Abstract
The mechanisms of abortive synthesis and promoter escape during initiation of transcription are poorly understood. Here, we show that, after initiation of RNA synthesis, non-specific interaction of σ70 region 1.2, present in all σ70 family factors, with the non-template strand around position −4 relative to the transcription start site facilitates unwinding of the DNA duplex downstream of the transcription start site. This leads to stabilization of short RNA products and allows their extension, i.e. promoter escape. We show that this activity of σ70 region 1.2 is assisted by the β-lobe domain, but does not involve the β′-rudder or the β′-switch-2, earlier proposed to participate in promoter escape. DNA sequence independence of this function of σ70 region 1.2 suggests that it may be conserved in all σ70 family factors. Our results indicate that the abortive nature of initial synthesis is caused, at least in part, by failure to open the downstream DNA by the β-lobe and σ region 1.2.
INTRODUCTION
Initiation of transcription roughly comprises four steps: promoter recognition, promoter melting, abortive synthesis and promoter escape. In bacteria, all these steps involve specificity factor σ, which joins the core enzyme of RNA polymerase (RNAP) to form holoenzyme. Most of the σ factors belong to a σ70 family and are structurally and functionally related to the housekeeping σ70. In the context of the holoenzyme, regions 2.3 and 4.2 of σ70 (σR2.3 and σR4.2) recognize the −10 and −35 promoter elements, respectively (1–3). A minor class of extended −10 promoters, instead of the −35 motif, rely on a TG motif located immediately upstream of the −10 element (4). The TG motif is recognized by σR3.1 (5).
The σR2.3 facilitates melting of the DNA duplex at the −10 element, thus initializing formation of the open promoter complex (6,7). The σR1.2 was shown to allosterically control single-stranded DNA binding by σR2.3 (8). Mutations in or deletion of σR1.2 resulted in an extremely slow rate of open complex formation on the λPr promoter (9). The σR1.2 was also shown to make sequence-specific interactions with a non-template nucleotide just downstream of the −10 element (position −7 or −6 or −5, depending on the distance between the −10 element and the transcription start site) (10,11), which affect the stability of the promoter open complex on a number of promoters. Consistently, in the crystal structure of RNAP with a pre-melted DNA fork mimicking the promoter open complex (and closely resembling the M13ori used in our study), this (−6) residue of the non-template strand is bound in a separate pocket of σR1.2 (11). The downstream residues −5, −4 also make contacts with σR1.2 (11). Y101 of σR1.2 was proposed to play a particularly important role in σR1.2 functions during open complex formation (8,12). The σR1.1 was shown to influence open complex formation though with diverse effects on different promoters (13–15). The σR1.1 is also required for the efficient inhibition of open complex formation by T7 Gp2 protein (16). Besides σ70 domains, parts of the core enzyme are also involved in open complex formation. The β-lobe, β′-rudder and β′-switch-2 (β′-SW-2) regions were shown to be important for formation of the downstream part of the transcription bubble (17–22). In the crystal structure of the promoter open complex, the β-lobe makes contacts with residues −2, +1 and +2 of the non-template strand (11).
In the presence of nucleotide triphosphates (NTPs), the promoter open complex is capable of de novo RNA synthesis. The σR3.2 and β′-SW-2 play particularly important roles in the initial synthesis. The σR3.2, in concert with β′-SW-2, decreases the Km for initiating nucleotides apparently by aligning the template DNA strand in the active centre of RNAP (23–25). The initial synthesis is unproductive, and short abortive RNA products are released from the complex. The reasons for the abortive nature of transcription initiation are not fully understood. It was suggested to be caused by the growing strain in the open complex during scrunching of DNA (26,27) or steric collision of RNAs with σR3.2 (28,29).
The efficient extension of abortive products, which leads to promoter clearance, was proposed to require σR3.2 and β′-SW-2, which apparently support binding of short RNAs through stabilizing the template strand (23,25). Other domains were also shown to influence abortive synthesis and promoter escape. Deletion of σR1.1 decreased the amount of abortive products without affecting their size distribution (13). Mutations in σR1.2 led to increased amount of abortive transcripts and decreased promoter escape (9). However, the roles of the core RNAP and σ70 domains in abortive initiation and promoter escape remain poorly understood. One of the reasons is that the investigation of these processes is complicated owing to the direct involvement of most of core and σ70 domains in the preceding steps of promoter recognition and opening; mutagenesis of these domains almost inevitably affects the obligatory upstream events of promoter utilization, thus obstructing analysis.
Here, we used a unique experimental system based on the promoter M13ori of M13 bacteriophage, which mimics the open promoter and does not require conventional steps of promoter recognition/opening and thus allows unbiased investigation of abortive initiation and promoter escape and the roles of core and σ70 domains in it. We show that, after open complex formation, σR1.2 and the β-lobe cooperate to promote the melting of the DNA duplex downstream of the transcription start site and thus facilitate stabilization of abortive products and their extension, i.e. promoter escape. Our results also suggest that the abortive nature of initial RNA synthesis is, at least in part, caused by the failure of σR1.2 and the β-lobe to open the DNA duplex downstream of the transcription start site.
MATERIALS AND METHODS
Proteins
Wild-type and mutant RNAP core enzymes were purified as described (30). EβΔ186−433 lacking β-lobe mutant RNAP was obtained as described (17). RNAP with a deletion in rudder segment (Eβ′Δ311−314) was obtained by site-directed mutagenesis. The wild-type and most of the mutant σ70 subunits were described by us earlier, and σY101A was constructed and purified as described (8).
In vitro transcription
Three pmols of wild-type or mutant RNAP core with or without 15 pmols of σ70 (wild-type or mutant) and 3 pmols of single-stranded M13ori promoter derivative (IDT) (Supplementary Figure S1) were incubated at 37°C for 10 min in 10 µl of transcription buffer [20 mM Tris–HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2]. Transcription was initiated by the addition of 1 mM ATP, 250 µM or 1 mM CTP and UTP, 40 µM or 1 mM GTP and 1 µl of α-[32P]GTP (10 mCi/ml) (Hartmann Analytic). Reactions were stopped after 30-min incubation at 37°C by the addition of formamide-containing loading buffer. Products were separated on denaturing (8 M urea) polyacrylamide gels, revealed by PhosphorImaging (GE Healthcare), and analysed using ImageQuant software (GE Healthcare).
M13ori photo cross-linking
Phosphorothiolated fragments of M13ori (IDT) (Supplementary Figure S1) were derivatized with p-azidophenacyl bromide (APAB; Sigma) and radiolabelled with γ-[32P]ATP (Hartmann Analytic) as described (31). In all, 200 nM of RNAP core, 1 µM of wild-type or mutant σ70 and 200 nM of DNA were incubated in 10 µl of transcription buffer at 37°C for 10 min and then ultraviolet irradiated at 365 nm for 120 s using a UVStratalinker (Stratagene) as described (31,32). Cross-linked complexes were resolved on 5% sodium dodecyl sulphate–polyacrylamide gel and analysed as aforementioned.
RESULTS
DNA melting downstream of the transcription start site is prerequisite of promoter escape
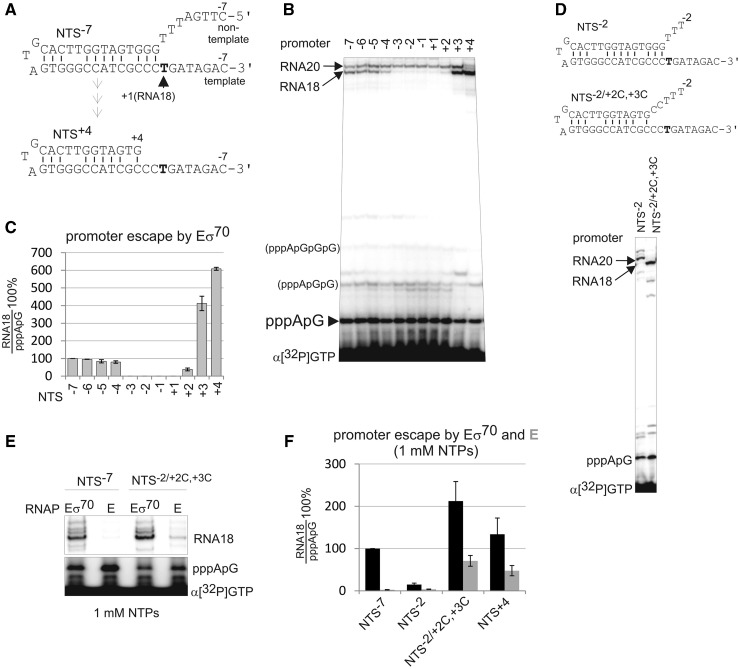
The single-stranded M13ori promoter adopts a hairpin structure, which mimics a double-stranded promoter with the transcription bubble pre-melted to position +1 (Figure 1A). The structure of the M13ori promoter is close to that of the pre-melted promoter in the recently published crystal structure of the bacterial open promoter complex (11). As on conventional double-stranded promoters, abortive synthesis on M13ori is followed by promoter escape and formation of a stable elongation complex, indistinguishable from elongation complexes formed on double-stranded DNA templates (33). The full size product (RNA18) on M13ori is an 18-nt long RNA primer, which is a result of priming complex formation (33). Although not influencing abortive synthesis, promoter escape and initial elongation, the formation of the priming complex, after synthesis of 18 nt (or 20-nt long RNA; see later in the text), ensures that transcription on M13ori is a single round event. The common promoter elements required for the recognition/melting of the double-stranded promoters by the σ70 subunit, −10 (or extended −10) and −35 elements, are absent and/or dispensable in the M13ori promoter under physiological ionic strength conditions (23) (in contrast to the mentioned crystallographic promoter). This property permits the investigation of functions of the core RNAP and the σ70 subunit during abortive initiation and promoter escape while omitting complicating steps of closed and open promoter complexes formation.
Figure 1.
The non-template strand between −4 and +2 is required for DNA melting downstream of the transcription start site and for promoter escape. (A). Structures of M13ori derivatives used in our study. At the top is the initial NTS−7 promoter, which was progressively shortened from the 5′-end to NTS+4 (bottom). (B) Synthesis of abortive and the full-length RNA18 on the NTS−7 promoter and its shorter derivatives by Eσ70 in the presence of all NTPs (no short primers were used). Note that the synthesis of RNA20, which originates upstream of the RNA18 start site, is not affected by non-template strand truncations. A number of minor bands running slower than the abortive dinucleotide correspond to tri- and tetra nucleotides as well as small amount of transcripts resulting from non-specific initiation. These products were not taken into consideration during quantification. (C) Quantification of the results in (B). Here and after, promoter escape was quantified as a ratio of RNA18 to the abortive products (pppApG) and normalized to promoter escape on the NTS−7 promoter. Here, and after, data presented as mean ± SEM from at least three independent experiments. (D) Transcription by Eσ70 on pre-melted promoter NTS−2/+2C,+3C. (E and F) Transcription and promoter escape by holo (Eσ70) (black) and core (E) RNAPs (grey) on templates indicated above the gel and below the histogram were analysed in 1 mM NTPs.
Dispensability of general promoter elements for promoter function allows shortening of the non-template strand (top part of the M13ori hairpin; Figure 1A) downstream of the −10 element. We investigated the role of the non-template strand upstream and around the start site of transcription (+1TSS) in promoter escape. To do so, we progressively shortened the non-template strand of the M13ori from position −7 (NTS−7 promoter, which already lacks the region corresponding to the −10 element; Figure 1A and Supplementary Figure S1) and analysed abortive synthesis and promoter escape by E. coli holoenzyme Eσ70 on resulting promoters. Unexpectedly, trimming of the non-template strand to position −3 led to abolishing of promoter escape and synthesis of RNA18 (Figure 1B and C). However, further truncation of the non-template strand to position +3 downstream of the +1TSS led to the restoration of RNA18 synthesis (Figure 1B and C). The effects of truncations on RNA18 synthesis were independent of the concentration of NTPs (Figure 2A, compare NTS−7 and NTS−2). Note that only extension of abortive products, but not the abortive synthesis per se, was significantly affected by the truncations (Figure 1B).
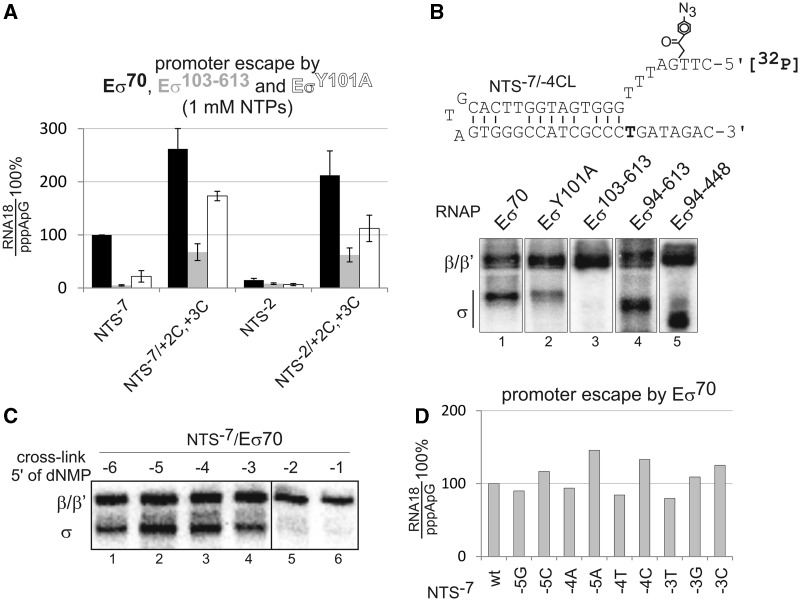
Figure 2.
Interaction of σR1.2 with the non-template strand at position −4 facilitates melting of DNA downstream of the transcription start site. (A) Promoter escape by Eσ70 (black), Eσ103–613 (grey) and EσY101A (white) on promoters depicted below the histogram was analysed in 1 mM NTPs. (B) UV-induced DNA/protein cross-link on radiolabelled NTS−7 promoter derivative (NTS−7/−4CL) bearing a cross-linkable group at position −4 (shown above the gels) in complexes formed by Eσ70, EσY101A, Eσ103–613, Eσ94–613 and Eσ94–448. Products were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis. (C) Cross-link with Eσ70 on NTS−7 promoters bearing cross-linkable groups at positions depicted above the gel. (D) Promoter escape on NTS−7 promoters carrying single nucleotide substitutions in positions −5, −4 or −3, depicted below the histogram.
In contrast to RNA18, synthesis of RNA20, which is initiated 2 nt upstream of RNA18 in the single-stranded region of the NTS−7 promoter, was not affected by non-template strand truncations (Figure 1B). Taken together with the aforementioned results, this observation suggests that promoter escape may require melting/opening of the DNA duplex just downstream of the +1TSS, which, for some reason, is deficient on NTS−3, NTS−2, NTS−1, NTS+1 and NTS+2 promoters. To test this hypothesis, we introduced mismatches in positions +2 and +3 of the NTS−2 promoter (NTS−2/+2C,+3C). As seen from Figure 1D, the promoter escape was restored on this derivative. We conclude that the non-template strand upstream of the +1TSS [referred to as Discriminator (DIS)] is required for melting of the DNA duplex downstream of the +1TSS, and this, in turn, is required for promoter escape.
Core RNAP cannot escape into elongation on the NTS−7 promoter, even though the latter contains DIS required for escape by the holo RNAP (23) (Figure 1E). This suggests that the DIS cooperates with σ70 to facilitate melting of the DNA downstream of the +1TSS. We hypothesized that core RNAP could be forced into promoter escape if the DNA downstream of the +1TSS was pre-melted. Transcription was performed at high NTP concentrations because core RNAP has a much lower affinity for initiating NTPs than Eσ70 (23,25). Indeed, on NTS−2/+2C,+3C and NTS+4 promoters, core acquired ability to synthesize RNA18 (Figure 1E and F). Note that no short RNA primers, which can stimulate promoter escape (23), were used in our work.
Non-specific interaction of σR1.2 with DIS facilitates promoter escape
The aforementioned results suggest that the DIS in cooperation with the σ70 factor facilitates promoter escape by unwinding the DNA downstream of the +1TSS. Previously, the upstream part of the DIS (two base pairs downstream the −10 element, i.e. position −7 or −6 or −5 depending on the distance between −10 and +1TSS) was shown to be sequence specifically recognized by σR1.2 to determine stability of the open promoter complex (10). We hypothesized that σR1.2 may also interact with the DIS further downstream and somehow facilitate melting downstream of the +1TSS. To test this hypothesis, we used a mutant variant of σ70 factor, in which σR1.1–1.2 (amino acids 1–102) were deleted, σ103−613 (8), and constructed a mutant σY101A bearing an alanine substitution of Y101, which was proposed to be important for interactions of σR1.2 with DIS (8,12). Transcription was performed in the presence of high NTP concentrations, as we found that deletion of σR1.2 or mutation of Y101 decreases the affinity of RNAP for initiating NTPs (unpublished). As seen in Figure 2A, Eσ103−613 was deficient in promoter escape, and EσY101A had a decreased promoter escape even in the presence of DIS. These defects, however, were rescued by pre-melting of the DNA duplex downstream of the +1TSS (Figure 2A).
To directly visualize the interaction of σR1.2 with the DIS, we introduced a UV-inducible cross-linkable group at position −4 of a radiolabelled NTS−7 promoter (NTS−7/−4CL derivative) and analysed DNA/protein cross-linking with wild-type and mutant holo RNAPs (Figure 2B). All the modified templates were active in transcription (Supplementary Figure S2A). We observed efficient cross-linking with σ subunit in the context of Eσ70, which, however, decreased when EσY101A was used and disappeared with Eσ103−613 (Figure 2B). The cross-link with σ was still present when either σR1.1 (Eσ94−613) or σR1.1 together with σR3 and σR4 (Eσ94−448) were deleted, suggesting that it is σR1.2 that interacts with the DIS at position −4. The DNA/σ70 cross-link decreased when the cross-linking group was at position −3 and disappeared when it was at positions −2 or −1 of the NTS−7 promoter (Figure 2C). This suggests that position −4 of the DIS may play the major role in the DIS interactions with σR1.2 that are required for downstream DNA melting. As expected, the cross-link decreased as RNAP was allowed to escape from the promoter in the presence of NTPs (Supplementary Figure S2B). Sequence changes in positions −5, −4 and −3 of the NTS−7 promoter did not have any significant effect on promoter escape by Eσ70 (Figure 2D), suggesting a requirement for only non-specific interaction of σR1.2 with this region. This also further distinguishes the σR1.2 interaction with position −4 from the earlier observed sequence-specific σR1.2/DIS interaction that determines stability of promoter opening (10). Our biochemical data are in full agreement with the recently published structure of the open promoter complex (11), which shows that residue −6 of the non-template strand is bound in a protein pocket of σR1.2 separately from residues −5, −4, which interact with σR1.2 non-specifically, whereas residue −3 loses contact with σR1.2 (Figure 3A).
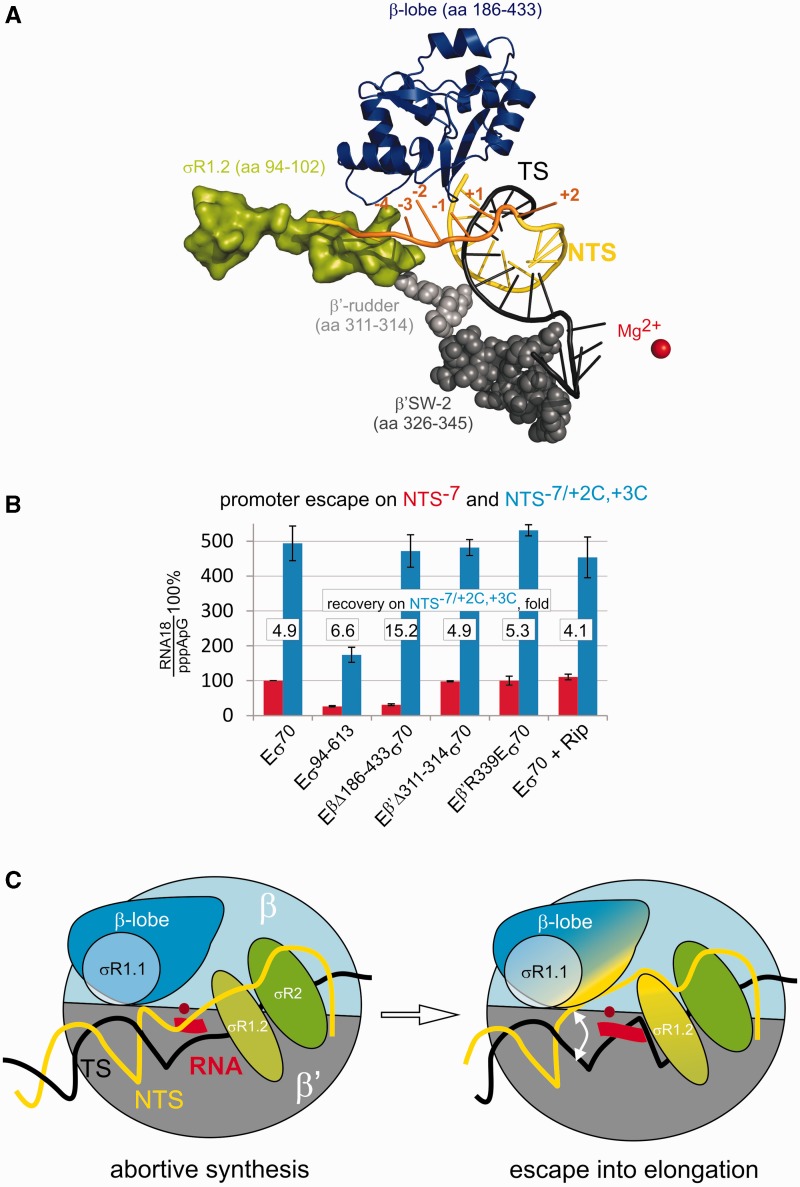
Figure 3.
The β-lobe along with σR1.2 is involved in melting of DNA downstream of the transcription start site and promoter escape. (A) The view of σ and core domains that could be involved in melting downstream of the start site, based on a recently determined structure of the promoter open complex (11). Template (TS) and non-template (NTS) strands are black and yellow, respectively. Positions of the truncations of the non-template strand that abolished promoter escape on the M13ori promoter are orange. (B) Promoter escape by holo enzymes depicted below the histogram and by Eσ70 in the presence of 200 µM Ripostatin on NTS-7 (red) and NTS−2/+2C,+3C (blue). Relative rescue of promoter escape on the pre-melted template is shown. (C) Scheme summarizing our results. Promoter open complex (left) is capable of abortive synthesis but fails to extend short RNA without DNA melting downstream the +1TSS. Interactions of the non-template strand with σR1.2 and the β-lobe (with possible participation of σR1.1) facilitate unwinding of DNA downstream of the transcription start site and, consequently, extension of abortive product and promoter escape.
β-lobe assists σR1.2 in promoter escape
The σR1.1, β-lobe, β′-rudder and β′-SW-2 were all proposed to be involved in melting of the promoter DNA close to the +1TSS and in promoter escape (Figure 3A) (13,17–22). To investigate involvement of these domains in promoter escape controlled by the DIS/σR1.2 interaction, we prepared mutant holo RNAPs lacking σR1.1 (Eσ94−613), the β-lobe (EΔβ186−433σ70), the part of the β′-rudder closest to the downstream DNA (EΔβ′311−314σ70) and an RNAP bearing a mutation that disrupts function of the β′-SW-2 (Eβ′R339Eσ70) (21,25). To investigate the function of β′-SW-2, we also used antibiotic Ripostatin, which targets and inactivates the β′-SW-2 (20). As seen from Figure 3B, the deletion of β′-rudder, the mutation in β′-SW-2 or addition of Ripostatin had no or little effect on promoter escape on the NTS−7 promoter. In contrast, deletion of the β-lobe significantly decreased promoter escape (Figure 3B). This defect, however, is fully rescued on the NTS−7/+2C,+3C promoter with pre-melted DNA downstream of the +1TSS, suggesting that the β-lobe, along with σR1.2, is involved in unwinding the DNA downstream of the +1TSS. Although deletion of σR1.1 also strongly affected promoter escape, this defect could be only partly rescued on the NTS−7/+2C,+3C promoter. This could be explained by a possible partial distortion of σR1.2 by the σR1.1 deletion, given that the defect of the σR1.2 deletion in promoter escape cannot be fully restored on the NTS−7/+2C,+3C promoter (Figure 2A). Note that β′-SW-2 is critical for promoter open complex formation. Therefore, it is difficult to separate its possible effects on promoter escape from its involvement in open complex formation on conventional double-stranded templates. This, presumably, was the reason for apparently mistaken suggestion of involvement of β′-SW-2 in promoter escape by the earlier study (25).
DISCUSSION
Our results reveal that interaction of σR1.2 with DIS around position −4 is one of the major determinants of the escape from abortive initiation into elongation. The trigger for promoter escape is the melting of the DNA duplex downstream of the +1TSS. This melting allows for further extension of short abortive products, which, otherwise, cannot be extended and are released from the complex. Unwinding of the downstream DNA is not an active process, and −4/σR1.2 interaction likely stabilizes a spontaneously (thermally) melted intermediate. Such unwinding is apparently a dynamic process, and our results suggest that formation of the abortive products, at least in part, is determined by the rewinding of the DNA duplex downstream of the +1TSS. Indeed, we observed much more efficient promoter escape by Eσ70 on the pre-melted promoter, especially in the low NTPs concentrations (Figure 3B). In the case of the M13ori promoter, extension of dinucleotide abortive product (which cannot happen without melting downstream the +1TSS) appears to be critical for efficient escape. This is consistent with our earlier observation that a trinucleotide RNA primer can force core RNAP into elongation (23). M13ori, however, has a mismatch at position +5 of the downstream DNA duplex, which likely destabilizes the helix, explaining such modest requirements for efficient promoter escape. On some double-stranded promoters, σR1.2-dependent melting further downstream, as well as synthesis of longer RNA, is likely required to overcome the stability of the downstream DNA duplex. Furthermore, on the conventional double-stranded promoters, requirement for scrunching and breaking of the upstream contact with the promoter may further decrease the probability of promoter escape, before the short RNA is released.
A number of domains of σ70 and core RNAP were suggested to be important for promoter escape. Their involvement in promoter escape, however, could not be separated on double-stranded promoters from the roles they play in open promoter complex formation, thus obstructing analysis. β′-SW-2 was suggested to facilitate promoter escape by melting and fixing the template strand around the +1TTS (25). Our results, however, reveal that β′SW-2 becomes dispensable for promoter clearance when the promoter is already melted to position +1 in the open promoter complex (though it possibly does participate in the formation of the open promoter complex). Instead, the σR1.2 in concert with β-lobe (and possibly σR1.1) are required for melting of the DNA downstream the +1TSS and as a result for promoter escape. Note that the role of σR1.2 in promoter escape also could not have been studied directly on conventional double-stranded promoters because of the deficiency of deleterious mutants in forming stable promoter complexes (9). The interactions of σR1.2 with the non-template strand around position −4 (DIS) and of the β-lobe further downstream [Core Recognition Element, CRE (11)] deduced in our work are in full agreement with the very recently solved structure of the promoter open complex (11) (Figure 3A). As follows from our results, interactions of either σR1.2 or β-lobe with their respective parts of the non-template strand are not enough for unzipping the downstream DNA, and only their concerted action leads to efficient promoter escape (although the σR1.2 interaction seems to be more critical). In contrast to the previously described sequence-specific σR1.2/DIS interaction (10), the interaction of position −4 of the DIS with σR1.2 uncovered here is non-specific, and thus may be critical on all promoters of σ70-family factors. Taken together with the involvement of σR1.2 in allosteric modulation of the −10-σR2.3 interaction (8) and in stabilization of the open complex (10), results presented here suggest that σR1.2 may serve as a major functional switch during transcription initiation and promoter escape.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: UK Biotechnology and Biological Sciences Research Council and the European Research Council [ERC-2007-StG 202994-MTP].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Peter E. Geiduschek and Rob van Nues for critical reading of the manuscript and advice.
REFERENCES
- 1.Helmann JD, deHaseth PL. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 2.Burgess RR, Anthony L. How sigma docks to RNA polymerase and what sigma does. Curr. Opin. Microbiol. 2001;4:126–131. doi: 10.1016/s1369-5274(00)00177-6. [DOI] [PubMed] [Google Scholar]
- 3.Borukhov S, Severinov K. Role of the RNA polymerase sigma subunit in transcription initiation. Res. Microbiol. 2002;153:557–562. doi: 10.1016/s0923-2508(02)01368-2. [DOI] [PubMed] [Google Scholar]
- 4.Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10' motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanderson A, Mitchell JE, Minchin SD, Busby SJ. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- 6.Panaghie G, Aiyar SE, Bobb KL, Hayward RS, de Haseth PL. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]
- 7.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the −10 promoter element. EMBO J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldwin NE, Dombroski AJ. Isolation and characterization of mutations in region 1.2 of Escherichia coli sigma70. Mol. Microbiol. 2001;42:427–437. doi: 10.1046/j.1365-2958.2001.02642.x. [DOI] [PubMed] [Google Scholar]
- 10.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haugen SP, Ross W, Manrique M, Gourse RL. Fine structure of the promoter-sigma region 1.2 interaction. Proc. Natl Acad. Sci. USA. 2008;105:3292–3297. doi: 10.1073/pnas.0709513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson C, Dombroski AJ. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- 14.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 15.Hook-Barnard IG, Hinton DM. The promoter spacer influences transcription initiation via sigma70 region 1.1 of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James E, Liu M, Sheppard C, Mekler V, Camara B, Liu B, Simpson P, Cota E, Severinov K, Matthews S, et al. Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2. Mol. Cell. 2012;47:755–766. doi: 10.1016/j.molcel.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl Acad. Sci. USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuznedelov K, Korzheva N, Mustaev A, Severinov K. Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 2002;21:1369–1378. doi: 10.1093/emboj/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodolin K, Zenkin N, Severinov K. Remodeling of the sigma70 subunit non-template DNA strand contacts during the final step of transcription initiation. J. Mol. Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belogurov GA, Vassylyeva MN, Sevostyanova A, Appleman JR, Xiang AX, Lira R, Webber SE, Klyuyev S, Nudler E, Artsimovitch I, et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tupin A, Gualtieri M, Leonetti JP, Brodolin K. The transcription inhibitor lipiarmycin blocks DNA fitting into the RNA polymerase catalytic site. EMBO J. 2010;29:2527–2537. doi: 10.1038/emboj.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenkin N, Severinov K. The role of RNA polymerase sigma subunit in promoter-independent initiation of transcription. Proc. Natl Acad. Sci. USA. 2004;101:4396–4400. doi: 10.1073/pnas.0400886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulbachinskiy A, Mustaev A. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 25.Pupov D, Miropolskaya N, Sevostyanova A, Bass I, Artsimovitch I, Kulbachinskiy A. Multiple roles of the RNA polymerase {beta}' SW2 region in transcription initiation, promoter escape, and RNA elongation. Nucleic Acids Res. 2010;38:5784–5796. doi: 10.1093/nar/gkq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 29.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 30.Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A. Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 31.Burrows PC, Severinov K, Buck M, Wigneshweraraj SR. Reorganisation of an RNA polymerase-promoter DNA complex for DNA melting. EMBO J. 2004;23:4253–4263. doi: 10.1038/sj.emboj.7600406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burrows PC, Wigneshweraraj SR, Buck M. Protein-DNA interactions that govern AAA+ activator-dependent bacterial transcription initiation. J. Mol. Biol. 2008;375:43–58. doi: 10.1016/j.jmb.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 33.Zenkin N, Naryshkina T, Kuznedelov K, Severinov K. The mechanism of DNA replication primer synthesis by RNA polymerase. Nature. 2006;439:617–620. doi: 10.1038/nature04337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.