |
koff (s−1) |
||
|---|---|---|---|
| 31T/21Pa | D38/25PGA | Lac46T/21P1 | |
| O_WT | 0.080 ± 0.010 | 0.103 ± 0.012 | 0.207 ± 0.014 |
| O_V75I | 0.051 ± 0.003 (0.6) | 0.093 ± 0.009 (0.9) | 0.210 ± 0.017 (1.0) |
| O_D443N | 0.187 ± 0.010 (2.3)** | 0.214 ± 0.011 (2.1)** | 0.264 ± 0.010 (1.3)** |
| O_E478Q | 0.123 ± 0.018 (1.5)* | 0.133 ± 0.006 (1.3)** | 0.311 ± 0.005 (1.5)** |
| O_V75I/D443N | 0.256 ± 0.025 (3.2)** | 0.232 ± 0.025 (2.3)** | 0.315 ± 0.020 (1.5)** |
| O_V75I/E478Q | 0.163 ± 0.001 (2.0)** | 0.169 ± 0.004 (1.6)** | 0.270 ± 0.018 (1.3)* |
The koff values were obtained with DNA/DNA duplexes of different sizes: 31/21mer (31T/21P), 38/25mer (D38/25PGA) and 46/21mer (Lac46T/21P1). Reported values are the averages ± standard deviations, obtained from at least three independent experiments. Numbers between parentheses represent the fold-increase relative to the koff value for the WT enzyme. **P < 0.01 and *P < 0.05 compared with the corresponding O_WT or O_V75I RTs by unpaired Student’s t test.
aDeterminations based on the incorporation of several nucleotides gave consistent results. Thus, the koff values obtained with O_WT, O_E478Q and O_V75I/D443N RTs, after elongating the primer 21P in the presence of a mixture of dTTP and dATP, were 0.092 ± 0.009, 0.138 ± 0.014 and 0.212 ± 0.020 s−1, respectively.
DISCUSSION
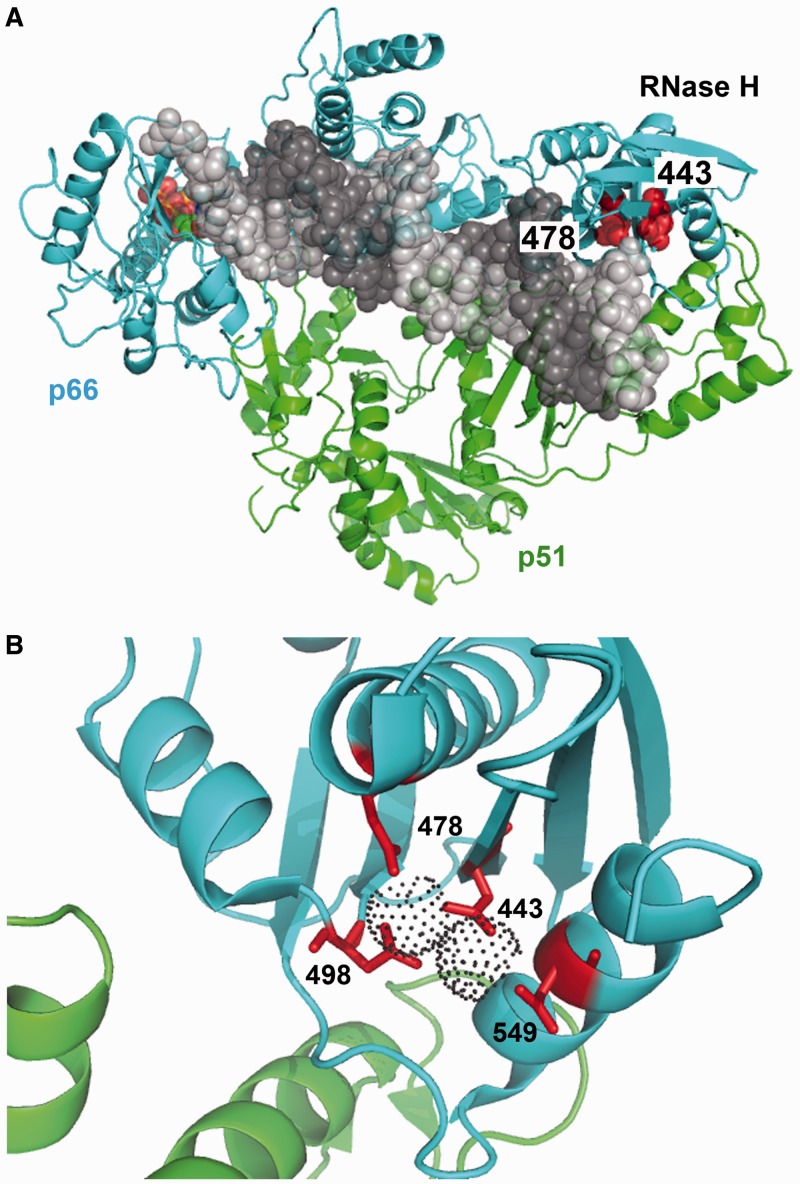
During reverse transcription, the HIV-1 RT RNase H activity is required to hydrolyse the RNA strand of RNA/DNA heteroduplex molecules. The hydrolytic cleavage is mediated by a two-metal-ion catalytic mechanism (4,5). The side chains of active site residues Asp443 and Glu478 participate in the coordination of two and one metal ions, respectively (Figure 4). In HIV-1 RT, mutations D443N and E478Q abolish RNase H activity while retaining wild-type DNA polymerase activity (7). Our results confirm those early findings but also showed that both RNase H-inactivating mutations have an impact on fidelity of DNA-dependent DNA synthesis. These findings are surprising, as RNase H active site residues are located away from the DNA polymerase nucleotide-binding site and DNA/DNA template–primers are not RNase H substrates.
Figure 4.
Crystal structure of HIV-1 RT showing the location of RNase H active site residues. (A) Ternary complex of HIV-1 group M subtype B RT, double-stranded DNA and dTTP [PDB coordinates from Huang et al. (39), PDB code 1RTD]. The RT subunits are represented by cyan and green ribbons. The template and primer strands are shown in light and dark grey sphere models. The incoming dNTP is shown in orange, and the side-chains at positions 443 and 478 are shown in red. (B) Close-up view of the RNase H active site showing the location of Asp443, Glu478, Asp498 and Asp549 and the coordinating metal ions (dot surfaces) [PDB coordinates from Su et al. (40), PDB code 3LP1].
The results of the forward mutation fidelity assays show that both HIV-1 group O and group M RTs bearing mutations D443N or E478Q are more accurate than the wild-type enzyme. Analyses of their mutational spectra indicate that this is a consequence of their lower tendency to generate base substitution errors, although HIV-1 group O RNase H-deficient RTs were also prone to introduce frameshifts, mostly by producing one-nucleotide deletions. Previously, we showed that the frameshift error rate of O_WT and O_V75I RTs was very low (<1.85 × 10−6), as determined with the M13mp2 lacZα-based assay (22). However, in the present study, RNase H-deficient HIV-1 group O RTs showed frameshift error rates as high as 1.4 × 10−5, with insertions and deletions representing >50% of all errors detected in their mutational spectra. In addition, these RTs often generated large insertions and deletions. These results suggested that inactivation of the RNase H could play a role in the dynamics of template–primer binding. In agreement with this proposal, we found that inactivation of the RNase H leads to RTs with decreased processivity and higher dissociation rate from the template–primer. However, RNase H mutations did not seem to affect template–primer affinity. Previous reports have demonstrated that the RNase H-inactivating mutation D443N decreases HIV-1 group M subtype B RT processivity (41), and similar effects have been reported for its equivalent residue in MLV RT (i.e. D524N) (42). However, in the case of mutant E478Q, differences with the wild-type HIV-1 (group M—subtype B RT) were not significant when assayed in the presence of 6 mM Mg2+ (43).
DNA polymerization processivity is defined as the number of nucleotides added per cycle of enzyme binding, synthesis and dissociation from the template–primer. Lower processivity is related to an increase in prematurely paused products and/or a higher dissociation rate (koff) of the RT from the template–primer (36) that would facilitate strand transfer and template or primer slippage. We did not observe differences in the distribution of termination sites during processive DNA synthesis, except in the case of mutants O_D443N and O_E478Q at position +102, where both enzymes showed an increased tendency to introduce large deletions. These observations suggest that those mutants could also have increased template switching activity with DNA/DNA duplexes. The higher frameshift error rates of mutants O_V75I/D443N and O_D443N RTs correlated well with their high koff values, in agreement with their expected contribution to slippage.
Previous studies have shown that one-nucleotide deletions or insertions represented ∼20–30% of all errors introduced by the wild-type HIV-1 group M subtype B RT (33,38,44,45). Mutations at the nucleotide-binding site (e.g. K65R, L74V, Q151N and M184I) have a relatively minor impact on error distribution, with frameshifts occurring mostly at nucleotide runs (44–46). Interestingly, these mutational patterns changed when the introduced mutations affected residues that interact with the template–primer. For example, substituting Ala for Gly262 or Trp266 renders RTs with lower processivity, higher koff and increased tendency to introduce frameshift errors (13,14). In contrast, the BH10_WT RT showed a higher frameshift error rate than the RNase H-deficient mutant E478Q (Table 4). These data correlated well with the higher dissociation rate constants obtained with the wild-type enzyme [(43); data not shown]. The different effects of E478Q on the dissociation rate constants of HIV-1 group O and group M RTs could be related to structural differences between the corresponding RNase H domains. The number of amino acid sequences differences between the RNase H domains of BH10 and HIV-1 group O RTs represents around 28%, while this figure drops to only 15% when the DNA polymerase domain is considered (47). Furthermore, these differences represent >50% of the sequence when the comparison is restricted to residues 460–483.
Interestingly, the substitution of Gly for Glu89 did not have a major effect on the overall fidelity of the HIV-1HXB2 RT as determined in forward mutation assays, but as observed with the HIV-1 group O RNase H-deficient mutants, the E89G RT showed a relatively high tendency to introduce frameshift errors (both run and non–run-associated), and a higher accuracy for base substitutions in comparison with the wild-type RT (44). Frameshift errors made by the E89G mutant accumulated at well-known hotspots such as the one located at positions −34 to −36 of the lacZα gene. E89G RT and RNase H-deficient mutants share an increased fidelity of mispair extension in comparison with the wild-type RT (48,49). However, unlike in the case of the mutants described in our study, E89G increased DNA polymerase processivity (50,51). Despite the mechanistic relationship between processivity, koff and fidelity, available studies do not provide strong evidence to support a correlation between fidelity and processivity (50,52).
Current models to explain HIV-1 genetic recombination assume a relevant role for RNase H in promoting template switching during minus-strand DNA synthesis and the initiation of plus-strand DNA synthesis (19,20,53). DNA polymerization catalysed by HIV-1 RT is 7–10 times faster than RNA degradation (27,54). During RNA-dependent DNA synthesis, the balance between DNA polymerase and RNase H activities plays a major role in controlling the frequency of RT template switching. RNase H degradation reduces the base pairing between the growing DNA strand and the RNA template, and thereby facilitates annealing of the nascent DNA to an acceptor template. In vitro and ex vivo experiments have shown that decreased RNase H activity leads to reduced template switching (55–58). In our assays, based on DNA-dependent DNA synthesis, we found that RNase H-deficient mutants can generate large deletions (and insertions), while these types of errors are rare in RTs with wild-type RNase H activity (22,31). Previous studies showed that RNase H minus HIV-1 RT (i.e. p66E478Q/p51) is able to perform DNA to DNA transfers in a process mediated by the strand displacement activity of the RT (59). However, sequence analyses of mutants containing large deletions did not reveal the existence of hairpin structures that could resemble the strand displacement substrate. On the contrary, most deletions involved repeated sequences of five to eight nucleotides (Supplementary Figure S3). The higher dissociation rate constants shown by RNase H-deficient HIV-1 group O RTs are likely major contributors to increased template switching and therefore to the generation of large deletions and insertions during DNA-dependent DNA synthesis.
The roles of the RNase H activity of RT provide support for the likelihood that most recombination events occur during minus-strand DNA synthesis. However, our results demonstrate that RNase H function can affect frameshift fidelity in DNA-dependent DNA synthesis while influencing processivity and the dissociation of the RT from the template–primer. Taken together, our data suggest that the influence of the RNase H domain on template switching might not be exclusively related to its enzymatic activity, but a result of its physicochemical properties affecting the interaction of the RT with the duplex DNA (i.e. koff). Therefore, mutations in the RNase H domain could also contribute to HIV variability during plus-strand DNA synthesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–3.
FUNDING
Ministry of Economy and Competitiveness (Spain) [BIO2010/15542]; Ministry of Health, Social Services and Equality (Spain) [EC11-025]; Fondo de Investigación Sanitaria (through the ‘Red Temática de Investigación Cooperativa en SIDA’) [RD06/0006]; Fundación Ramón Areces (institutional grant to Centro de Biología Molecular ‘Severo Ochoa’). Funding for open access charge: Research grant [BIO2010/15542].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cristina Jiménez Sánchez for technical assistance with forward mutation assays, and Kasia Bebenek and Thomas Kunkel for helpful advice.
REFERENCES
- 1.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matamoros T, Álvarez M, Barrioluengo V, Betancor G, Menéndez-Arias L. Reverse transcriptase and retroviral replication. In: Kušić-Tišma J, editor. DNA Replication and Related Cellular Process. 2011. InTech, Rijeka, Croatia, pp. 111–142. [Google Scholar]
- 3.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 6.DeStefano JJ, Wu W, Seehra J, McCoy J, Laston D, Albone E, Fay PJ, Bambara RA. Characterization of an RNase H deficient mutant of human immunodeficiency virus-1 reverse transcriptase having an aspartate to asparagine change at position 498. Biochim. Biophys. Acta. 1994;1219:380–388. doi: 10.1016/0167-4781(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 7.Mizrahi V, Brooksbank RL, Nkabinde NC. Mutagenesis of the conserved aspartic acid 443, glutamic acid 478, asparagine 494, and aspartic acid 498 residues in the ribonuclease H domain of p66/p51 human immunodeficiency virus type I reverse transcriptase: expression and biochemical analysis. J. Biol. Chem. 1994;269:19245–19249. [PubMed] [Google Scholar]
- 8.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SFJ. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 9.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menéndez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 11.Menéndez-Arias L. Mutation rate and intrisic fidelity of retroviral reverse transcriptases. Viruses. 2009;1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamburgh ME, Curr KA, Monaghan M, Rao VR, Tripathi S, Preston BD, Sarafianos S, Arnold E, Darden T, Prasad VR. Structural determinants of slippage-mediated mutations by human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2006;281:7421–7428. doi: 10.1074/jbc.M511380200. [DOI] [PubMed] [Google Scholar]
- 13.Beard WA, Stahl SJ, Kim HR, Bebenek K, Kumar A, Strub MP, Becerra SP, Kunkel TA, Wilson SH. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an α-helix in the thumb subdomain. J. Biol. Chem. 1994;269:28091–28097. [PubMed] [Google Scholar]
- 14.Bebenek K, Beard WA, Casas-Finet JR, Kim HR, Darden TA, Wilson SH, Kunkel TA. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- 15.Cases-González CE, Menéndez-Arias L. Increased G→A transition frequencies displayed by primer grip mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 2004;78:1012–1019. doi: 10.1128/JVI.78.2.1012-1019.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansky LM, Le Rouzic E, Benichou S, Gajary LC. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 2003;77:2071–2080. doi: 10.1128/JVI.77.3.2071-2080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang WH, Svarovskaia ES, Barr R, Pathak VK. Y586F mutation in murine leukemia virus reverse transcriptase decreases fidelity of DNA synthesis in regions associated with adenine-thymine tracts. Proc. Natl Acad. Sci. USA. 2002;99:10090–10095. doi: 10.1073/pnas.152186199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbisa JL, Nikolenko GN, Pathak VK. Mutations in the RNase H primer grip domain of murine leukemia virus reverse transcriptase decrease efficiency and accuracy of plus-strand DNA transfer. J. Virol. 2005;79:419–427. doi: 10.1128/JVI.79.1.419-427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menéndez-Arias L, Abraha A, Quiñones-Mateu ME, Mas A, Camarasa MJ, Arts EJ. Functional characterization of chimeric reverse transcriptases with polypeptide subunits of highly divergent HIV-1 group M and O strains. J. Biol. Chem. 2001;276:27470–27479. doi: 10.1074/jbc.M104342200. [DOI] [PubMed] [Google Scholar]
- 22.Álvarez M, Matamoros T, Menéndez-Arias L. Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 2009;392:872–884. doi: 10.1016/j.jmb.2009.07.081. [DOI] [PubMed] [Google Scholar]
- 23.Matamoros T, Kim B, Menéndez-Arias L. Mechanistic insights into the role of Val75 of HIV-1 reverse transcriptase in misinsertion and mispair extension fidelity of DNA synthesis. J. Mol. Biol. 2008;375:1234–1248. doi: 10.1016/j.jmb.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 25.Boretto J, Longhi S, Navarro JM, Selmi B, Sire J, Canard B. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 2001;292:139–147. doi: 10.1006/abio.2001.5045. [DOI] [PubMed] [Google Scholar]
- 26.Matamoros T, Deval J, Guerreiro C, Mulard L, Canard B, Menéndez-Arias L. Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by α-phosphate-modified thymidine analogues. J. Mol. Biol. 2005;349:451–463. doi: 10.1016/j.jmb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 28.Gutiérrez-Rivas M, Menéndez-Arias L. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 2001;29:4963–4972. doi: 10.1093/nar/29.24.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricchetti M, Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990;9:1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martín-Hernández AM, Gutiérrez-Rivas M, Domingo E, Menéndez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrioluengo V, Álvarez M, Barbieri D, Menéndez-Arias L. Thermostable HIV-1 group O reverse transcriptase variants with the same fidelity as murine leukaemia virus reverse transcriptase. Biochem. J. 2011;436:599–607. doi: 10.1042/BJ20101852. [DOI] [PubMed] [Google Scholar]
- 32.Roberts JD, Preston BD, Johnston LA, Soni A, Loeb LA, Kunkel TA. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol. Cell. Biol. 1989;9:469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckert KA, Kunkel TA. Fidelity of DNA synthesis catalyzed by human DNA polymerase α and HIV-1 reverse transcriptase: effect of reaction pH. Nucleic Acids Res. 1993;21:5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menéndez-Arias L. Studies on the effects of truncating α-helix E’ of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry. 1998;37:16636–16644. doi: 10.1021/bi981830g. [DOI] [PubMed] [Google Scholar]
- 35.Creighton S, Huang MM, Cai H, Arnheim N, Goodman MF. Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J. Biol. Chem. 1992;267:2633–2639. [PubMed] [Google Scholar]
- 36.Bibillo A, Eickbush TH. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 2002;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- 37.Gao L, Hanson MN, Balakrishnan M, Boyer PL, Roques BP, Hughes SH, Kim B, Bambara RA. Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J. Biol. Chem. 2008;283:9196–9205. doi: 10.1074/jbc.M710148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 39.Huang H, Chopra R, Verdine G, Harrison S. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 40.Su HP, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, et al. Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J. Virol. 2010;84:7625–7633. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudding LR, Nkabinde NC, Mizrahi V. Analysis of the RNA- and DNA-dependent DNA polymerase activities of point mutants of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry. 1991;30:10498–10506. doi: 10.1021/bi00107a019. [DOI] [PubMed] [Google Scholar]
- 42.Telesnitsky A, Goff SP. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc. Natl Acad. Sci. USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cristofaro JV, Rausch JW, Le Grice SFJ, DeStefano JJ. Mutations in the ribonuclease H active site of HIV-RT reveal a role for this site in stabilizing enzyme-primer-template binding. Biochemistry. 2002;41:10968–10975. doi: 10.1021/bi025871v. [DOI] [PubMed] [Google Scholar]
- 44.Drosopoulos WC, Prasad VR. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J. Virol. 1998;72:4224–4230. doi: 10.1128/jvi.72.5.4224-4230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah FS, Curr KA, Hamburgh ME, Parniak M, Mitsuya H, Arnez JG, Prasad VR. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2000;275:27037–27044. doi: 10.1074/jbc.M002881200. [DOI] [PubMed] [Google Scholar]
- 46.Rezende LF, Curr K, Ueno T, Mitsuya H, Prasad VR. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J. Virol. 1998;72:2890–2895. doi: 10.1128/jvi.72.4.2890-2895.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quiñones-Mateu ME, Soriano V, Domingo E, Menéndez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 48.Rubinek T, Bakhanashvili M, Taube R, Avidan O, Hizi A. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74→Val and Glu89→Gly. Eur. J. Biochem. 1997;247:238–247. doi: 10.1111/j.1432-1033.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 49.Hamburgh ME, Drosopoulos WC, Prasad VR. The influence of 3TC-resistance mutations E89G and M184V in the human immunodeficiency virus reverse transcriptase on mispair extension efficiency. Nucleic Acids Res. 1998;26:4389–4394. doi: 10.1093/nar/26.19.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avidan O, Hizi A. The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res. 1998;26:1713–1717. doi: 10.1093/nar/26.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quan Y, Inouye P, Liang C, Rong L, Götte M, Wainberg MA. Dominance of the E89G substitution in HIV-1 reverse transcriptase in regard to increased polymerase processivity and patterns of pausing. J. Biol. Chem. 1998;273:21918–21925. doi: 10.1074/jbc.273.34.21918. [DOI] [PubMed] [Google Scholar]
- 52.Curr K, Tripathi S, Lennerstrand J, Larder BA, Prasad VR. Influence of naturally occurring insertions in the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and mutation frequencies in vitro. J. Gen. Virol. 2006;87:419–428. doi: 10.1099/vir.0.81458-0. [DOI] [PubMed] [Google Scholar]
- 53.Delviks-Frankenberry K, Galli A, Nikolaitchik O, Mens H, Pathak VK, Hu W-S. Mechanisms and factors that influence high frequency retroviral recombination. Viruses. 2011;3:1650–1680. doi: 10.3390/v3091650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeStefano JJ, Buiser RG, Mallaber LM, Myers TW, Bambara RA, Fay PJ. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J. Biol. Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 55.Pfeiffer JK, Georgiadis MM, Telesnitsky A. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct repeat deletion frequencies. J. Virol. 2000;74:9629–9636. doi: 10.1128/jvi.74.20.9629-9636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svarovskaia ES, Delviks KA, Hwang CK, Pathak VK. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 2000;74:7171–7178. doi: 10.1128/jvi.74.15.7171-7178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brincat JL, Pfeiffer JK, Telesnitsky A. RNase H activity is required for high-frequency repeat deletion during Moloney murine leukemia virus replication. J. Virol. 2002;76:88–95. doi: 10.1128/JVI.76.1.88-95.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 2004;78:8761–8770. doi: 10.1128/JVI.78.16.8761-8770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuentes GM, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis in the central polypurine tract region of HIV-1 promotes DNA to DNA strand transfer recombination. J. Biol. Chem. 1996;271:29605–29611. doi: 10.1074/jbc.271.47.29605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.