Abstract
A key signature of module exchange in the genome is phase symmetry of exons, suggestive of exon shuffling events that occurred without disrupting translation reading frame. At the protein level, intrinsic structural disorder may be another key element because disordered regions often serve as functional elements that can be effectively integrated into a protein structure. Therefore, we asked whether exon-phase symmetry in the human genome and structural disorder in the human proteome are connected, signalling such evolutionary mechanisms in the assembly of multi-exon genes. We found an elevated level of structural disorder of regions encoded by symmetric exons and a preferred symmetry of exons encoding for mostly disordered regions (>70% predicted disorder). Alternatively spliced symmetric exons tend to correspond to the most disordered regions. The genes of mostly disordered proteins (>70% predicted disorder) tend to be assembled from symmetric exons, which often arise by internal tandem duplications. Preponderance of certain types of short motifs (e.g. SH3-binding motif) and domains (e.g. high-mobility group domains) suggests that certain disordered modules have been particularly effective in exon-shuffling events. Our observations suggest that structural disorder has facilitated modular assembly of complex genes in evolution of the human genome.
INTRODUCTION
The intron/exon structure of genes bears witness to the evolutionary history of their genesis, often revealing their assembly from pre-existing genetic elements (exons) encoding for functional units at the protein level (structural modules and domains). The assembly procedure requires that effective genetic mechanisms operate for exon exchange and insertion (exon shuffling), and that the module is incorporated into the recipient protein without much structural (and functional) conflict. A key signature of this assembly mechanism is a bias in exon symmetry, i.e. the enrichment for exons flanked by introns of the same phase (1–4). In principle, introns can split the reading frame between codons (Phase 0) or within codons (Phases 1 and 2), which results in nine different exonic phase types. It is generally thought that the observed genomic bias in exon phases is explained by the evolutionary preference for exchanging symmetric exons (0,0, 1,1 and 2,2), which do not disrupt the reading frame downstream. The successful integration of such exons also implies the structural and functional compatibility of the encoded regions with the recipient proteins.
Our traditional view of protein structure and function assumes that proteins have a well-defined 3D structure; therefore, such compatibility is thought to infer that successful modules correspond to domains and/or secondary structural building blocks (5–7). With the advent of recognizing structural disorder in proteins (8–10), this view needs to be re-examined and extended. Bioinformatic predictions suggest that ∼50% of human proteins have at least one long disordered region (11), and structural disorder plays important roles in proteins of signalling and regulatory functions (12). In light of the diverse functional advantages of structural disorder (13,14) and the noted preference for disordered modules in alternative splicing (15), we decided to re-visit modularity of human protein-coding genes in light of exon symmetry and structural disorder.
Structural disorder is usually higher in eukaryotes than in prokaryotes (12,16,17), although it varies highly in both phylogenetic groups (18,19). If evolutionary expansion of disorder has been driven by module exchange, this general trend of disorder may also result from the advance of exon shuffling in metazoa (7), which has increased complexity and functional diversity in multicellular organisms. As underscored by its power-law genomic distribution (13), structural disorder encompasses modularity at different scales (20), corresponding to short-linear motifs (SLiMs) or eukaryotic linear motifs (ELMs) (21–23), domains (24) or linker regions (25). The intimate connection of structural disorder and modularity is perhaps best exemplified by scaffolds, complex multi-domain signalling proteins (26).
To address the involvement of structural disorder in modular evolution by exon shuffling, we generated a library of human exons and determined their flanking intron phases (exon phase) by analysing transcriptome data. We observed a significant bias for exon symmetry (1–3,5) and elevated levels of disorder in the protein regions encoded by symmetric exons. We also found that exons encoding for largely disordered regions tend to be symmetric, and the genes of mostly disordered proteins tend to be assembled from symmetric exons. Successive symmetric exons show significant homology to each other, and they encode for regions of elevated disorder. The facility of molecular assembly from such elements is also underlined by a bias for phase symmetry and enhanced encoded disorder in alternative (versus constitutive) exons. The length distribution of exons encoding for disordered regions is much broader than that of exons encoding for ordered regions, in accord with previously observed power-law distribution of disorder (13), which may suggest that these exons may encode for either short- (motifs) or long- (domains or linkers) functional elements (21,22,24). A significant enrichment of certain functional motif types in symmetric exons demonstrates that shuffling of symmetric exons can incorporate short-functional modules into proteins. In all, symmetric exons encoding for disordered regions seem to have been amply exploited in the generation of multi-exon genes of modular proteins, probably contributing to the explosive spread of structural disorder early in eukaryotic evolution.
MATERIALS AND METHODS
Data preparation
Human mRNA sequences containing the locations of coding sequences and exons were retrieved from the NCBI Refseq database. To filter out redundancy, only the longest sequences (splice variants) were selected for every gene identifier. We calculated the phases of the N- and C-terminal flanking introns for every exon: Phase 0 introns split the reading frame between two codons, whereas Phase 1 and Phase 2 introns follow the first and second nucleotide of the codon, respectively. Coding mRNA sequences were translated into protein sequences, and regions corresponding to exons were assumed to start with the first complete codon and end with last (even if interrupted) codon. We only took exons into consideration if they had determined phases at both termini because of lying entirely in the coding region (termed complete exons). Our data set contains 8552 protein sequences with boundaries and phases of 78 502 complete exons (of 94 471 total exons).
Prediction of structural disorder and disorder definitions
Structural disorder of proteins and the regions encoded by individual exons were predicted with the IUPred algorithm (14,27). A residue was classified as locally disordered if its disorder score is ≥0.5. For the disorder of exons, disorder was predicted for the whole protein, which was then split into regions encoded by the individual exons and their average disorder was calculated. The average disorder of a protein or a region corresponding to an exon is the per cent of disordered residues in the sequence. The level of disorder for a class of proteins or exon-encoded regions is meant as the mean of individual values: it is termed ‘mostly ordered’ or ‘mostly disordered’ if <30% or >70% of the residues are predicted to be disordered, respectively.
Analysis of exon phases
The expected frequency of exons with any of the nine possible exon-phase combinations was calculated on the basis of the observed frequencies of flanking introns of the three possible phases. If introns limit exons by pure chance, exon-phase combination (i,j) would occur at a frequency Ni*Nj/Ntotal, where Ni and Nj are the occurrence of phase i and phase j introns, respectively, and Ntotal is the total number of exons. The statistical significance of the difference between expected and observed frequencies of each exon phase combinations was tested by the χ2 test, applying Bonferroni correction.
Analysis of adjacent exons
To test whether the sequence of subsequent exons of the same symmetric phase type tends to be more similar to each other than that of randomly selected exons, we ran BlastP search with the sequences of individual exons. We recorded the occurrence of significant similarity identity hits and calculated their per cent frequency of neighbouring exons of identical symmetrical phase type versus neighbouring exons of different phase type. We omitted exon pairs that contain exons without self-identity hit. For checking significance, we performed χ2 test with the concrete occurrence numbers. We also randomly selected 5000–5000 identity values (including zero values) belonging to neighbouring exons of the same or different phase types and performed unpaired t-test. The frequency of significant similarities and also the sets of identity values were found to be significantly different. To detect cases of exon duplication, we compared each exon with its neighbouring exon using also Blastp similarity search.
Identification of alternative and constitutive exons
To compare phase preferences and disorder of alternative and constitutive exons, we used all the human mRNA sequences in NCBI Refseq database of genes encoding for more than one protein product (not only the longest sequence for every gene identifier). Exon phases, exon boundaries and structural disorder were calculated as described earlier in the text. The resulting data set contains 10 245 protein sequences with boundaries, phases and predicted disorder of 108 006 complete exons. An exon was classified as ‘constitutive’, if it can be found in all protein isoforms generated from the same gene (having the same gene identifier), otherwise it was classified as ‘alternative’. To determine whether an exon occurs in a certain isoform, we used BlastP with a 100% identity threshold.
Linear motif and domain prediction
Linear motifs available in the ELM database (28) were predicted for all proteins. For each exon, we then calculated what portion of its sequence is covered by linear motif(s), termed motif coverage. Occurrence and location of domains were computed by the hmmsearch algorithm using the PFAM A seed alignment database (29) for all proteins. For each exon, we calculated what portion of its sequence is covered by PFAM domain(s), termed domain coverage. We also looked for the most significantly overrepresented cases of motifs and domains in disordered regions of proteins encoded by symmetric exons. We counted the occurrence of motifs and domains in different types of (e.g. all, symmetric, symmetric disordered and so forth) exons, and the resulting values were normalized to the total length (number of amino acids) of exons of the proper type. Actually, we normalized the occurrence numbers to 1000 and 10 000 amino acids for motifs and for domains, respectively (because of the high proportion of false-positive hits, there are lot more predicted motifs than domains). Overrepresentation is the ratio of the normalized occurrence of a motif or domain in a certain exon type versus all exons. We only took into account motifs that occur >10 times in symmetric, disordered, short exons and domains that occur >5 times in symmetric disordered exons.
Search for select examples of proteins assembled from disordered modules
To select proteins of high disorder that are assembled from modules encoded by symmetric exons, we ranked all human proteins by their number of successive exons with the same (symmetric) phase and selected those that have a predicted disorder >40%. From these, we selected and discuss in detail some biologically interesting examples.
Statistical analysis and programming
All programs were written in Perl. The software IUPred was obtained from the authors and was compiled and executed locally. For checking significance, when we compared sets of values, we randomly selected 5000 values belonging to each exon sets and performed unpaired t-test. In the case of comparing occurrences, we used χ2 test with Bonferroni correction, if needed.
RESULTS
Exon-phase bias in the genome
First, we asked whether the intron-phase combinations of our selected exons show the characteristic bias previously observed (3,5). Our data set contains 78 502 complete exons flanked by 86 487 introns on both sides in 8552 genes/proteins, i.e. 9.2 on average. First, we counted different types of introns (0, 1, 2) and calculated their per cent occurrence; in accordance with the literature (1,2), the frequency of introns of different phases is significantly different: 46.28% (40 023) for Phase 0, 32.65% (28 238) for Phase 1 and 21.07% (18 226) for Phase 2. From these figures, we calculated the expected occurrences of exons of the nine different phase classes and compared them to the actual observations (Table 1): all three symmetric exon types (0,0, 1,1 and 2,2) occur more frequently than expected by chance. The differences are highly significant (P < 0.0001), except for class 2,2, probably because of its low incidence, as shown by χ2 statistical analysis with Bonferroni correction.
Table 1.
Observed and expected occurrence (of exons), and average disorder and length of regions (encoded by exons) of different phase classes
| Phase type | Observed occurrence | Expected occurrence | Average disorder per cent | Average length |
|---|---|---|---|---|
| (0,0) | 18 646 (23.75%) | 16 815 (21.42%) | 20.36 | 46.04 |
| (0,1) | 9949 (12.67%) | 11 862 (15.11%) | 19.31 | 49.62 |
| (0,2) | 7697 (9.80%) | 7654 (9.75%) | 14.24 | 46.95 |
| (1,0) | 10 158 (12.94%) | 11 862 (15.11%) | 18.58 | 48.49 |
| (1,1) | 10 164 (12.95%) | 8368 (10.66%) | 24.16 | 58.33 |
| (1,2) | 5312 (6.77%) | 5401 (6.88%) | 17.18 | 51.28 |
| (2,0) | 7792 (9.93%) | 7654 (9.75%) | 14.42 | 46.07 |
| (2,1) | 5033 (6.41%) | 5401 (6.88%) | 18.84 | 47.90 |
| (2,2) | 3751 (4.78%) | 3485 (4.44%) | 17.64 | 45.15 |
| All | 78 502 (100%) | 78 502 (100%) | 18.85 | 48.93 |
| Symmetric | 32 561 (41.48%) | 28 668 (36.52%) | 21.23 | 49.78 |
| Asymmetric | 45 941 (58.52%) | 49 834 (63.48%) | 17.17 | 48.32 |
| Successive (0,0) | 9764 (65.80%) | N/A | 25.12 | 43.14 |
| Successive (1,1) | 4405 (29.68%) | N/A | 26.17 | 60.01 |
| Successive (2,2) | 671 (4.52%) | N/A | 25.34 | 45.90 |
| Successive all | 14 840 (100%) | N/A | 25.44 | 48.27 |
Exon-phase symmetry and structural disorder
Next, we asked whether structural disorder of encoded regions distinguishes symmetric and asymmetric exons. To this end, we predicted disorder for whole proteins, then split them into regions encoded by the individual exons and calculated their average disorder (Table 1). The average disorder of all regions corresponding to symmetric exons, 21.23% (24.16% of phase 1,1 exons), is significantly higher (P < 0.0001, using unpaired t-test) than those of asymmetric exons 17.17% (Figure 1A). These values suggest that the evolutionary mechanism that preferred symmetric versus asymmetric exon types also favoured protein disorder. We also calculated the average length of exons of the different phase classes. Here, no conspicuous differences in the averages are observed, with the exception of exons 1,1, which are significantly longer than all others. The reason of this difference is not clear.
Figure 1.
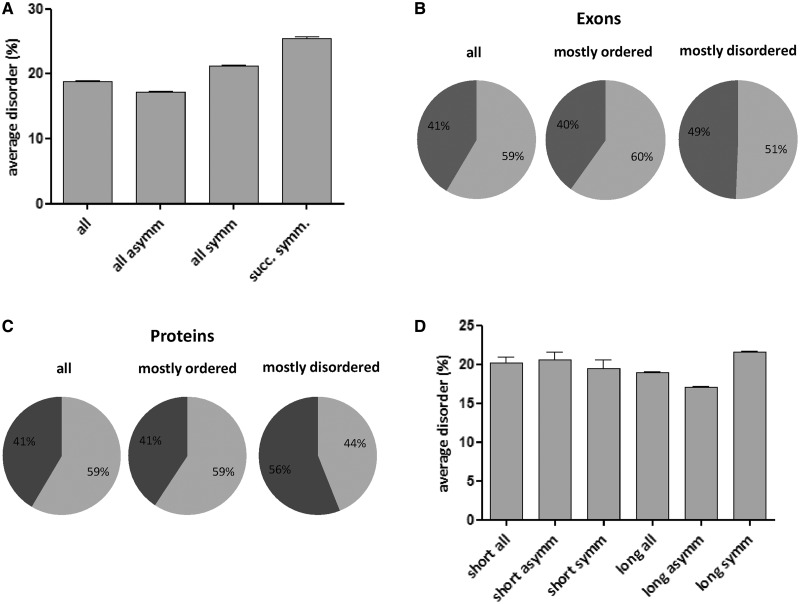
Correlation of structural disorder and exon-phase bias. Symmetric exons have a preference for structural disorder. (A) Average structural disorder (per cent of amino acids predicted to be disordered) of all human exons, of all the six classes of asymmetric exons, of three classes of symmetric exons (phases 0,0; 1,1; and 2,2) and of consecutive symmetric exons of the same phase. All four values are significantly different from each other (with unpaired t-test). (B) Occurrence of symmetric (dark grey) and asymmetric (light grey) exons in humans [all exons, mostly ordered (<30% disorder) and mostly disordered (>70% disorder) exons]. (C) Occurrence of symmetric (dark grey) and asymmetric (light grey) exons in all human proteins and mostly ordered (<30% disorder) and mostly disordered (>70% disorder) proteins. (D) Average disorder of regions encoded by asymmetric and symmetric exons in short (encoded by a gene of maximum two exons) and long (encoded by a gene of at least five exons) proteins. Error bars represent standard errors of mean.
Although the observed differences in the disorder content of exons of different phase classes are significant, they all tend to have values <25%, which do not reveal the potential use of shuffling exons of largely disordered regions modular assembly. To address this issue, we distinguished exons of mostly ordered and mostly disordered regions by a threshold of predicted disorder (<30 and >70%, respectively) and calculated their per cent occurrence (Table 2 and Figure 1B). A large proportion of exons of mostly disordered regions are symmetric (0,0 and 1,1), exceeding even the biased occurrence of all symmetric exons (all symmetric, expected: 36.52%; all symmetric, observed: 41.48%; all symmetric, mostly disordered: 49.37%). The difference (between the occurrence of all symmetric and mostly disordered symmetric exons) is highly significant (P < 0.0001), as shown by χ2 statistical analysis.
Table 2.
Per cent occurrence and average length of mostly ordered and mostly disordered regions encoded by exons of different phase classes, or exons located in the genes of mostly ordered and mostly disordered proteins
| Phase type | Exon disorder <30% |
Exon disorder >70% |
Protein disorder <30% |
Protein disorder >70% |
||||
|---|---|---|---|---|---|---|---|---|
| Occurrence per cent | Average length | Occurrence per cent | Average length | Occurrence per cent | Average length | Occurrence per cent | Average length | |
| (0,0) | 23.36 | 45.25 | 27.09 | 42.93 | 23.00 | 45.93 | 35.72 | 39.13 |
| (0,1) | 12.56 | 46.93 | 12.61 | 56.77 | 12.55 | 48.07 | 11.22 | 62.48 |
| (0,2) | 10.44 | 44.54 | 6.63 | 59.65 | 10.16 | 45.25 | 5.87 | 64.04 |
| (1,0) | 12.93 | 45.73 | 11.93 | 55.60 | 13.00 | 46.76 | 10.18 | 61.72 |
| (1,1) | 11.95 | 55.05 | 17.77 | 63.97 | 12.98 | 54.76 | 15.90 | 68.94 |
| (1,2) | 6.91 | 47.84 | 6.06 | 67.30 | 6.83 | 48.84 | 5.39 | 71.77 |
| (2,0) | 10.58 | 43.43 | 6.87 | 64.55 | 10.21 | 44.27 | 6.57 | 68.66 |
| (2,1) | 6.43 | 45.29 | 6.52 | 56.24 | 6.51 | 45.63 | 4.78 | 59.17 |
| (2,2) | 4.85 | 42.44 | 4.51 | 61.07 | 4.74 | 42.36 | 4.37 | 73.40 |
| All | 100 | 46.47 | 100 | 55.68 | 100 | 47.22 | 100 | 56.41 |
| Symmetric | 40.16 | 47.83 | 49.37 | 52.16 | 40.72 | 48.32 | 55.99 | 50.27 |
| Asymmetric | 59.84 | 45.56 | 50.63 | 59.12 | 59.28 | 46.46 | 44.01 | 64.21 |
We were also curious to know whether the increased disorder associated with symmetric exons reflects the disorder of only the region corresponding to the exon, the entire protein or both because there are several possible scenarios for the use of symmetric disordered modules in the assembly of proteins encoded by multi-exon genes. It is possible that exons encoding for these are incorporated into the gene of a mostly ordered or mostly disordered protein, or both. To this end, we correlated exon-phase preferences with the predicted disorder of the entire protein, distinguishing mostly ordered (<30% disorder) and mostly disordered (>70% disorder) proteins (Table 2). The preference for symmetric exons is even more striking here than in the previous cases: 55.99% of the exons of mostly disordered proteins are symmetric, highly significantly more than 40.72% of exons of mostly ordered proteins (P < 0.0001) (Table 2 and Figure 1C), suggesting prevalent modular evolutionary assembly mechanism relying on the shuffling of symmetric exons (see also ‘Select Examples of Proteins Assembled from Disordered Modules Encoded by Symmetric Exons’ section). Interestingly, the level of disorder of regions of symmetric exons strongly correlates with the overall level of disorder of the protein (P < 0.0001) (Supplementary Figure S1). Actually, disorder corresponding to all/asymmetric exons also strongly correlates with the overall level of disorder of the protein (P < 0.0001), but the average disorder is always higher in the case of symmetric exons, which may infer that in the evolutionary construction of ordered proteins, mostly ordered symmetric exons have been used (5,6), whereas in the evolutionary construction of disordered proteins, mostly disordered symmetric exons have been preferred.
Modular assembly of proteins
The previous results indicate that two complementary strategies might have been used for the construction of modular proteins, either dominated by the assembly of ordered modules or disordered modules. Of course, the slight preferences do not exclude the complementary use of the two types of modules, which would occur if modular proteins composed of ordered domains and disordered linkers (26) are assembled, for example. In light of the preference of disordered proteins for the presence of repetitive regions (30), however, this observation is compatible with internal tandem duplications of exons. To check whether the combination of phase symmetry and structural disorder has played a role in generating multi-exon genes by this mechanism, first we asked whether structural disorder associated with successive symmetric exons (necessarily of the same phase class) tends to correlate. We found that this is the case: when at least three symmetric exons are found in succession, the corresponding level of predicted disorder is significantly higher than the average (P = 0.0001; using unpaired t-test) (Table 1, Figures 1A and 2A and Supplementary Table S1). Moreover, both among exons of local disorder and exons encoding for disordered proteins, successive symmetric exons occur with a significantly elevated frequency (P < 0.0001 in both cases, with χ2 test) (Figure 2B).
Figure 2.
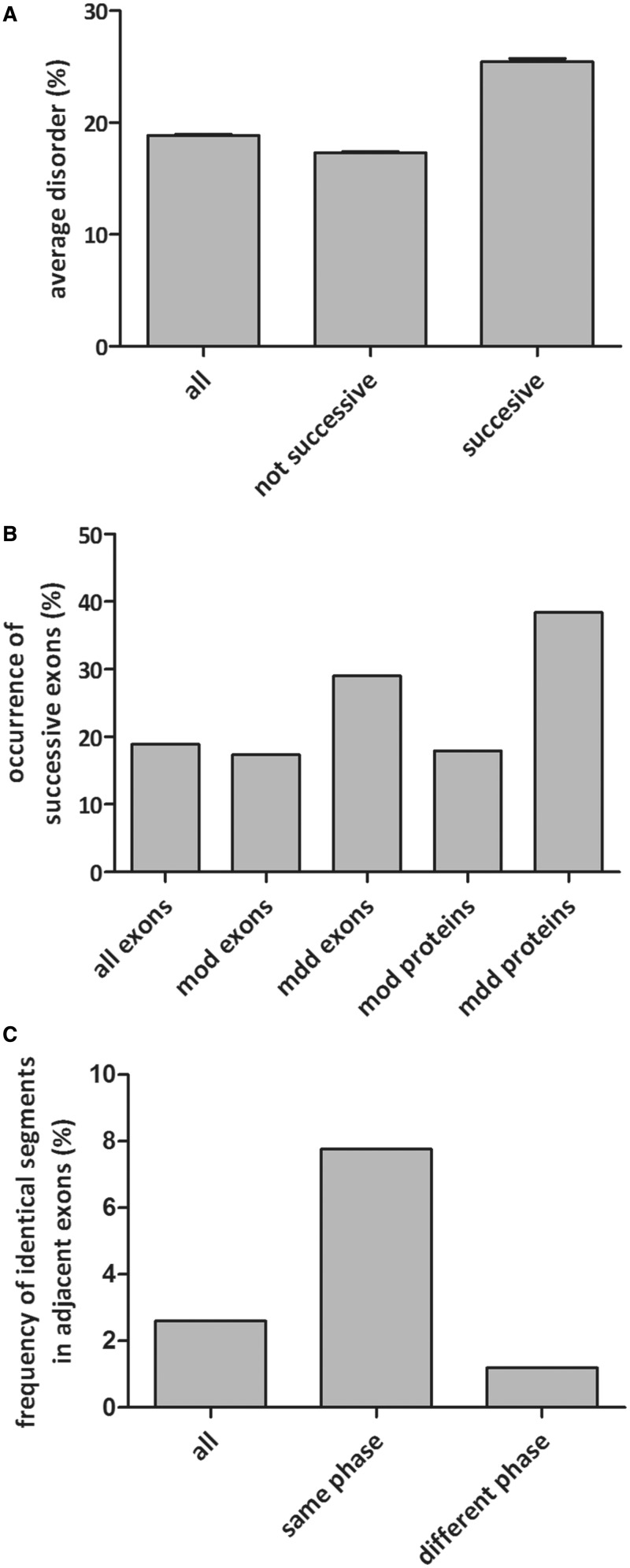
Disorder and occurrence of adjacent exons. (A) Average structural disorder (per cent of predicted disordered amino acids) encoded by all human exons, at least three consecutive (successive) exons with the same phase and all other, not consecutive exons. All three values are significantly different (with unpaired t-test). (B) Occurrence of at least three successive exons with the same phase for all exons, mostly ordered (mod, <30% disorder) exons, mostly disordered (mdd, >70% disorder) exons and such exons located in mostly ordered (mod) or mostly disordered (mdd) proteins. (C) Frequency of sequentially similar segments in all adjacent exons and in adjacent exons with the same or different phase types.
To provide evidence that internal duplications have been preferred in these cases, we have looked whether the sequences of subsequent exons of the same symmetric phase class tend to be more similar than those of random consecutive exons. We compared the per cent occurrence of segments of significant similarity by BlastP search in different exon sets (Figure 2C and Supplementary Table S2). We found that among consecutive symmetric exons, similarity occurs much more frequently than among random consecutive exons (7.8 versus 2.6%) and even more than among asymmetric consecutive exons (1.19%). These values are highly significantly different from each other (see ‘Materials and Methods’ section). This preference is even more evident if we compare the phase classes of neighbouring exons having highly similar segments (identity is >70%): in 108 of 118 cases (91.5%), consecutive exons have the same (symmetric) phase type.
These results are indicative of the preferred evolution of complex genes by internal duplications of exons, especially disordered symmetric exons, which would suggest that it can be observed much more frequently in genes constituted of more exons. First we checked, whether the long genes are long because of exon duplication. Blastp searches were carried out to identify homology, and we compared the frequency of similarity of adjacent exons in ‘short’ (≤2 exons) and ‘long’ (≥5 exons) genes. Of course, among short genes, only two-exon genes can have exon duplication. We found that long genes have higher frequency of similarity of adjacent exons indicative of exon duplication (2.6% of neighbouring exons in 9.3% of proteins versus only 1.8% of two-exon genes, the differences are significant, P < 0.0001, shown by χ2 analysis). Sixty-five per cent of these presumably duplicated exons have the same phase type. Long genes can arise not only by internal duplications but also by shuffling of exons between distinct genes, both of which can signalled by exon-phase symmetry. To this end, we compared the occurrence of symmetric exons associated with disorder in ‘short’ (≤2 exons) and ‘long’ (≥5 exons) genes (Supplementary Table S3 and Figure 1D). In the case of short genes, symmetric exons encoded for more order than asymmetric exons (but, the difference is not significant between the disorder of symmetric and asymmetric exons in the case of short genes), which is in contrast to long genes (where the difference is significant with unpaired t-test, P = 0.001). In the case of the latter, the difference is most conspicuous if genes of mostly symmetric exons (≥ 80% symmetric exons, 45.21% disorder) are compared with genes constructed of preferentially asymmetric exons (≤20% symmetric exons, 16.04% disorder).
Phase and disorder of alternative and constitutive exons
The evolutionary (genomic) process of exon shuffling in the assembly of a gene has strong conceptual parallels with alternative splicing in mRNA maturation, when an exon is inserted into (or removed from) a mature transcript. Although the underlying mechanisms are entirely different, the possible consequence of the event on the translational frame and the structural integrity of the protein are the same, actually the two processes might be evolutionarily connected (31–33). In accord, we expected a similar evolutionary preference for phase symmetry and structural disorder in exons subject to alternative splicing. Here, we identified the phase class of all exons, which could be clearly identified to be expressed constitutively or alternatively (cf. ‘Materials and Methods’ section) and predicted their disorder (Table 3, please note values here somewhat differ from ‘all’ exons studied previously). In accord with previous studies (15,23), alternative exons correspond to significantly more disordered regions than constitutive exons (29.7 versus 18.8%). The difference is even more pronounced in case of symmetric exons (32.46 versus 19.79%). Differences are significant (P < 0.0001) in both cases, using unpaired t-test.
Table 3.
Disorder and occurrence of regions encoded by alternative and constitutive exons
| Phase type | Disorder (%) all | Disorder (%) alternative | Disorder (%) constitutive | Occurrence (%) all | Occurrence (%) alternative | Occurrence (%) constitutive |
|---|---|---|---|---|---|---|
| (0,0) | 21.48 | 29.95 | 19.25 | 24.3 | 22.38 | 24.87 |
| (0,1) | 22.13 | 29 | 20.15 | 12.43 | 12.3 | 12.47 |
| (0,2) | 16.47 | 24.18 | 14.78 | 9.65 | 7.68 | 10.23 |
| (1,0) | 21.45 | 28.35 | 19.94 | 12.64 | 10.04 | 13.4 |
| (1,1) | 26.77 | 34.46 | 22.46 | 13.4 | 21.28 | 11.09 |
| (1,2) | 20.2 | 28.28 | 17.83 | 6.69 | 6.7 | 6.69 |
| (2,0) | 16.89 | 22.33 | 15.8 | 9.89 | 7.34 | 10.63 |
| (2,1) | 21.98 | 28.46 | 19.95 | 6.17 | 6.49 | 6.07 |
| (2,2) | 21.26 | 34.8 | 16.22 | 4.83 | 5.79 | 4.55 |
| All | 21.26 | 29.7 | 18.8 | 100 | 100 | 100 |
| Symmetric | 23.12 | 32.46 | 19.79 | 42.5 | 49.4 | 40.5 |
| Asymmetric | 19.89 | 27.01 | 18.12 | 57.5 | 50.6 | 59.5 |
Interestingly, the preference for structural disorder is much more pronounced in the case of alternative versus constitutive exons than symmetric versus asymmetric constitutive exons. As will be discussed in detail, this difference comes from the pressure of the gene (product) to be viable both with and without the inclusion of the exon in the case of alternative splicing, which makes the situation much more demanding than in the case of a constitutive exon, which was only created and fixed once.
Symmetric exons as functional modules
The observed bias for phase symmetry and structural disorder may not necessarily infer a functional role for the region encoded by the exon, only that these exons are used because they do not disrupt either the translation reading frame or the structural integrity of the recipient protein. Structural disorder, however, is known for its manifold functional advantages (8–10), modularity (20) and association with motifs (22,23,34) and domains (24), which all suggest its potential for functional integration into proteins. To address this feature, we first scrutinized the length distribution of symmetric exons, which apparently have a preference to be used in the construction of modular genes (Figure 3). Exons encoding for mostly ordered, symmetric (<30% disorder) and mostly disordered, symmetric (>70% disorder) regions have a rather similar average length [46 and 56 residues, respectively, there is no significant difference (P = 0.11), cf. Table 2], but they differ in length distribution, with symmetric exons corresponding to disordered regions having significantly higher frequency in the short (<25 residues) and long (>75 residues) length range (Figure 3A). A similar difference can be seen when symmetric and asymmetric exons in this latter class are compared, with an excess of symmetric exons in the short length range (Figure 3B). These differences might infer that exons of order represent more uniform building blocks [domains, in accord with prior inferences (3,5,6)], whereas exons of disorder are much more spread out because they represent three different types of functional modules (motifs, linkers and domains). Although comparison of the motif and domain ‘content’ of exons of different lengths is not conclusive enough, the overrepresentation of certain motif and domain types points to this direction (see later in the text).
Figure 3.
Length distribution of exons. (A) The length distribution of symmetric, mostly ordered (<30% disorder, light grey) and symmetric, mostly disordered (>70% disorder, dark grey) exons. The two distributions have similar means (46 and 56 residues, respectively), with exons corresponding to symmetric disordered regions displaying a broader distribution that has a significant excess in the regions <25 and >75 residues. (B) The length distribution of mostly disordered symmetric (dark grey) and mostly disordered asymmetric (light grey) exons shows the same difference, asymmetric disordered exons having a tail in the long region.
To address the possibility of these associations, we elaborated on the tendency of short-symmetric exons of disorder (<25 residues) to encode short-functional motifs ELMs (28) or SLiMs (35), and that of longer ones (>70 residues) to be either linker regions or domains (24). To this end, we have searched whether there is a length dependence of the occurrence of motifs and domains in these exons, based on the ELM database and PFAM families, respectively. Apparently, predicting linear motifs is fraught with very high–false-positive rates (average coverage is 64% for all exons and 71% for exons of mostly disordered regions, without any clear length preferences), which precludes straightforward generalizations. On analysing PFAM data, we found that occurrence of domains in symmetric exons of disorder is more frequent within short exons, especially in the phase 0,0 type (Supplementary Figure S2).
That is, based solely on coverage, no clear distinction can be made between the occurrence of motifs and domain in short- versus long-symmetric exons encoding for disordered regions. The power of modularity in the assembly process, however, can be clearly demonstrated by the significant overrepresentation (compared with their expected occurrence) of certain functional motifs (Table 4 for top 10 hits, for further examples, see Supplementary Table S4) and PFAM domains (Table 5 for top 10 hits, for further examples, see Supplementary Table S5) in symmetric exons encoding for mostly disordered regions. In case of motifs, SH3-binding regions, a range of phosphorylation and some other post-translational modification sites are found to preferentially occur in these regions. Most of these motifs contribute novel protein–protein interaction sites (partner of SH3 domain and nuclear localization receptor) or post-translational modification site (sumoylation site), i.e. they extend the functionality of the recipient protein in a simple but straightforward way, in accord with recent results showing that disordered regions are often involved in rewiring protein–protein interaction networks (36). That is, their enrichment is a strong indication that their presence provided an adaptive advantage to the gene after shuffling, in accord with our conjecture that not only structural but also functional compatibility with the recipient protein drive the fixation of a shuffled exon. In case of domains, the picture is even more varied because domains contribute novel functionality to the recipient protein in a more subtle and complex way: often they enable the complex extension of the function of the protein, such as enabling chromatin remodelling (HMG14 and HMG17), inhibition of an enzyme (calpain inhibitor), regulation of ribosomal DNA gene transcription (e.g. Treacher Collins syndrome protein) or the elastic function of titin [PPAK motif (37)]. They may also be involved in protein–protein interactions, as exemplified by the collagen triple helix repeat, which is a robust example of changing the oligomerization status of the given protein. In all, these examples also confirm that evolutionary fixation of a novel shuffled exon not only relies on its compatibility with the gene and structural integrity of the order but also at lest as much on the functional extension of the resulting gene product (30).
Table 4.
List and occurrence of the top 10 ELM motifs overrepresented in short symmetric exons encoding for disordered regions
| ELMIdentifier | Overrepresentation of motif in symmetric, disordered, short exons | Motif regex | Motif description |
|---|---|---|---|
| LIG_SH3_1 | 7.86 | [RKY]..P..P | This is the motif recognized by class I SH3 domains. |
| LIG_RGD | 4.65 | RGD | The RGD motif can be found in many proteins of the extracellular matrix, and it is recognized by different members of the integrin family. The structure of the 10th type III module of fibronectin has shown that the RGD motif lies on an exposed flexible location. |
| LIG_SH3_3 | 4.12 | …[PV]..P | This is the motif recognized by those SH3 domains with a non-canonical class I recognition specificity. |
| LIG_EVH1_1 | 4.04 | [FILVY].{0,1}P.[PAILSK]P | Proline-rich motif binding to signal transduction class I EVH1 domains. |
| LIG_SH3_2 | 3.79 | P..P.[KR] | This is the motif recognized by class II SH3 domains. |
| MOD_SUMO | 3.4 | [VILMAFP](K).E | Motif recognized for modification by SUMO-1. |
| LIG_TRAF6 | 3.33 | ..P.E..[FYWHDE]. | TRAF6-binding site. Members of the tumour necrosis factor receptor (TNFR) superfamily initiate intracellular signalling by recruiting the C-domain of the TNFR-associated factors (TRAFs) through their cytoplasmatic tails. |
| LIG_SH3_4 | 3.22 | KP..[QK]… | This is the motif recognized by those SH3 domains with a non-canonical class II recognition specificity. |
| TRG_NLS_MonoCore_2 | 3.11 | [^DE]((K[RK])|(RK)) [KRP][KR][^DE] | Monopartite variant of the classical basically charged NLS. Strong core version. |
| TRG_NLS_Bipartite_1 | 2.73 | [KR][KR].{7,15}[^DE]((K[RK])|(RK))(([^DE][KR])|([KR][^DE]))[^DE] | Bipartite variant of the classical basically charged NLS. |
Table 5.
List and occurrence of the top 10 PFAM domains of predicted disorder, encoded in symmetric exons
| Pfam ID | Overrepresentation of domain in symmetric disordered exons | Domain description |
|---|---|---|
| PF01101.12 | 15.11 | HMG14 and HMG17 |
| PF03546.8 | 15.07 | Treacher Collins syndrome protein Treacle |
| PF00748.13 | 15.07 | Calpain inhibitor |
| PF12301.2 | 14.13 | CD99 antigen like protein 2 |
| PF01391.12 | 13.65 | Collagen triple helix repeat (20 copies) |
| PF02818.9 | 13.58 | PPAK motif |
| PF05279.5 | 12.59 | Aspartyl β-hydroxylase N-terminal region |
| PF12235.2 | 9.07 | Fragile X-related 1 protein C terminal |
| PF06583.6 | 9.07 | Neogenin C-terminus |
| PF06464.5 | 8.13 | DMAP1-binding domain |
Select examples of proteins assembled from disordered modules encoded by symmetric exons
Behind all the predictions, correlations and general observations, there are individual proteins, the structural disorder of which is experimentally characterized and is shown to be involved in function. Studying the gene structure of these proteins provides further evidence to the evolutionary agility of symmetric exons encoding for disordered region. Here, we present a few select examples that demonstrate how these modularity principles apply to the assembly of disordered proteins (Figure 4 and Supplementary Table S6).
Figure 4.
Select examples of proteins assembled from disordered modules encoded by symmetric exons. (A) A few long disordered proteins, encoded by genes that have an exon/intron structure indicative of modular assembly from symmetric disordered exons. In each case, the domain structure of the protein based on structural and functional data is outlined, and phases of introns separating exons are indicated above the domain structure. CST is the inhibitor of calpain, a calcium-activated cysteine protease. The inhibitor has four inhibitory domains (I, II, III and IV) (olive), each having three conserved subdomains (black) and an additional L-domain (purple) of unrelated function. MAP2 belongs to the microtubule-associated protein family. Its N-terminal domain (dark green) exhibits chaperone-like activity, it has a proline-rich region (P) (light blue) and four tubulin-binding repeats (R1-R4) (red) in the C-terminal region, connected by a middle projection domain (orange). MBD1 is member of a family of nuclear proteins. This protein contains multiple domains: MBD (yellow) three CXXC-type zinc-finger domains (dark blue) that mediate binding to non-methylated CpG dinucleotides and a transcriptional repression domain (TRD) (pink). (B) Modular assembly of the family of fully disordered secretory Ca-binding phosphoproteins, expressed in bone and teeth: DSPP, DMP1 and BSP. Enamel matrix proteins are AMEL, AMBN and ENAM regulate the deposition of inorganic phase in mineralized tissues (47). The family has a common ancestor from whom gene duplication led to two clusters: the enamel protein genes (AMBN-ENAM) and the bone-dentin protein genes (DSPP, DMP1, IBSP, MEPE and SPP1), in which diversification occurred by the insertion of functional [signal peptide, SP (orange); kinase phosphorylation site, SXE (yellow); proline-rich, PP (blue); a proline-rich phosphorylation site (olive) and an integrin-binding tripeptide, RGD (red)] regions and/or tandem duplications of the exons.
In three cases (Figure 4A), we show fully disordered proteins encoded by multi-exon genes with a strong bias for symmetric exons of the same phase class. Their exons correlate with functional regions/domains of the proteins, which argue that the success of shuffling of the exon also relied on the productive incorporation of the encoded disordered structural/functional module into the protein. (i) Calpastatin (CST) is the fully disordered (38,39) inhibitor of the calcium-activated cysteine protease calpain that undergoes limited induced folding on inhibition (38). The gene of the protein is assembled from class 1,1 symmetric exons. The protein has four calpain-inhibitory (I through IV) domains and an N-terminal L domain of unrelated function, perfectly matched by the exonic structure of the gene. (ii) Microtubule-associated protein 2 (MAP2) is a fully disordered protein (40) that belongs to the protein family regulating microtubule assembly. Tubulin-bound MAP2 exhibits chaperone-like activity likely because of the N-terminal domain containing several patches of acidic amino acids. The protein also has a projection domain, a proline-rich region (P) and four tubulin-binding repeats (R1–R4) in the C-terminal region (41). Its gene has been assembled from type 1,1 symmetric exons matching the functional regions of the protein. The two 1,0–0,1 exon pairs at the C-terminus can probably be also explained by the insertion of a Phase 0 intron into an ancestral symmetric 1,1 exon followed by the duplication of this exon pair. (iii) Methyl-CpG–binding protein 1 (MBD1) is the member of a family of nuclear proteins that have a methyl-CpG–binding domain (MBD). The protein is fully disordered (42), it can bind methylated DNA, and it can repress transcription from methylated gene promoters. MBD1 contains multiple domains: an N-terminal MBD, three CXXC-type zinc-finger domains that can bind non-methylated CpG dinucleotides and a transcriptional repression domain (TRD) at the C-terminus (43). Almost the entire protein has been assembled from type 0,0 exons (some tend to be ordered), reflecting the domain organization of the protein.
In Figure 4B, we show an entire family of homologous fully disordered (44–46) proteins that diverged from an ancestral gene by the module exchange mechanisms based on symmetric exons. These proteins are involved in biomineralization in the formation of teeth and/or bone. Enamel matrix proteins amelogenin (AMELX/Y), ameloblastin (AMBN) and enamelin (ENAM) organize and regulate hydroxyapatite crystallization in the enamel organ (47). Bone and teeth proteins dentin sialophosphoprotein (DSPP), dentin matrix acidic phosphoprotein 1 (DMP1) and bone sialoprotein (BSP) belong to the SIBLING (small integrin-binding ligand, N-linked glycoprotein) family (44). At the primary amino acid sequence level, these proteins show little similarity, but their functional relatedness, modular organization and exon/intron structure point to their common origin. It is thought that the entire family diverged from a common ancestor, SPARC (osteonectin, not included in our analysis). Apparently, the family evolved by gene duplication, the addition of class 0,0 modules and internal duplications, which have led to two clusters: the enamel protein genes (AMBN, ENAM and AMEL) and the bone-dentin protein genes (DSPP, DMP1, integrin-binding sialoprotein (IBSP), matrix extracellular phosphoglycoprotein (MEPE) and secreted phosphoprotein 1 (SPP1)) (47,48). Functional modules and exons are in close correspondence and show the gradual evolution and diversification of the family. The first coding exon is the signal peptide and the first two amino acids of the mature protein (SP + AA), whereas exon 2 usually contains the consensus sequence (SXE) for casein kinase II phosphorylation. Exon 3 in the SIBLING family is usually somewhat proline-rich (PP), and among the acidic proteins it is the only significantly positive-charged domain. Exon 4 usually contains another casein kinase II site, and a unique integrin-binding Arg-Gly-Asp (RGD) motif has been added on the last exon. Enamel matrix proteins, however, contain only the first two domains, followed by the exon homologous to exon 2 of DSSP repeated as many as 10 times (in AMBN) (47,48).
DISCUSSION
Modularity is a powerful principle in the evolution of proteins of novel/altered activity because it facilitates the generation of novel combinations of already existing structural and functional elements. In accord, modularity is apparent both at the genome (exons, genes) and proteome (structural building blocks, motifs, domains) levels. It has been suggested that the split nature of genes facilitates the creation of novel genes through re-shuffling of exons via intronic recombination (49). Although the correlation of exons and domains does not always hold, exon shuffling is undoubtedly a prevalent mechanism of the generation of complex genes encoding for multi-domain proteins, as evidenced by the genomic bias for exon-phase symmetry (1–3,5). Because inclusion/exclusion of symmetric exons does not impair the translation reading frame, they are favoured in shuffling reactions. Further, their incorporation into a recipient gene may result in a protein product of altered/improved functionality, provided they encode for autonomous structural/functional units of proteins (domains) (5,6).
It occurred to us that this principle may be extended to structurally disordered proteins/regions. Because modularity in the form of motifs, linkers and disordered domains is often encountered in disordered proteins (13,22–24,26), and the incorporation of a disordered segment into a host protein might not impair the structural integrity of the whole protein (15,34), we expected that the phase symmetry of exons and the structural disorder of the protein regions they encode correlate. By analysing the human genome, we found significant correlations, which suggest that the genetic potential of exon shuffling and the insertion/functional potential of structural disorder act in synergy in the evolution of novel modular genes/proteins.
The first explanation for this observed bias might be the preferential shuffling of symmetric exons encoding for structural disorder (e.g. because of functions associated with these regions or particular base frequency bias). Of course, ‘exon shuffling’, as we see it today, is a net result of two mechanisms, the exchange of genetic material (which we may call the actual shuffling, let it occur by gene conversion, meiotic recombination and tandem duplication) and subsequent selection (for or against) the new exon (new gene), which is independent of the original genetic mechanism and largely works on the viability/functional fitness of the new protein product. It is reasonable to ask whether shuffling of an exon preferably occurs if the exon is symmetric and/or encodes for a disordered region. It seems symmetric and asymmetric exons are shuffled alike because the mechanism of shuffling does not care about the position of the beginning and end of an exon (which is only defined in RNA, at the stage of splicing). Structural disorder also does not seem to matter much because recombination hot spots, as defined by transposable elements (TEs), such as Long interspersed elements (LINEs) and Short interspersed elements, e.g. Alu repeats (SINEs), correlate with genes that are involved in processes of external stimuli, immunity, cellular signalling, transport and signalling (50), or metabolism, transport and signalling (51). GC richness in the genome also seems to correlate with TEs, none of these previous features are strongly correlated with structural disorder, i.e. preferential recombination driven by TEs is probably not the primary mechanism responsible for the observed preference of exon-phase symmetry and structural disorder.
An integrated novel genetic element is much more likely to be selected because of its structural and functional compatibility with the recipient gene and encoded protein. In the case of folded proteins, this dilemma is thought to be solved if the exon encodes for a domain (or secondary structure element) inserted at an appropriate point (most often in a loop) (3,5,6). In the case of disordered proteins (regions), this is not that much of an issue because they can easily accommodate multiple conformations (9,52) imposed by different end-point positions in the host protein. This is witnessed in alternative splicing, the conceptual equivalent of exon shuffling, which also inserts a novel segment into a protein and is facilitated by structural disorder (15,34). Novel symmetric exons brought into the gene by exon shuffling may equally well benefit, in an evolutionary sense, from encoded disorder.
It is of equal importance, however, that the encoded region functionally integrates into the protein. Structural disorder has been linked with many functional attributes, such as uncoupling specificity from binding strength, adaptation to different partners, regulation by post-translational modifications and rapid association in binding reactions (8–10) and disordered regions often harbour functional elements, such as SLiMs/ELMs, linkers and domains (13,22,25,34,53). This is also suggested by the observed length distribution of symmetric exons encoding for disordered regions: they apparently have the capacity to encode distinct functional elements ranging from motifs to domains. Incorporation of these elements might add to the functional repertoire of the protein (changing activity, subcellular localization, protein–protein interactions and phase transitions) and advance the mechanism of exon shuffling. This is clearly seen in the case of the select examples, when a symmetric exon can encode for a SLiM/ELM (e.g. integrin-binding RGD motif in bone-dentin proteins), a disordered domain (e.g. inhibitory domain in CST) or a linker region (e.g. the projection domain in MAP2). By looking at domains and motifs overrepresented in symmetric disordered exons, we also found that most of them contribute a novel protein–protein interaction site that mediates interaction with SH3 domains, nuclear transport receptors or some other modular interaction domain (enabled/VASP homology 1 domain (EVH1), TRAF domain) or serve as post-translational modification site (sumoylation). Introduction of a disordered domain via a symmetric exon may contribute more complex functionality (chromatin rearrangement and enzyme inhibition). In all, it is rather clear from the examples of overrepresentation that their evolutionary inclusion provides functional advantage, which contributes to fixation of the shuffled exon because it modulates the function of the protein by either affecting activity of the protein or its interactions with partner proteins. This is fully in line with recent observations based on comparing the human, fly and yeast interactomes (36), in which disordered proteins/regions are preferentially involved in rewiring interaction patterns of proteins. In all, all these examples suggest that shuffling enabled by phase symmetry and structural compatibility with the recipient protein because of structural disorder are necessary but not sufficient conditions for the evolution fixation of the shuffled exon: functional compatibility of the novel element of motif/domain must also come into picture for lasting fixation.
Although its molecular mechanism is entirely different from exon shuffling, strong parallels of the structural and functional implications make alternative splicing pertinent to these points. Our results also show that alternative splicing is significantly correlated with exon-phase symmetry and structural disorder. Previous studies also verified the correlation of alternative splicing with structural disorder (15,34). Not unexpectedly, alternatively spliced regions are (also) enriched in functional motifs (23,34), and their absence/presence also promotes functional diversity of proteins and rewiring of the interactome (23,54,55). Intriguingly, structural disorder is a stronger feature distinguishing alternative exons from constitutive exons than symmetric exons from asymmetric ones, which suggests an even stronger influence in the case of alternative splicing (Table 3). The most likely reason is that constitutive exons have been shuffled and fixed only once, which is far less demanding on protein structure and function than alternative splicing, in the case of which both gene products have to be viable at the same time. This actually highlights the strength of structural disorder in making a region acceptable in a new gene product that arises as a result of the inclusion of a new exon. The significantly less influence on symmetry/asymmetry status than on alternative/constitutive status may also be compatible with the toleration of alternatively spliced asymmetric exons because of the compatibility of structural disorder with frame shift, as established earlier (56).
These strong parallels, despite unrelated molecular mechanisms, might even imply an evolutionary link between exon shuffling and alternative splicing. An exon selected for following exon shuffling—because of its phase symmetry and encoded structural disorder—may also have a better chance to be alternatively spliced. In fact, it was observed that a large proportion of species-specific exons (i.e. human exons that arose rather recently in evolution) are also alternatively spliced (31–33). Apparently, these younger exons have weaker splice-sites and a lower abundance of splicing regulators, which might point to a deeper underlying correlation between exon shuffling and alternative splicing, which remains to be seen.
A further interesting aspect of exon shuffling by virtue of exon symmetry is the effective generation of tandem repeats. This is clearly the case in our select examples (e.g. AMBN; Figure 4B) and is statistically verified by the increased homology of subsequent symmetric exons. This mechanism is probably also promoted by functional selection because of the statistical overrepresentation of encoded SLiMs. It was found that the same SLiM often reappears in a protein, and SLiMs also often occur in tandem repeats (30,53). The ensuing functional advantages of this arrangement are apparent because cognate-binding domains might also occur in tandem (57); thus, repetition of the motif may result in an increased avidity, specificity, even complex regulatory phenomena based on cross-talk between tandem binding and post-translational modification sites (55). Specific functional attributes of multiple adjacent post-translational modification sites have been observed, for example, ultrasensitivity of binding of yeast Sic1 cell-cycle regulator to Cdc4, the substrate-recognition subunit of its cognate E3 ubiquitin ligase (58). A recent exciting development in the field of intrinsically disordered proteins even suggests a physical perspective to this phenomenon because the interaction of repetitive motifs and repeated domains [for example, in the binding of Wiskott-Aldrich syndrome protein (WASP) to non-catalytic region of tyrosine kinase adaptor protein 1 (NCK) (59) or between low-complexity regions of RNA-binding proteins (60)] can cause a phase transition in the form of micrometre-sized liquid droplets. This transition depends on the valency of both partners, and it can help bridge the length scales of proteins (angstrom) to that of organelles and cells (micrometres). The transition can be regulated by post-translational modification(s), and it can regulate protein activity.
In all, the effective operation of the mechanism put forward in this article may also shed some light on the advance of protein disorder in eukaryotic evolution. It has been often stated that the occurrence of structural disorder is higher in eukaryotes than in prokaryotes (12,16–19), which is associated with their function in signalling and regulation (12,16). The underlying genetic mechanisms, however, have hardly ever been addressed. Here, we can add one prevalent mechanism, module exchange based on exon shuffling, which may have contributed to the eukaryotic success story of structural disorder. In effect, we might actually suggest that the potential of evolutionary creation of novel genes, as outlined in this article, can be added to the ‘functional’ advantages of structural disorder.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: FWO Odysseus [G.0029.12]; Korean–Hungarian Joint Laboratory grant from Korea Research Council of Fundamental Science and Technology (KRCF); FP7 Infrastructures [261863] (BioNMR) from the European Commission.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Fedorov A, Fedorova L, Starshenko V, Filatov V, Grigor'ev E. Influence of exon duplication on intron and exon phase distribution. J. Mol. Evol. 1998;46:263–271. doi: 10.1007/pl00006302. [DOI] [PubMed] [Google Scholar]
- 2.Long M, Rosenberg C, Gilbert W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl Acad. Sci. USA. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patthy L. Intron-dependent evolution: preferred types of exons and introns. FEBS Lett. 1987;214:1–7. doi: 10.1016/0014-5793(87)80002-9. [DOI] [PubMed] [Google Scholar]
- 4.Patthy L. Exon shuffling and other ways of module exchange. Matrix Biol. 1996;15:301–310. doi: 10.1016/s0945-053x(96)90131-6. discussion 311–302. [DOI] [PubMed] [Google Scholar]
- 5.Kaessmann H, Zollner S, Nekrutenko A, Li WH. Signatures of domain shuffling in the human genome. Genome Res. 2002;12:1642–1650. doi: 10.1101/gr.520702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee B. Comparison of exon-boundary of old and young domains during metazoan evolution. Genomics Inform. 2009;7:131–135. [Google Scholar]
- 7.Patthy L. Introns and exons. Curr. Opin. Struct. Biol. 1994;4:383–392. [Google Scholar]
- 8.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 9.Tompa P. Unstructural biology coming of age. Curr. Opin. Struct. Biol. 2011;21:419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 12.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Tompa P, Kalmar L. Power law distribution defines structural disorder as a structural element directly linked with function. J. Mol. Biol. 2010;403:346–350. doi: 10.1016/j.jmb.2010.07.044. [DOI] [PubMed] [Google Scholar]
- 14.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and instrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 15.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tompa P, Dosztanyi Z, Simon I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J. Proteome. Res. 2006;5:1996–2000. doi: 10.1021/pr0600881. [DOI] [PubMed] [Google Scholar]
- 17.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burra PV, Kalmar L, Tompa P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS One. 2010;5:e12069. doi: 10.1371/journal.pone.0012069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancsa R, Tompa P. Structural disorder in eukaryotes. PLoS One. 2012;7:e34687. doi: 10.1371/journal.pone.0034687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pentony MM, Jones DT. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78:212–221. doi: 10.1002/prot.22504. [DOI] [PubMed] [Google Scholar]
- 21.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 23.Weatheritt RJ, Davey NE, Gibson TJ. Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res. 2012;40:7123–7131. doi: 10.1093/nar/gks442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31:328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 25.Daughdrill GW, Narayanaswami P, Gilmore SH, Belczyk A, Brown CJ. Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino Acid sequence conservation. J. Mol. Evol. 2007;65:277–288. doi: 10.1007/s00239-007-9011-2. [DOI] [PubMed] [Google Scholar]
- 26.Balazs A, Csizmok V, Buday L, Rakacs M, Kiss R, Bokor M, Udupa R, Tompa K, Tompa P. High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, CASK-interactive protein 1. FEBS J. 2009;276:3744–3756. doi: 10.1111/j.1742-4658.2009.07090.x. [DOI] [PubMed] [Google Scholar]
- 27.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 28.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 29.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays. 2003;25:847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]
- 31.Corvelo A, Eyras E. Exon creation and establishment in human genes. Genome Biol. 2008;9:R141. doi: 10.1186/gb-2008-9-9-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XH, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 2010;38(Suppl.):W534–W539. doi: 10.1093/nar/gkq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosca R, Pache RA, Aloy P. The role of structural disorder in the rewiring of protein interactions through evolution. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014969. M111.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greaser M. Identification of new repeating motifs in titin. Proteins. 2001;43:145–149. doi: 10.1002/1097-0134(20010501)43:2<145::aid-prot1026>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 38.Kiss R, Bozoky Z, Kovacs D, Rona G, Friedrich P, Dvortsak P, Weisemann R, Tompa P, Perczel A. Calcium-induced tripartite binding of intrinsically disordered calpastatin to its cognate enzyme, calpain. FEBS Lett. 2008;582:2149–2154. doi: 10.1016/j.febslet.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 39.Kiss R, Kovacs D, Tompa P, Perczel A. Local structural preferences of calpastatin, the intrinsically unstructured protein inhibitor of calpain. Biochemistry. 2008;47:6936–6945. doi: 10.1021/bi800201a. [DOI] [PubMed] [Google Scholar]
- 40.Hernandez MA, Avila J, Andreu JM. Physicochemical characterization of the heat-stable microtubule-associated protein MAP2. Eur. J. Biochem. 1986;154:41–48. doi: 10.1111/j.1432-1033.1986.tb09356.x. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar T, Mitra G, Gupta S, Manna T, Poddar A, Panda D, Das KP, Bhattacharyya B. MAP2 prevents protein aggregation and facilitates reactivation of unfolded enzymes. Eur. J. Biochem. 2004;271:1488–1496. doi: 10.1111/j.1432-1033.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- 42.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 43.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem. Biophys. Res. Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 45.Kalmar L, Homola D, Varga G, Tompa P. Structural disorder in proteins brings order to crystal growth in biomineralization. Bone. 2012;51:528–534. doi: 10.1016/j.bone.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Kaplon TM, Rymarczyk G, Nocula-Lugowska M, Jakob M, Kochman M, Lisowski M, Szewczuk Z, Ozyhar A. Starmaker exhibits properties of an intrinsically disordered protein. Biomacromolecules. 2008;9:2118–2125. doi: 10.1021/bm800135m. [DOI] [PubMed] [Google Scholar]
- 47.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc. Natl Acad. Sci. USA. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sire JY, Delgado S, Fromentin D, Girondot M. Amelogenin: lessons from evolution. Arch. Oral Biol. 2005;50:205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Gilbert W. The exon theory of genes. Cold Spring Harb. Symp. Quant. Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 50.Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- 51.Grover D, Majumder PP, Rao CB, Brahmachari SK, Mukerji M. Nonrandom distribution of alu elements in genes of various functional categories: insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol. 2003;20:1420–1424. doi: 10.1093/molbev/msg153. [DOI] [PubMed] [Google Scholar]
- 52.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 53.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol. Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 54.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Weatheritt RJ, Gibson TJ. Linear motifs: lost in (pre)translation. Trends Biochem. Sci. 2012;37:333–341. doi: 10.1016/j.tibs.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Kovacs E, Tompa P, Liliom K, Kalmar L. Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc. Natl Acad. Sci. USA. 2010;107:5429–5434. doi: 10.1073/pnas.0907841107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 58.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl Acad. Sci. USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.