Abstract
Studying complex biological processes such as cancer development, stem cell induction and transdifferentiation requires the modulation of multiple genes or pathways at one time in a single cell. Herein, we describe straightforward methods for rapid and efficient assembly of bacterial marker free multigene cassettes containing up to six complementary DNAs/short hairpin RNAs. We have termed this method RecWay assembly, as it makes use of both Cre recombinase and the commercially available Gateway cloning system. Further, because RecWay assembly uses truly modular components, it allows for the generation of randomly assembled multigene vector libraries. These multigene vectors are integratable, and later excisable, using the highly efficient piggyBac (PB) DNA transposon system. Moreover, we have dramatically improved the expression of stably integrated multigene vectors by incorporation of insulator elements to prevent promoter interference seen with multigene vectors. We demonstrate that insulated multigene PB transposons can stably integrate and faithfully express up to five fluorescent proteins and the puromycin-thymidine kinase resistance gene in vitro, with up to 70-fold higher gene expression compared with analogous uninsulated vectors. RecWay assembly of multigene transposon vectors allows for widely applicable modelling of highly complex biological processes and can be easily performed by other research laboratories.
INTRODUCTION
As our understanding of complex biological pathways and networks becomes more thorough, the genetic engineering needed to model biological phenomenon becomes more complicated and challenging. To further study and model the details of these complex phenomena, the ability to modulate multiple genes and/or pathways at one time in a single cell is needed. For instance, the discovery that fully differentiated cells, such as skin fibroblasts, can be reprogrammed to multipotent-induced pluripotent stem cells (iPS) by overexpressing four genes in a single cell (OCT3/4, SOX2, KLF4 and c-MYC) clearly demonstrates the power of modulating multiple genes simultaneously (1). Even a phenotype as complex as cancer development, or cellular transformation, has been induced by the modulations of many genes in human primary cells (2–5). Thus, having the ability to modulate many genes in a single cell allows for more accurate modelling and subsequent understanding of a wide variety of complex biological processes.
Commonly used methods for modulating many genes in a single cell are currently laborious and time consuming. Most experiments use serial or concurrent addition of each gene of interest using lentiviral/retroviral transduction or stably integrating linearized plasmid DNA (1–4,6). Integration of plasmid DNA is inefficient and frequently produces less than optimal results, as partial fragments of the plasmid may be integrated (7). Though lentiviral/retroviral transduction is highly efficient, the small cargo capacity (∼10–12 kb) is a limiting factor (8). Furthermore, when more than one gene is modulated in a single vector, there is typically a single promoter combined with viral elements such as internal ribosomal entrance sequences (IRES), ribosomal skip sequences or self-cleaving peptides (9–11). These viral elements and vectors typically do not allow for stoichiometric expression, require very precise cloning procedures to produce, are not amenable to very large genes and can even be repressed in some cell types by triggering cellular immunity systems (12,13).
Ideally, each gene of a multigene vector would be expressed from its own promoter to achieve high-level, reliable expression as is seen in mammalian cells. One of the largest hurdles to this strategy is the actual construction of the multigene vector, as they become very large and repetitive, as each promoter-gene element is added, making traditional restriction enzyme (RE) cloning difficult or impossible. Nevertheless, systems to generate multigene vectors using recombinases (e.g. MulitiLable, Multisite Gateway) or type IIS REs (e.g. GoldenBraid, Modular Cloning) have been described (14–17). The obvious downfall of using REs to assemble multigene vectors is that any desired gene or element containing the RE recognition site cannot be used. The use of site-specific recombinases overcomes this issue, but all previously described recombinase systems have one or more of the following issues: complicated to use, inefficient, use non-modular components for assembly or produce final vectors that contain large amounts of bacterial sequence.
Beyond the shortcomings of simple and efficient multigene vector assembly, the validation of gene expression and delivery of correctly assembled multigene vectors is also substantially lacking in all prior publications. The assembled multigene vectors intended for mammalian gene expression have only been validated by transient transfection in human cells. Transient transfection masks a major issue with multigene vectors, namely promoter interference (18). It has been demonstrated by numerous publications that tandem promoters suffer from promoter interference (18–22). However, it has been shown that the addition of a tandem copy of the chicken beta-globin insulator (cHS4) alleviates promoter interference substantially when placed between two stably integrated tandem EF1A promoters (23,24).
Owing to the limitations of the aforementioned multigene assembly methods, we developed a system to quickly and efficiently assemble multigene vectors in which each complementary DNA (cDNA)/short hairpin RNA (shRNA) is under the control of a single ubiquitous promoter with a single poly adenylation (pA) signal. This method, which we have termed RecWay assembly, uses both Cre recombinase and the Gateway cloning technology, removing the need for any traditional RE-based cloning. We also engineered our vectors both with and without tandem copies of the cHS4 insulator between each promoter-gene-pA element to assess and eliminate potential promoter interference. Because the general use of multigene vectors is also, in part, hindered by the ability to reliably deliver the large vectors intact into mammalian cells, we used the piggyBac (PB) DNA transposon system. PB has been shown to be highly active in mammalian cells and capable of mobilizing large vectors of >100 kb (25,26). Thus, we designed our multigene cassettes to be flanked by the PB inverted terminal repeats to allow for integration via transposon transposition.
Herein, we describe methods for highly efficient assembly of transposon integratable, multigene vectors using RecWay assembly. We validated the system using multigene vectors containing up to five unique fluorescent proteins in addition to the puromycin-thymidine kinase fusion (puro/tk) in human and mouse cells by both transient and stable expression. Further, we demonstrated that the use of insulators dramatically improves gene expression of stably integrated multigene vectors and increased the frequency of generating stable integration clones. Finally, we show a four-gene RecWay vector works in primary hepatocytes in situ in living mice to induce liver tumours. RecWay assembly combined with PB transposon delivery will allow for expedited development of multigene vectors and efficient delivery for experiments requiring stable and reversible overexpression or knockdown of multiple genes in a single cell.
MATERIALS AND METHODS
Cell culture and transfections
Normal immortalized human schwann cells, human embryonic kidney 293T (HEK293T) and NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium media (Cellgro) supplemented with 10% fetal bovine serum and 1% Penicillin/Streptomycin. All cells were grown at 37°C with 5% CO2. The immortalized human schwann cell and NIH 3T3 cells were electroporated using the NEON electroporation system (Invitrogen), following manufacturer’s protocols. Briefly, one million cells were resuspended in 100 µL of R buffer and 4 µg of transposon and 4 µg Super PB transposase (System Biosciences) added. Cells were then loaded into 100 µL of electroporation tips and electroporated using pre-optimized parameters (Invitrogen). HEK293T cells were transfected using X-tremeGENE HP DNA Transfection Reagent (Roche). Cells were plated 1 day before transfection in 6-well plates to obtain 80–90% confluency at time of transfection. One microgram of transposon and 1 µg Super PB transposase (System Biosciences) were mixed with 3 µL of Lipofection reagent (2:3 ratio) in 100 µL of Opti-MEM serum free media (Invitrogen) and allowed to incubate for 15 min and added to the media in a drop-wise fashion. Cells were allowed to incubate with the transfection reagent for 48 h before fluorescent imaging. For colony formation assays, 250 000 cells were plated in triplicate 48 h after transfection.
Microscopy and fluorescence imaging
Cells expressing fluorescent proteins were imaged using the Olympus IX70 Inverted Microscope (Arcturus) equipped with a ProgRes C12 Plus camera at the University of Minnesota imaging centre. Images were taken using the ProgRes Capture Pro v2.8.8 software package (Jenoptik Optical Systems).
RNA extraction and quantitative reverse transcriptase-polymerase chain reaction
RNA was extracted from cell culture pellets using the RNA mini-prep kit (Invitrogen) and reverse transcribed into cDNA using the Superscript III reverse transcriptase kit (Invitrogen), following manufacturer’s instructions. Quantitative reverse transcriptase-polymerase chain reaction (QRT-PCR) was performed using FastStart Universal SYBR Green Master (Rox) mix (Roche) using a Eppendorf Realplex2 Mastercycler EP Gradient S. Primer sequences are listed in Supplementary Table S2.
Mice
Hydrodynamic injections for in vivo gene delivery to the liver were performed as previously described (27). Fah−/− mice were maintained with 7.5 µg/ml of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione drinking water and replaced with normal drinking water after hydrodynamic injection of Fah cDNA expressing PB transposons. Ten micrograms of transposon vector and 10 ug of pCMV-PB7 transposase were used for hydrodynamic injections. Luciferase imaging was performed 2 weeks post injection and performed as previously described (28).
Vectors construction and LR clonase reactions
Plasmids containing the ccdb gene were maintained in One Shot® ccdB Survival™ 2 T1R bacteria (Invitrogen). All REs used in this study were purchased from New England BioLabs. All pENTR1 vectors were derived originally from pENTR221-GUS (Invitrogen). The pENTR2 vector was constructed using attL4 and attL3, and the Destination (DEST)2 cassette was constructed using attR4 and attR3. These alternative att sites are from the Gateway multifragment assembly kit and were taken from the Tol2 kit (29). Briefly, p3E-EGFP-pA was digested with MfeI and SspI to produce a fragment containing attL3 and part of the kanamycin resistance gene, which was then cloned into p5E-MCS at MfeI and SspI sites flanking the attR1 and the same fragment of the kanamycin coding sequence, repairing the kanamycin resistance gene. The Tetracycline resistance gene was cloned into pENTR2 (Kan) from pBR322 (Invitrogen) using blunted BamHI sites flanking the Tetracycline gene into blunted EcoRI and AvaI sites in pENTR2 (Kan) to create pENTR2 (Kan/Tet). The kanamycin resistance gene was then removed by BspHI digestion and self-ligation to create pENTR2 (Tet). The DEST2 cassette was generated by replacing the chloramphenicol resistance gene of pDEST-TOL2-CG2 with the Tetracycline resistance gene from pBR322. The Tetracycline resistance gene was cut out of pBR322 with BamHI and blunted; this fragment was then cloned in place of chloramphenicol by blunting EcoRI- and AvaI-digested pDEST-TOL2-CG2. The DEST2 cassette was then removed using blunted KpnI and SfiI sites and cloned into R26 (−) Shuttle at a blunted NheI site.
Shuttle vectors were assembled from a multistep cloning process starting with R26 (−) Shuttle plasmid. The pDual-DEST (Kan) was constructed by first PCR amplifying EF1A-DEST1 from pKO-EF-1A-DEST1 with flanking EcoRV sites engineered in the PCR primers and cloned into the EcoRV site of R26 (−) Shuttle to produce Shuttle-EF1A-DEST1 (Kan). EF1A-DEST2-pA was PCR amplified from R26 (−) Shuttle-EF1A-DEST2 with SbfI and NsiI sites engineered into the PCR primers and cloned into the SbfI site of Shuttle-EF1A-DEST1 (Kan) to produce pDual-DEST (Kan). The pDual-DEST Shuttle (Cm) and pDual-DEST Shuttle (Spec) were cloned using the same method as pDual-DEST (Kan) but started with the R26 (−) Shuttle-FRT3-Cm or R26 (−) Shuttle-FRT14-Spec, respectively. Shuttle vectors containing the tandem cHS4 insulators were constructed using NsiI-flanked cHS4 from PCR-II-TOPO-cHS4 cloned into the SbfI site of all three pDual DEST vectors. The second cHS4 insulator was cloned into a single ZraI site of the (Kan) and (Spn) vectors using SspI- and HincII-flanking cHS4 from the pTraffic vector. PB-Dual-DEST (Kan) and all other dual expression transposon vectors were constructed using the flanking I-SceI sites of pDual-DEST (Kan) cloned into a single I-SceI site of PB-I-SceI transposon vectors. Single BP and LR Clonase reactions were performed using manufacturer’s protocols with Clonase II enzyme mixes (Invitrogen). Dual LR Clonase reactions were performed in the same fashion by replacing the appropriate amount of Tris-EDTA(Ethylenediamine Tetraacetic Acid) (TE) buffer with pENTR2 plasmid.
Immunohistochemistry and H&E staining
H&E and immunohistochemistry performed on liver tumours from hydrodynamic injections were carried out using standard techniques, as previously described (27).
Cre recombinase retrofitting
Removal of shuttle vector ColE was performed by using 10 µg of plasmid cut with 20 units of I-SceI (New England BioLabs) at 37°C for 2 h in a total volume of 400 µL followed by heat inactivation at 65°C for 20 min. Forty-four microliters of 10× T4 ligation buffer and 4 µL of T4 ligase was then added and incubated at 16°C for 1 h. Self-ligated shuttle vector was then purified using QIAquick PCR Purification Kit (Qiagen). Cre-mediated retrofitting was performed using 100 ng of self-ligated shuttle vector and 100 ng of expression vector in a 20 µL of reaction using 1 Unit of Cre recombinase (New England BioLabs) incubated at 37°C for 1 h. Ten microliters of the Cre reaction was then transformed into top 10 bacteria (Invitrogen) or electroporated into 10-beta Electrocompetent Escherichia coli (New England BioLabs).
FLP recombination
For removal of flippase recognition target (FRT)-flanked kanamycin, chloramphenicol, and spectinomycin cassettes, plasmids were transformed into Arabinose-induced flippase (FLP) recombinase-expressing bacteria EL250, as previously described (30).
RESULTS
Efficient dual LR Clonase reaction to produce dual cDNA/shRNA vectors
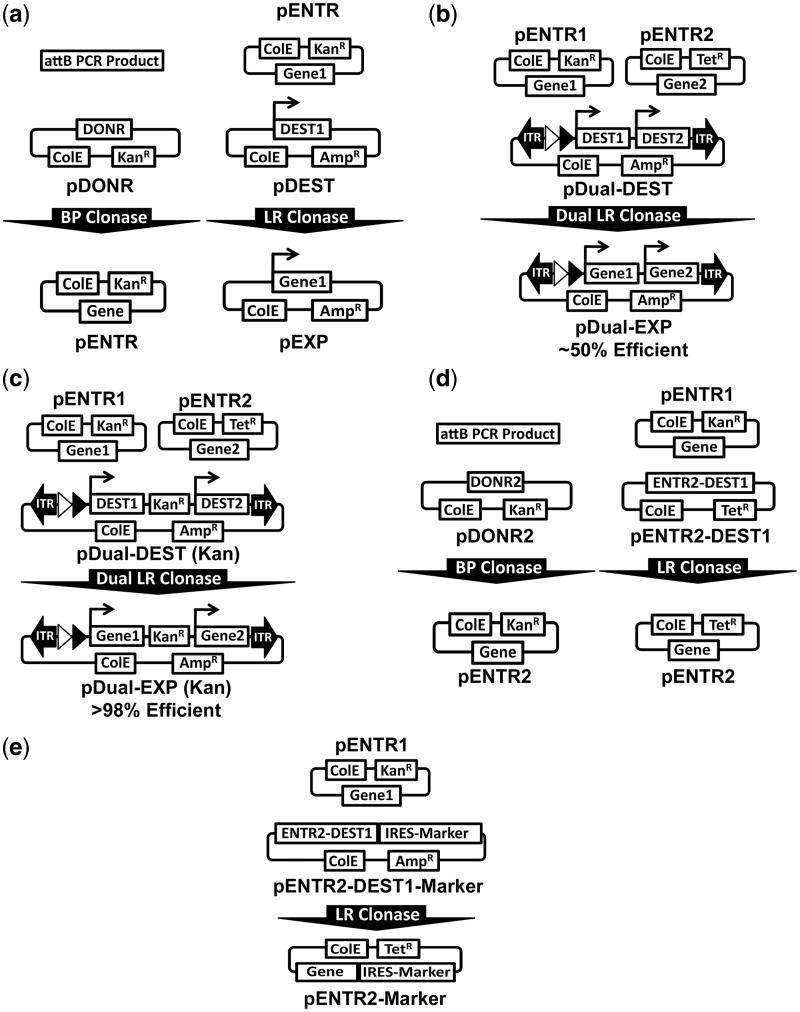
To facilitate the generation of multigene vectors of up to six cDNAs/shRNAs, we first developed a novel recombination-based method for generation of dual gene vectors. Gateway cloning typically consists of a single cDNA in an entry vector (pENTR) being recombined into a single destination expression vector (pDEST) in place of the ccdb bacterial suicide gene, termed an LR Clonase reaction (Figure 1a) (31,32). This reaction is carried out using a commercially available mix of purified lambda phage enzymes in 1 h with >99% efficiency. The recombination is initiated between a flanking pair of attL sites and attR sites of the entry and destination vector, respectively. The result of this LR Clonase reaction produces a larger pair of attP sites flanking the suicide gene and a smaller inert pair of attB sites flanking the gene of interest (Figure 1a).
Figure 1.
Assembly of dual expression vectors via dual LR Clonase reaction. (a) Canonical Gateway cloning using the BP Clonase reaction to generate entry vectors (Left) and LR Clonase reaction to generate expression vectors (Right). (b) Initial dual DEST architecture that produces ∼50% correctly recombined clones on dual LR Clonase reaction with pENTR1 and pENTR2 plasmids. (c) Improved dual DEST architecture that produces >98% correctly recombined clones on dual LR Clonase reaction with pENTR1 and pENTR2 plasmids by addition of a kanamycin resistance gene between DEST1 and DEST2. (d) Diagram of BP Clonase reaction to generate pENTR 2 vectors from attB PCR products analogous to canonical Gateway cloning (Left). Diagram of the entry transfer system developed to move cDNAs/shRNAs from pENTR1 vectors into pENTR2 vectors via LR Clonase reaction (Right). This system was generated by inserting the DEST1 cassette with the attL sites of pENTR2 and replacing the kanamycin resistance with Tetracycline in pENTR2. (e) In addition to transferring cDNA/shRNAs between entry vectors, we also developed pENTR2-DEST1-IRES-Marker vectors to include useful selectable markers, such as puromycin, neomycin, hygromycin and fluorescent proteins.
To allow for the recombination of two genes into a single plasmid in one reaction, we used two independently functioning attL and attR pairs to create two entry and destination vectors, respectively (Figure 1b and c) (29). To validate the independent functionality of the two Gateway cassettes in a single reaction, we constructed a plasmid with tandem EF1A-DEST expression cassettes (DEST1 and DEST2) and performed a dual LR Clonase reaction using mYFP and mKATE in pENTR1 and pENTR2, respectively (Figure 1b). The dual LR clonase reaction was found to be functional with 50% of the resultant colonies (four/eight) screened, having perfect assembly of each gene into its respective expression cassette. We hypothesized that the low efficiency was due to misrecombination between the attL and attR sequences of the two DESTs. Thus, we inserted a kanamycin resistance gene between the two DEST cassettes to negatively select against misrecombination clones (Figure 1c). Using this design, the dual LR Clonase reaction was found to be >98% efficient (40 independent reactions using cDNA/shRNAs varying from 0.75 to 4.5 kb in size) (Table 1). Further, we flanked the kanamycin resistance marker with FRT1 sites to allow for removal of the bacterial sequences if desired. These results demonstrate that the dual clonase reaction of two independent cDNA/shRNAs into a single dual destination vector is highly efficient using the particular attL and attR sites and vector configuration of the RecWay system.
Table 1.
Dual clonase efficiencies: results of all dual clonase reactions using vectors designed to prevent misrecombination by virtue of the bacterial drug resistance marker positioned between the two tandem DEST cassettes
| Vector | pENTR1 | Size (kb) | pENTR2 | Size (kb) | Correct Clones | % Correct |
|---|---|---|---|---|---|---|
| pDual DEST Shuttle (Cm) | DsRED2 | 0.82 | EGFP | 0.78 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | Venus | 0.78 | AmCyan | 0.75 | 3/4 | 75 |
| pPB-Dual DEST (Kan) | EGFR-GFP | 4.53 | shP53-GFP | 3.23 | 4/4 | 100 |
| pPB-Dual DEST (Kan) | DsRED2 | 0.82 | EGFP | 0.78 | 4/4 | 100 |
| pPB-Dual DEST (Kan) | EGFP | 0.78 | DsRED2 | 0.82 | 4/4 | 100 |
| pPB-Dual DEST (Kan) | DsRED2 | 0.82 | EGFP-IRES-Puro/TK | 3.32 | 3/3 | 100 |
| pPB-Dual DEST (Kan) | LargeT Antigen | 2.81 | NRASV12G | 0.61 | 4/4 | 100 |
| pPB-Dual DEST (Kan) | shP53-GFP | 3.23 | rtTA-IRES-Luc | 3.22 | 4/4 | 100 |
| pDual DEST Shuttle (Cm) | EGFP | 0.78 | Luciferase | 1.91 | 3/3 | 100 |
| pPB-Dual DEST (Kan) | EGFR | 3.70 | shP53-GFP | 3.23 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | EGFP | 0.78 | DsRED2 | 0.82 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | DsRED2 | 0.82 | EGFP-IRES-Puro/TK | 3.32 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | EGFP | 0.78 | DsRED2 | 0.82 | 2/2 | 100 |
| pDual DEST Shuttle (Spec) | mYFP | 0.73 | AmCyan | 0.75 | 1/2 | 50 |
| pIDual DEST Shuttle (Spec) | mYFP | 0.73 | AmCyan | 0.75 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | DsRED2 | 0.82 | EGFP-IRES-Puro/TK | 3.32 | 2/2 | 100 |
| pPB-Dual DEST (Kan) | shCREBBP-RFP | 1.26 | shPTEN-GFP | 3.23 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | mKate | 0.73 | Puro-TK | 1.99 | 2/2 | 100 |
| pPB-Dual DEST (Kan) | EGFR-GFP | 4.53 | FOXR2-IRES-Puro/TK | 3.56 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | mKATE | 0.73 | Puro-TK | 1.99 | 2/2 | 100 |
| pDual DEST Shuttle (Spec) | mKATE | 0.73 | Puro-TK | 1.99 | 2/2 | 100 |
| pPB-Dual DEST (Kan) | mKATE | 0.73 | Puro-TK | 1.99 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | LargeT Antigen | 2.81 | NRASV12G-IRES-Luciferase | 3.42 | 2/2 | 100 |
| pDual DEST Shuttle (Spec) | eBFP2 | 0.73 | mOrange2 | 0.90 | 2/2 | 100 |
| pIDual DEST Shuttle (Spec) | eBFP2 | 0.73 | mOrange2 | 0.90 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | M2-rtTA | 0.77 | Puro-TK | 1.99 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | EGFP | 0.78 | mKATE | 0.73 | 2/2 | 100 |
| pPB-Dual DEST (Kan) | mKATE | 0.73 | EGFP | 0.78 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | mKATE | 0.73 | EGFP | 0.78 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | mKATE | 0.73 | EGFP | 0.78 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | EGFP | 0.78 | mKATE | 0.73 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | EGFR-GFP | 4.53 | FOXR2-IRES-Puro/TK | 3.56 | 2/2 | 100 |
| pIDual DEST Shuttle (Cm) | shTrp53-GFP | 3.63 | NRASV12G | 0.61 | 2/2 | 100 |
| pIDual DEST Shuttle (Cm) | EGFP | 0.78 | Luciferase | 1.91 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | Fah | 1.30 | Luciferase | 1.91 | 1/1 | 100 |
| pPB-IDual DEST (Kan) | LargeT Antigen | 2.81 | NRASV12G | 0.61 | 2/2 | 100 |
| pDual DEST Shuttle (Cm) | mYFP | 0.73 | AmCyan | 0.75 | 2/2 | 100 |
| pIDual DEST Shuttle (Cm) | mYFP | 0.73 | AmCyan | 0.75 | 2/2 | 100 |
| pPB-Dual DEST (Kan) | mYFP | 0.73 | mKATE | 0.73 | 2/2 | 100 |
| pPB-IDual DEST (Kan) | mYFP | 0.73 | mKATE | 0.73 | 2/2 | 100 |
| 91/93 | 98.10% | |||||
Generation of pENTR2 vectors by LR or BP Clonase reaction
For simple generation of pENTR2 vectors, a pDONR2 vector was constructed to allow for BP Clonase-mediated generation of pENTR2 vectors from PCR-amplified cDNAs or shRNAs, analogous to how pENTR1 vectors are made. To further simplify the dual Clonase system, we sought to design a transfer system to shuttle genes in pENTR1 vectors into pENTR2 vectors via an LR Clonase reaction. This was accomplished by inserting the DEST1 cassette in between the attL sites of pENTR2 to generate pENTR2-DEST1 (Figure 1d). The entry vector transfer system was found to be very robust with an efficiency of >96% and all correct clones having perfect recombination events on sequencing (Supplementary Table S1). This method eliminates the need for PCR amplification and subsequent sequence confirmation, which would be required if a BP clonase reaction or traditional cloning were used. In addition to simply transferring a cDNA from pENTR1 to pENTR2, numerous pENTR2-DEST1 vectors with IRES markers were generated to expand the versatility of the system (Figure 1e).
Efficient assembly of dual cassettes to form multigene cassettes
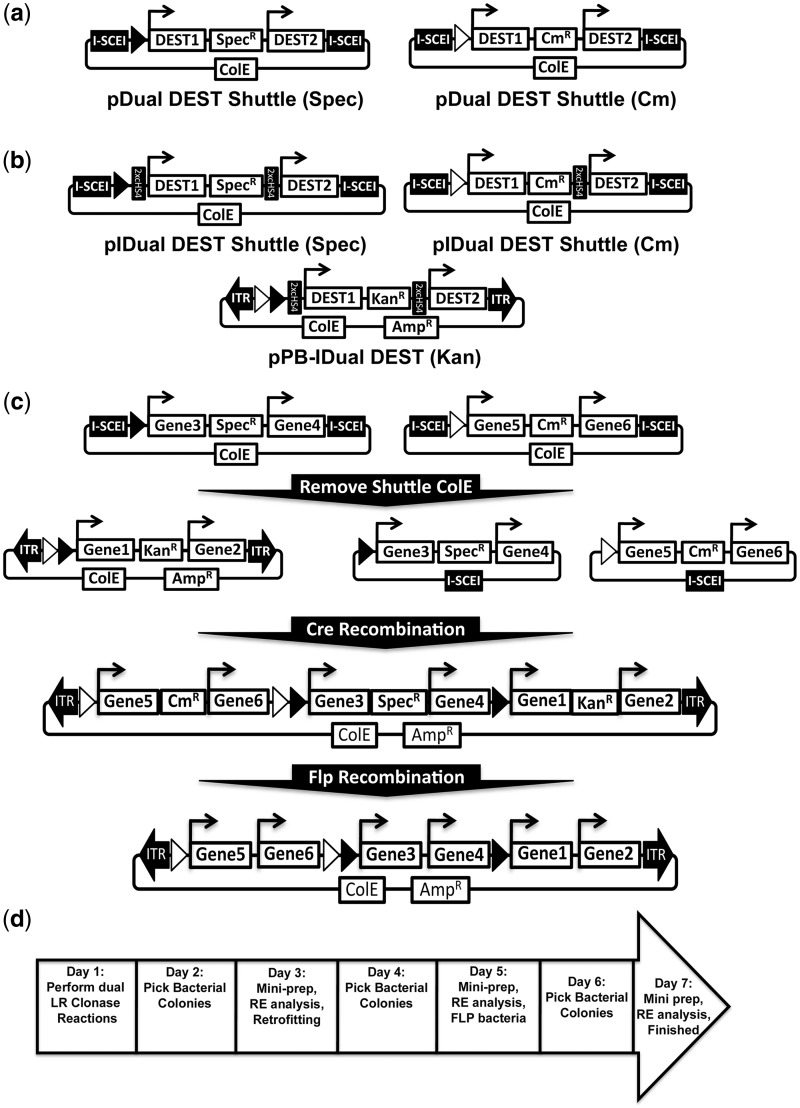
To produce multigene cassettes of four or six cDNAs/shRNAs, the widely used Cre recombination system was implemented. Cre recombinase is typically used to remove a sequence flanked by LoxP sites. However, Cre recombinase can also be used to combine two circular plasmids that each contain a LoxP site, a process termed retrofitting (33,34). To this end, two ‘shuttle’ vectors containing the dual DEST cassette described earlier in the text were generated to allow for Cre recombinase-mediated retrofitting with a final expression vector (Figure 2a). Further, to both assess and prevent potential promoter interference, additional dual DEST vectors with two tandem copies of the cHS4 insulator positioned between each promoter-DEST-pA elements were also generated (Figure 2b). To allow for reproducible and specific retrofitting of two dual expression shuttle plasmids into the final expression vector, the canonical LoxP and independently functioning Lox2722 mutants were used (35). The final expression vector therefore contains both LoxP and Lox2722 sites, whereas the two dual DEST shuttle vectors contain either LoxP or Lox2722. Correctly retrofitted clones are selected for by virtue of either a chloramphenicol or spectinomycin resistance gene in the dual DEST shuttle vectors.
Figure 2.
Assembly of up to 6 gene vectors using Cre Recombinase. (a) Diagram of the two dual DEST Shuttle vectors used to subsequently generate four or six gene vectors after dual LR Clonase reaction. The Cre recombinase LoxP and Lox2722 mutant are represented as black and white triangles, respectively. (b) Diagram of vector layout of the insulated versions of the shuttle vectors and final PB dual DEST vectors are also shown. (c) Schematic of steps to generate bacterial marker-free six gene transposon vectors using RecWay assembly. Three dual clonase reactions are performed using two shuttle vectors and a single transposon dual DEST vector. I-SceI digestion and self-ligation of shuttle vectors containing the cDNAs/shRNAs of interest is performed followed by retrofitting via Cre recombination of all three dual vectors. Once assembled, vectors can be transformed into FLP recombinase-expressing bacteria to remove the KanR (kanamycin), CmR (chloramphenicol) and SpecR (spectinomycin) bacterial markers flanked by unique FRT sites (FRT1, 3 and 14, respectively). The FRT sites and insulators are not shown for simplicity; see Supplementary Figure S1a and b for a more highly detailed schematic representation. (d) Seven-day timeline for the assembly of bacterial marker-free six gene transposon vectors using RecWay assembly.
To allow for plasmid replication and maintenance in E. coli, the final expression plasmid can only contain a single origin of replication; therefore, the origin of replication in the shuttle vectors must be removed before retrofitting. The origins of replication of the dual cDNA/shRNA shuttle vectors are removed by I-SceI homing endonuclease digestion and self-ligation (Figure 2c). With the origins of replication removed, the 2 shuttle vectors can be combined with the final expression vector using Cre-mediated retrofitting (Figure 2c). Retrofitting to generate four or six gene vectors was extremely efficient with >92% clones analysed found to be correct (20 independent reactions using shuttle vectors varying from 7.5 to 24.9 kb) (Table 2). Six gene vectors can also be efficiently assembled in a single step with this method (12/12 clones correct, n = 3); though the much larger insulated vectors need to be generated by two sequential retrofitting reactions in most cases.
Table 2.
Cre recombinase efficiencies: results of all retrofitting reactions performed between final transposon vectors and shuttle vectors with ColE elements removed by I-SceI digest
| Expression Vector | Size (kb) | Shuttle Vector | Size (kb) | Final Size (kb) | Correct Clones | % Correct |
|---|---|---|---|---|---|---|
| pPB-Dual-EXP-EGFP/DsRED2 (Kan) | 9.75 | pDual Shuttle-Venus/AmCyan (Cm) | 7.59 | 17.34 | 8/8 | 100 |
| pPB-Dual-EXP-LgT/NRASV12G (Kan) | 15.20 | pDual Shuttle-EGFP/Luciferase (Cm) | 8.74 | 23.95 | 3/4 | 75 |
| pPB-Dual-EXP-EGFP/DsRED2 (Kan) | 9.75 | pDual Shuttle-EGFP/DsRED2 (Cm) | 10.23 | 19.99 | 2/2 | 100 |
| pPB-Dual-EXP-EGFP/DsRED2 (Kan) | 9.75 | pDual Shuttle-mYFP/AmCyan (Spec) | 7.53 | 17.28 | 1/2 | 50 |
| pPB-IDual-EXP-mKATE/Puro-TK (Kan) | 16.63 | pIDual Shuttle-mYFP/AmCyan (Spec) | 12.98 | 29.61 | 1/1 | 100 |
| pPB-IDual-EXP-mKATE/Puro-TK (Kan) | 16.63 | pIDual Shuttle-EGFP/DsRED2 (Cm) | 10.23 | 26.87 | 2/2 | 100 |
| pPB-IDual-EXP-mKATE/Puro-TK (Kan) | 16.63 | pIDual Shuttle-EGFP/DsRED2 (Cm) | 10.23 | 26.87 | 2/2 | 100 |
| pPB-Dual-EXP-mKATE/Puro-TK (Kan) | 11.53 | pDual Shuttle-EGFP/DsRED2 (Cm) | 10.23 | 21.77 | 2/2 | 100 |
| pPB-Dual-EXP-mKATE/Puro-TK (Kan) | 11.53 | pDual Shuttle-mYFP/AmCyan (Spec) | 7.53 | 19.06 | 1/1 | 100 |
| pPB-Quad-EXP-mYFP/AmCyan/mKATE/Puro-TK (Kan)(Spec) | 17.64 | pDual Shuttle-EGFP/DsRED2 (Cm) | 10.23 | 27.88 | 2/2 | 100 |
| PB-IQuad-EXP-EGFP/DsRED2/mKATE/Puro-TK | 22.58 | pIDual Shuttle-mYFP/AmCyan (Spec) | 12.98 | 35.56 | 1/1 | 100 |
| pPB-Dual-EXP-mKATE/Puro-TK | 9.82 | pDual Shuttle-eBFP2/mOrange2 (Spec) | 7.84 | 17.66 | 2/2 | 100 |
| pPB-IDual-EXP-mKATE/Puro-TK | 16.63 | pIDual Shuttle-eBFP2/mOrange2 (Spec) | 12.94 | 29.58 | 2/2 | 100 |
| PB-Quad-EXP-eBFP2/mOrange2/mKATE/Puro-TK | 14.68 | pDual Shuttle-mYFP/AmCyan (Cm) | 7.53 | 22.21 | 2/2 | 100 |
| PB-IQuad-EXP-eBFP2/mOrange2/mKATE/Puro-TK | 24.88 | pIDual Shuttle-mYFP/AmCyan (Cm) | 10.10 | 34.98 | 1/2 | 50 |
| pPB-Dual-EXP-mKATE/Puro-TK | 9.82 | pDual Shuttle-mYFP/AmCyan (Cm) | 7.53 | 17.35 | 2/2 | 100 |
| pPB-IDual-EXP-mKATE/Puro-TK (Kan) | 16.63 | pIDual Shuttle-mYFP/AmCyan (Cm) | 10.10 | 26.74 | 2/2 | 100 |
| 36/39 | 92.60% | |||||
All clones were analysed and confirmed as correct or incorrect by standard RE digest analysis.
Once the final multigene expression vector is assembled, all of the bacterial drug-resistant genes, except the ampicillin resistance in the plasmid backbone, can be removed by exposure to FLP recombinase-expressing bacteria, as the resistance markers are flanked by independently functioning FRT sites (Figure 2c) (34). The entire process of RecWay assembly of six cDNA/shRNA vectors takes <1 week and can be performed by anyone with basic molecular biology training (Figure 2d) (See Supplementary Figure S1a and b for highly detailed vector layout).
Multigene expression by transient transfection and stable integration
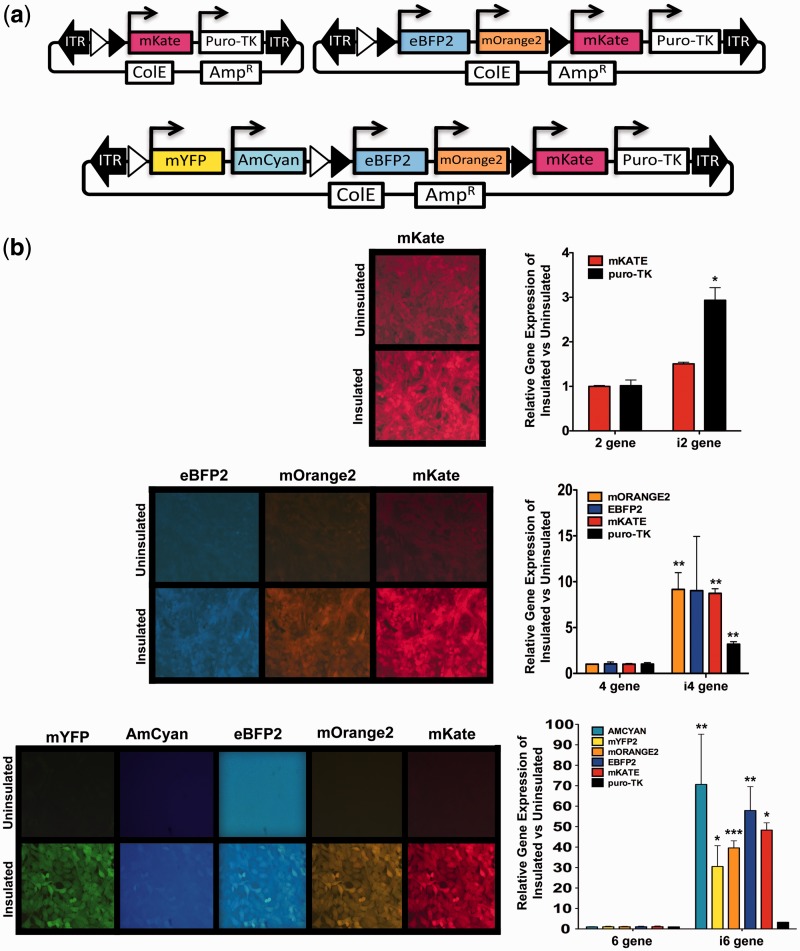
To validate that RecWay-assembled multigene vectors express all genes appropriately, multigene vectors expressing multiple fluorescent proteins that can be observed by simple fluorescence microscopy were assembled (Figure 3a). The puro-tk selection marker was also included to allow for generation of stable integration using the PB transposon system. These vectors were first transiently transfected into HEK293T cells and analysed for gene expression using fluorescence microscopy (Supplementary Figure S2). Single gene vectors were also assembled and transiently transfected to assess the amount of spectral overlap of the fluorescent proteins used in our multigene vectors, which was found to be acceptable for these initial studies (Supplementary Figure S3). It was found that all fluorescent proteins were expressed as expected with all vectors tested. These findings demonstrate that multigene vectors generated by RecWay assembly express all genes appropriately, regardless of insulator use, in a transient transfection setting.
Figure 3.
Expression of stably integrated multigene transposon vectors in NIH 3T3 cells. (a) Diagram of vectors containing one, three or five fluorescent proteins along with the puromycin-tk (puro-tk) fusion. (b) Representative fluorescence photomicrographs and QRT-PCR analysis of stably integrated insulated and uninsulated multigene vectors. Expression levels are normalized to uninsulated vector gene expression for direct comparison of insulated and uninsulated vectors. P-values were calculated using a two-tailed unpaired t-test (P > 0.0001***, P > 0.001**, P > 0.01*).
To properly assess and quantify the amount of promoter interference in RecWay-assembled multigene vectors, expression must be assessed after stable vector integration. To this end, NIH 3T3 was generated with stably integrated multigene vectors with and without cHS4 insulators. The efficiency of PB transposase integration of multigene vectors of various sizes (∼7–30 kb) was also investigated using HEK293T cells. Stable lines were efficiently obtained with all vectors tested in both cell lines. Stable lines were puromycin resistant and expressed the appropriate fluorescent proteins by fluorescence microscopy (Figure 3b). It was clear that uninsulated vectors suffered substantially from promoter interference based on fluorescence levels. The amount of interference also increased with the number of genes in the vector. To quantify this promoter interference, QRT-PCR was performed for all genes in each multigene vector tested. Promoter interference measurably increased as the number of genes increased with up to a 70-fold lower gene expression with the uninsulated six-gene vector (Figure 3b).
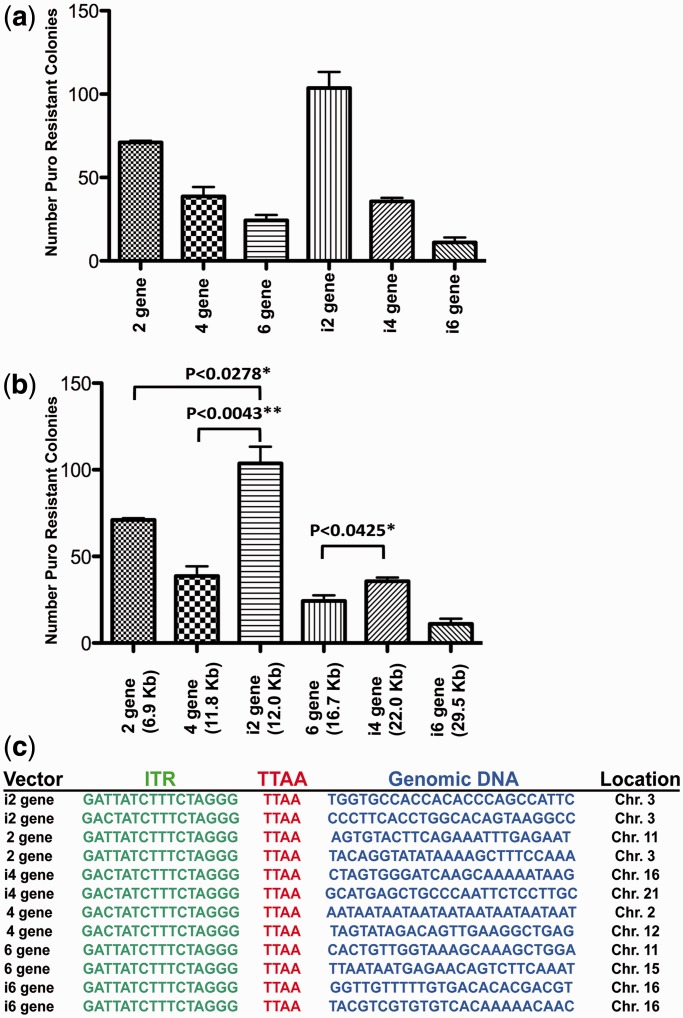
Although insulated multigene vectors produce higher gene expression levels than their uninsulated counterparts, they also have a substantially larger cargo size. As it has been previously shown that increased cargo size reduces rates of transposition, and therefore rates of stable integration clone generation, colony-forming assays were performed to assess the effect of size on transposition efficiencies (26,36). As with these previous reports, we observed reduced transposition efficiencies, as cargo size increased with both insulated and uninsulated vectors (Figure 4a). Unexpectedly, we found that insulated vectors consistently generated more puromycin-resistant colonies compared with uninsulated vectors of similar size (Figure 4b). Further, using a normal immortalized Schwann cell line, we found that uninsulated vectors were not capable of generating stable integrant clones with more than two genes (data not shown).
Figure 4.
Transposition efficiency of multigene vectors. (a,b) Bar graph demonstrating the number of puromycin-resistant stable integration clones generated in HEK293T cells after transfection of transposase and insulated or uninsulated transposon vectors. (c) Representative sequencing results of PB splinkerette analysis of stable transposon integration cell lines with each vector tested. All cloned insertion sites contained inverted terminal repeat sequence, canonical TTAA insertion site sequence and adjacent genomic sequence.
No puromycin resistant clones were generated without PB transposase in any NIH 3T3 or normal immortalized Schwann cells with the DNA concentration and number of plated cells we used. Also, splinkerette PCR confirmed that drug-resistant clones were generated by transposition, and not random plasmid integration (Figure 4c). However, puromycin-resistant clones were observed at very low frequencies in HEK293T cells without transposase. These clones were inconsistent in gene expression and were the result of random plasmid integration, which was confirmed by splinkerette PCR analysis. Collectively, these data demonstrate that the use of insulators to prevent promoter interference is critical, and necessary in less transformed cells, for robust gene expression and efficient clone generation using multigene vectors of ≥4 genes. Further, PB is capable of efficiently integrating multigene transposons, even with the largest transposon tested (∼30 kb), and all genes are appropriately and robustly expressed when insulators are used.
Modelling cancer development in mice using multigene transposon vectors
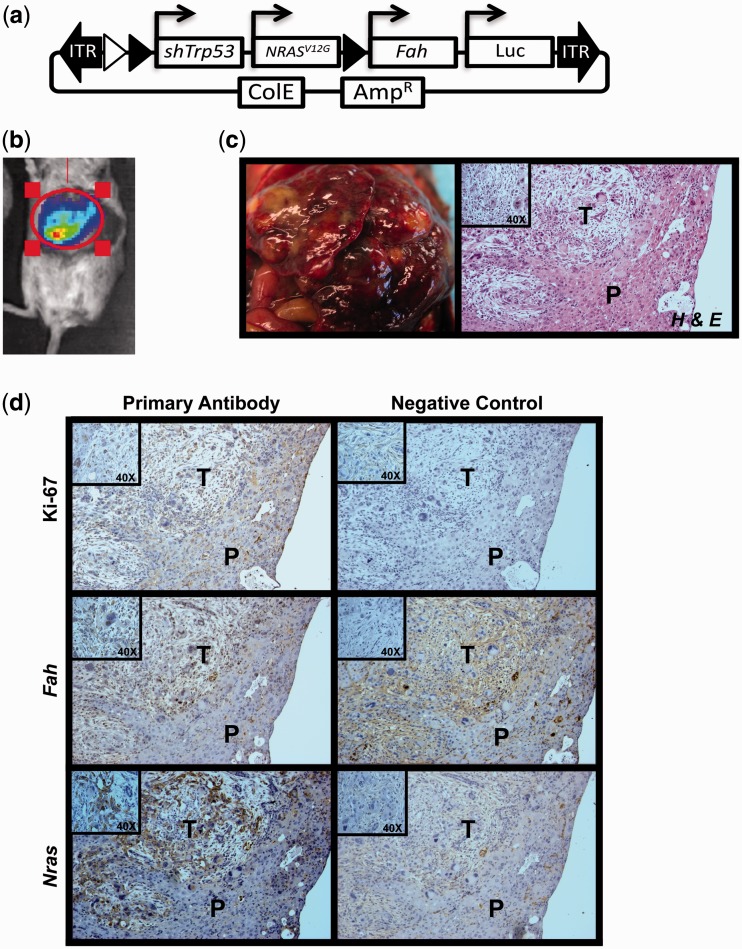
To demonstrate that RecWay-assembled multigene vectors can be used to model the complex biological process of cancer development, a multigene PB transposon vector to induce hepatocellular carcinoma was generated. The four-gene vector was engineered with activated NRASG12V, short hairpin against Trp53, fumarylacetoacetate hydrolase (Fah) and Luciferase. This combination of genes has previously been shown to generate liver tumours using multiple Sleeping Beauty transposons for stable integration (37). The Fah gene was also included to use the previously described hydrodynamic injection and in vivo selection method for liver cancer development (27,38). Nine Fah−/− animals received hydrodynamic injections of the multigene transposon vector along with PB7 transposase plasmid (Figure 5a). Luciferase expression was detectable in seven of the nine animals 14 days after injection (Figure 5b). Tumour development was observed in all animals that survived hydrodynamic injections and liver repopulation (five of nine) with an average latency of 45.5 days (Figure 5c). This penetrance is lower than typical results using individual Sleeping Beauty transposons as previously reported (38). This lower penetrance with PB could be increased by optimizing the amount of transposase or transposon used, in addition to the use of insulators to increase gene expression.
Figure 5.
Hydrodynamic injection of PB multigene vectors induce liver cancer in the Fah-deficient mouse model. (a) Multigene PB transposon vector used for hydrodynamic tail vein injection. PB-Quad-EXP-shTrp53/NRASG12V/Fah/Luc, short hairpin to Trp53, constitutively active NRASG12V cDNA, fumarylacetoacetate hydrolase (Fah) cDNA and Luciferase cDNA all under the control of independent human elongation factor 1 alpha (EF1A) promoters. (b) Representative luciferase imaging of a mouse two weeks post-hydrodynamic injection. (c) Gross examination of the liver on necropsy identified numerous tumour nodules that were formalin fixed for H&E and immunohistochemical analysis. (d) Immunohistochemical staining results for NRAS, Fah and Ki-67 with appropriate no primary controls. T, tumour; P, parenchyma; higher magnification shown as insert box.
Analysis of tumour-bearing livers by H&E demonstrated hyperplastic nodules in all samples analysed. Further, we performed Immunohistochemistry (IHC) for Fah and NRASG12V and found both genes to be expressed in the hyperplastic nodules, but not in the surrounding normal tissue (Figure 5d). Ki67 staining also demonstrated high levels of cell proliferation in the tumour nodules (Figure 5d). These results demonstrate that multigene transposon vectors are capable of inducing tumour formation in vivo after somatic gene transfer in mice. To our knowledge, this is the first report of the use and validation of multigene PB vectors in vivo.
DISCUSSION
We report the development of RecWay assembly, a modular recombinase-based system to quickly and easily produce multigene vectors capable of expressing up to six cDNAs or shRNAs in a single cell. The advent of a dual clonase reaction and a modified Cre recombinase-mediated retrofitting method makes the assembly of multigene vectors nearly 100% efficient at every step of construction. These vectors can be produced using no REs, are assembled from truly modular entry vector components and contain no bacterial sequence in the final vector. Further, these multigene vectors, even of very large size, can easily be stably integrated into the genome using the piggyBac DNA transposon system in vitro and in vivo. Although other multigene expression systems have been reported, our system is by far the most straightforward and thoroughly validated (14–17).
Previously reported multigene systems have been validated primarily by transient transfection and analysis by fluorescent protein expression or western blot (14,15). Importantly, validation by transient transfection completely masks the issue of promoter interference. It has been previously reported that promoter interference in the context of multigene vectors can be a major problem (18,20,23). Indeed, RecWay-assembled uninsulated multigene vectors suffered promoter interference substantially when stably integrated, which was not evident by transient transfection (Supplementary Figure S2). Stable integration is critical for modelling cancer or cellular transdifferentiation where transgene expression must be maintained for long periods with many cell doublings, which cannot be attained by transient transfection.
Interestingly, the amount of promoter interference seen with uninsulated RecWay-assembled vectors appeared to vary among the cell lines tested. HEK293T demonstrated the lowest amount of promoter interference, followed by NIH 3T3 cells, and the most seen in normal immortalized human Schwann cells, where promoter interference actually prevented establishment of stable integration lines, likely owing to promoter interference of the puro-tk resistance gene. Our experience suggests that transgene expression is highly promiscuous in HEK293T cells compared with less transformed cell lines such as NIH 3T3 or normal immortalized Schwann cells. Thus, it seems that promoter interference potentially becomes more robust, as cells are less transformed or more primary in nature. This suggests that multigene vectors intended for use in primary or ‘normal’ immortalized cells may require insulators for uninhibited high-level gene expression.
Our system overcomes the issue of promoter interference by the inclusion of two tandem copies of the cHS4 insulator between each expression element. It has been previously shown that two tandem copies of cHS4 can alleviate promoter interfere by providing binding sites for several proteins (CBP, CTCF and USF1) that maintain an open chromatin state by recruiting necessary chromatin-modifying host factors (23). Importantly, the addition of large tandem cHS4 insulators (2.5 kb) did not affect RecWay-assembly efficiencies. We constructed and validated 2, 4 and 6 gene vectors using this insulated design to express fluorescent proteins and the puro-tk selection marker. Although the use of insulators adds a considerable amount of size to the multigene cassette vectors, it is well within the capabilities of piggyBac’s ability to faithfully transpose them into the genome. We found that insulated vectors produced more stable integrant clones than uninsulated vectors of the same size, likely owing to promoter interference effecting the expression of the puro-tk gene.
Another method used to express multiple genes in a single cell without the concern of promoter interference uses multiplex transposon integration (27,39). Though this method likely overcomes the promoter interference seen with uninsulated multigene vectors, it has some down sides that insulated multigene RecWay vectors overcome. For instance, the rate of integration of multiple transposons in a single cell has been shown to be less efficient than integration of a single multigene transposons described herein. Kahlig et al. reported multiplex transposon integration rates of 31% for four transposons and 60% for three transposons after drug selection, whereas 100% of drug-selected cells contain the RecWay-assembled multigene transposon vector. Beyond the improved rate of drug-resistant cells expressing all desired transgenes, the use of RecWay-assembled multigene vectors also maintains stoichiometric ratios of transgenes and reduces the likelihood of insertional mutagenesis.
Beyond the improved gene expression and delivery of multigene expressing vectors, the most powerful aspect of RecWay assembly is that it uses widely available modular entry vector components. There have been numerous reports using Gateway cloning to assemble modular ORFeome cDNA libraries in entry vectors from various organisms using the BP Clonase reaction (40–44). These libraries can then be used to produce cDNA expression libraries en masse using the LR Clonase reaction, which can then be used to perform forward genetic screens. In addition to cDNA libraries, there are also >36 000 human and 17 500 mouse cDNAs commercially available in Gateway ready entry vectors (available through Invitrogen and Geneacopia). As RecWay assembly uses modular Gateway entry vectors, it is possible to make randomly assembled 2 gene vectors from pooled cDNA or shRNA libraries to screen for phenotypes of interest in vitro or in vivo.
The development and validation of our truly modular, highly efficient RecWay assembly method to produce insulated multigene PB transposon vectors will likely lead to new discoveries of complex biological phenomena in numerous systems in vitro and in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
NIH/NIAMS AR050938 Musculoskeletal Training Grant. Funding for open access charge: National Cancer Institute [R01 CA113636 to D.A.L.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Adam J. Dupuy for the R26(−)Shuttle and pTraffic plasmids and Dr. Allen Bradley for piggyBac ITR plasmids used in this study. Dr. Brian Rabinovich also kindly provided multiple fluorescent protein cDNAs.
B.S.M. and D.A.L. designed the research. J.B.B. and V.W.K performed hydrodynamic injections and immunohistochemical staining analysis. E.P.R. and N.K.W. performed the QRT-PCR analysis. B.S.M., L.S.M. and D.A.B. constructed all vectors. B.S.M. and T.K. performed all transfections and generated all stable cell lines. B.S.M. wrote the article.
REFERENCES
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Naini S, Etheridge KT, Adam SJ, Qualman SJ, Bentley RC, Counter CM, Linardic CM. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res. 2008;68:9583. doi: 10.1158/0008-5472.CAN-07-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–4495. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 4.Kendall SDS, Linardic CM, Adam SJ, Counter CM. A network of genetic events sufficient to convert normal human cells to a tumorigenic state. Cancer Res. 2005;65:9824. doi: 10.1158/0008-5472.CAN-05-1543. [DOI] [PubMed] [Google Scholar]
- 5.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 6.Li N, Yang R, Zhang W, Dorfman H, Rao P, Gorlick R. Genetically transforming human mesenchymal stem cells to sarcomas. Cancer. 2009;115:4795–4806. doi: 10.1002/cncr.24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuchbury G, Münch G. Optimizing the generation of stable neuronal cell lines via pre-transfection restriction enzyme digestion of plasmid DNA. Cytotechnology. 2010;62:189–194. doi: 10.1007/s10616-010-9273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 9.De Felipe P. Skipping the co-expression problem: the new 2A. Genet. Vaccines Ther. 2004;2:13. doi: 10.1186/1479-0556-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 11.Szymczak AL, Vignali DAA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert. Opin. Biol. Ther. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- 12.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 2006;80:3147–3156. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sone T, Yahata K, Sasaki Y, Hotta J, Kishine H, Chesnut JD, Imamoto F. Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: Modular construction of multiple cDNA expression elements using recombinant cloning. J. Biotechnol. 2008;136:113–121. doi: 10.1016/j.jbiotec.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Kriz A, Schmid K, Baumgartner N, Ziegler U, Berger I, Ballmer-Hofer K, Berger P. A plasmid-based multigene expression system for mammalian cells. Nat. Commun. 2010;1:120. doi: 10.1038/ncomms1120. [DOI] [PubMed] [Google Scholar]
- 16.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PloS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PloS One. 2011;6:e21622. doi: 10.1371/journal.pone.0021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yahata K, Kishine H, Sone T, Sasaki Y, Hotta J, Chesnut JD, Okabe M, Imamoto F. Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: Conditional gene expression at near physiological levels. J. Biotechnol. 2005;118:123–134. doi: 10.1016/j.jbiotec.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Curtin J, Dane A, Swanson A, Alexander I, Ginn S. Bidirectional promoter interference between two widely used internal heterologous promoters in a late-generation lentiviral construct. Gene Ther. 2008;15:384–390. doi: 10.1038/sj.gt.3303105. [DOI] [PubMed] [Google Scholar]
- 20.Proudfoot N. Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–365. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K, Ikenaka K, Nakahira K, Morita N, Mikoshiba K. An improved retroviral vector for assaying promoter activity: analysis of promoter interference in pIP211 vector. FEBS Lett. 1993;315:129–133. doi: 10.1016/0014-5793(93)81148-s. [DOI] [PubMed] [Google Scholar]
- 22.Emerman M, Temin HM. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984;39:459–467. [PubMed] [Google Scholar]
- 23.Yahata K, Maeshima K, Sone T, Ando T, Okabe M, Imamoto N, Imamoto F. cHS4 insulator-mediated alleviation of promoter interference during cell-based expression of tandemly associated transgenes. J. Mol. Biol. 2007;374:580–590. doi: 10.1016/j.jmb.2007.09.054. [DOI] [PubMed] [Google Scholar]
- 24.Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. USA. 2002;99:16433. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson MH, Coates CJ, George AL. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 26.Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148–e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KAT, Sarver A, Starr TK, Akagi K. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belur LR, Podetz-Pedersen K, Frandsen J, McIvor RS. Lung-directed gene therapy in mice using the nonviral sleeping beauty transposon system. Nat. Protoc. 2007;2:3146–3152. doi: 10.1038/nprot.2007.460. [DOI] [PubMed] [Google Scholar]
- 29.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 30.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard P, Kezdy KE, Van Melderen L, Steyaert J, Wyns L, Pato ML, Higgins PN, Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 1993;234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 32.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SY, Horrigan SK, Altenhofen JL, Arbieva ZH, Hoffman R, Westbrook CA. Modification of bacterial artificial chromosome clones using cre recombinase: introduction of selectable markers for expression in eukaryotic cells. Genome Res. 1998;8:404. doi: 10.1101/gr.8.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J. Mol. Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 36.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Keng VW, Tschida BR, Bell JB, Largaespada DA. Modeling hepatitis B virus X–induced hepatocellular carcinoma in mice with the sleeping beauty transposon system. Hepatology. 2011;53:781–790. doi: 10.1002/hep.24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wangensteen KJ, Wilber A, Keng VW, He Z, Matise I, Wangensteen L, Carson CM, Chen Y, Steer CJ, McIvor RS. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology. 2008;47:1714–1724. doi: 10.1002/hep.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahlig KM, Saridey SK, Kaja A, Daniels MA, George AL, Jr, Wilson MH. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl Acad. Sci. USA. 2010;107:1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandner CJ, Maier RH, Henderson DS, Hintner H, Bauer JW, Önder K. The ORFeome of staphylococcus aureus v 1.1. BMC Genomics. 2008;9:321. doi: 10.1186/1471-2164-9-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bürkle L, Meyer S, Dortay H, Lehrach H, Heyl A. In vitro recombination cloning of entire cDNA libraries in arabidopsis thaliana and its application to the yeast two-hybrid system. Functional & Integrative Genomics. 2005;5:175–183. doi: 10.1007/s10142-005-0134-5. [DOI] [PubMed] [Google Scholar]
- 42.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P. hORFeome v3. 1: A resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuyama A, Yoshida M. Systematic cloning of an ORFeome using the gateway system. Methods Mol. Biol. 2009;577:11–24. doi: 10.1007/978-1-60761-232-2_2. [DOI] [PubMed] [Google Scholar]
- 44.Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14:2064–2069. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.