Abstract
Therapeutic small interfering RNAs (siRNAs) are composed of chemically modified nucleotides, which enhance RNA stability and increase affinity in Watson–Crick base pairing. However, the precise fate of such modified nucleotides once the siRNA is degraded within the cell is unknown. Previously, we demonstrated that deoxythymidine release from degraded siRNAs reversed the cytotoxicity of thymidylate synthase (TS)-targeted siRNAs and other TS inhibitor compounds. We hypothesized that siRNAs could be designed with specific nucleoside analogues that, once released, would enhance siRNA cytotoxicity. TS-targeted siRNAs were designed that contained 5-fluoro-2′-deoxyuridine (FdU) moieties at various locations within the siRNA. After transfection, these siRNAs suppressed TS protein and messenger RNA expression with different efficiencies depending on the location of the FdU modification. FdU was rapidly released from the siRNA as evidenced by formation of the covalent inhibitory ternary complex formed between TS protein and the FdU metabolite, FdUMP. These modified siRNAs exhibited 10–100-fold greater cytotoxicity and induced multiple DNA damage repair and apoptotic pathways when compared with control siRNAs. The strategy of designing siRNA molecules that incorporate cytotoxic nucleosides represents a potentially novel drug development approach for the treatment of cancer and other human diseases.
INTRODUCTION
Since their discovery over 10 years ago, chemically synthesized small interfering RNAs (siRNAs) have become the standard molecular biology tool for gene function studies. Their potential clinical application as therapeutic molecules is slowly becoming a reality owing to improved delivery options. Although significant challenges remain for the systemic delivery of siRNAs, several clinical trials have already documented the biological activity of siRNAs in target human tissues (1,2). The traditional approach has been to design a 19-mer double-stranded siRNA molecule consisting of two deoxythymidine (dT) nucleotide overhangs on either 3′-end (3). While dTdT overhangs have remained the standard overhang in siRNA synthesis, nearly any nucleotide can be used without incurring a deleterious effect on gene silencing (4,5). To enhance siRNA stability against nuclease degradation, nucleotides are often modified on the phosphate backbone and/or the ribose sugar moiety (6). These modifications are able to significantly extend the half-life of siRNAs in serum from minutes to days. In addition, these modifications are associated with a reduced number of off-target effects such as immune stimulation, passenger strand inactivation and microRNA-like regulation (7,8). One issue that has yet to be addressed is the potential effect of these modified nucleotides on cellular metabolism following eventual intracellular degradation of the siRNA. Many anticancer and antiviral agents currently used in the clinical setting are nucleoside analogues (9,10). It is conceivable then that the modified nucleotides of siRNAs, once released from the siRNA molecule, might have potential impact on various cellular metabolic and signalling pathways.
Previous studies from our laboratory identified an siRNA molecule that potently and specifically inhibited thymidylate synthase (TS) expression (11). TS is a folate-dependent enzyme that catalyses the reductive methylation of deoxyuridine monophosphate (dUMP) by the reduced folate 5,10-methylenetetrahydrofolate to thymidylate (dTMP) and dihydrofolate (12). dTMP is then metabolized to dTTP, an essential precursor for DNA biosynthesis. Although dTMP can be formed by phosphorylation of thymidine via the thymidine kinase (TK)-catalysed pathway, the TS-mediated formation of dTMP provides for its sole intracellular de novo synthesis. Given its central role in DNA biosynthesis and given the observation that TS inhibition results in suppression of cellular proliferation, TS represents an important target for cancer chemotherapy (13,14).
One of the hallmarks of a TS inhibitor compound, such as raltitrexed, pemetrexed and 5-fluoro-2′-deoxyuridine (FdU), is the ability of exogenous thymidine to rescue against its cytotoxic and antitumor effects (15,16). We have previously demonstrated that the growth inhibitory effects of a specific TS-targeted siRNA was completely reversed by thymidine, suggesting that the siRNA specifically targets TS with minimal off-target effects on other genes that might impact cell growth and proliferation (11). Recent studies from our laboratory have shown that the intracellular degradation of siRNA released dT nucleosides from the 3′-end overhang, which, in turn, rescued against the cytotoxicity resulting from TS inhibition (17). This dT release was able to reverse the growth inhibitory effects of TS siRNA as well as the cytotoxic effects of small molecule inhibitors of TS, such as raltitrexed and FdU.
Given the observation that the released nucleosides from siRNAs have biological effects, we hypothesized that siRNA molecules could be rationally designed to contain specific nucleosides that, once degraded intracellularly, would release cytotoxic analogues and thereby enhance the therapeutic potential of the siRNA. Herein, we demonstrate that the fluoropyrimidine nucleoside FdU can be directly incorporated into the siRNA backbone, leading to enhanced cytotoxic and apoptotic effects.
MATERIALS AND METHODS
RNA
RNAs, siRNAs and ‘sticky end’ siRNAs (ssiRNAs) were synthesized by Dharmacon Research (ThermoScientific; Lafayette, CO), the University of Calgary Core DNA Services and the W.M. Keck Oligonucleotide Synthesis Facility at Yale University. RNAs were resuspended in RNase-free water and allowed to anneal for 30 min at room temperature before aliquots were stored at −80°C. Synthesis of TS6 ssiRNA has been previously described (17). All TS6 siRNA/ssiRNA sequences are listed in Table 1. A TK-targeted siRNA was purchased from Dharmacon Research (Cat#D-006787-01; Sense strand sequence: 5′-GCACAGAGAAGGAGGUCGA-UU-′3).
Table 1.
ssiRNA sequences
| ssiRNA | Nucleotide sequencse |
|---|---|
| TS6-UU | 5′-GGAUAUUGUCAGUCUUUAGG-UU-′3 |
| 3′-UU-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-dU/dA | 5′-GGAUAUUGUCAGUCUUUAGG-dUdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-1xF(4)-dU/dA | 5′-GGAFAUUGUCAGUCUUUAGG-dUdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-1xF(21)-dU/dA | 5′-GGAUAUUGUCAGUCUUUAGG-FdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-FdU/dA | 5′-GGAUAUUGUCAGUCUUUAGG-FFFFF-′3 |
| 3′-dAdAdAdAdA-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-dA/FdU | 5′-GGAUAUUGUCAGUCUUUAGG-dAdAdAdAdA-′3 |
| 3′-FFFFF-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-FdU/FdU | 5′-GGAUAUUGUCAGUCUUUAGG-FFFFF-′3 |
| 3′-FFFFF-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-3xF-dU/dA | 5′-GGAUAUUGUCAGUCUUUAGG-dUdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCFAFAACAGFCAGAAAUCC-′5 | |
| TS6-2xF-dU/dA | 5′-GGAUAUUGUCAGUCUUUAGG-dUdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCFAFAACAGUCAGAAAUCC-′5 | |
| TS6-dA/dA | 5′-GGAUAUUGUCAGUCUUUAGG-dAdAdAdAdA-′3 |
| 3′-dAdAdAdAdA-CCUAUAACAGUCAGAAAUCC-′5 | |
| TS6-U/A | 5′-GGAUAUUGUCAGUCUUUAGG-UUUUU-′3 |
| 3′-AAAAA-CCUAUAACAGUCAGAAAUCC-′5 | |
| Mis-dU/dA | 5′-GGAUACUGCCAAUCUCUAGG-dUdUdUdUdU-′3 |
| 3′-dAdAdAdAdA-CCUAUGACGGUUAGAGAUCC-′5 | |
| Mis-FdU/dA | 5′-GGAUACUGCCAAUCUCUAGG-FFFFF-′3 |
| 3′-dAdAdAdAdA-CCUAUGACGGUUAGAGAUCC-′5 |
The top strand is the sense (passenger) strand with the bottom strand being the antisense (guide) strand of the ssiRNA. Bolded and underlined nucleotides (F) are FdU. Nucleotides that are only bolded are mismatched nucleotides.
Cell culture
The human colon cancer RKO cell line (ATCC; Manassas, VA) was maintained in RPMI-1640 (Invitrogen; Carlsbad, CA) with 10% (v/v) dialysed fetal bovine serum at 37°C in a humidified incubator with 5% CO2 (11). The TK-deficient GC3/TK- cells were a gift from Dr. Janet Houghton (18). Cells were tested monthly for mycoplasma by the MycoAlert Mycoplasma detection assay (Cambrex BioScience; Rockland, ME).
Transfection
Cells were plated at a density of 1.5 × 105 cells/well. On the following day, ssiRNAs were complexed with Lipofectamine 2000 (LF2000, Invitrogen) in serum-free RPMI-1640 medium and added to the plated cells. After 48 h, cells were rinsed with phosphate buffered saline (PBS) and scraped in cell lysis buffer [10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% octylphenyl-polyethylene glycol (IGEPAL), 0.5% sodium deoxycholate and 0.1% sodium dodecyl sulphate] containing freshly added Protease Inhibitor Cocktail (Sigma; St. Louis, MO), 1 mM phenylmethylsulfonyl fluoride and phosphatase inhibitors (Sigma). Lysates were sonicated and then centrifuged at 14 000 rpm for 30 min at 4°C to remove debris. Cell lysates were stored at −80°C. Total RNA was extracted using Trizol (Invitrogen) and stored at −80°C.
Cell proliferation assay
RKO cells were plated in 96-well plates at a density of 800 cells/well. On the following day, cells were incubated with ssiRNA/LF2000 complexes for 6 h, after which time, the growth medium was replaced, and cells were grown for an additional 96 h. Similarly, RKO cells were treated with FdU (#F0503, Sigma) for 6 h followed by medium replacement and then allowed to grow for an additional 96 h. Cell proliferation/viability was quantified by the WST-1 assay (Roche; Indianapolis, IN).
Clonogenic assay
RKO cells were plated in six-well plates at a density of 600 cells/well. On the following day, cells were transfected with ssiRNA/LF2000 complexes for 6 h, after which time, the growth medium was replaced. Similarly, FdU was added to wells for 6 h followed by medium replacement. After 8 days, cell colonies were fixed with Trypan blue solution (75% methanol/25% acetic acid/0.25% trypan blue), washed and air-dried before counting colonies >50 cells.
Luciferase assay
A DNA oligonucleotide corresponding to the TS6 siRNA target sequence [TS messenger RNA (mRNA) nt 1058–1077: 5′-GGAUAUUGUCAGUCUUUAGG-3′] was annealed to its complimentary antisense sequence and ligated into the pGL3-Promoter plasmid (Promega; Madison, WI) at restriction sites Hind III and Nco I. The luciferase mRNA expressed from this plasmid was targeted by the antisense strand of TS6 siRNA. A second plasmid was constructed with the TS mRNA sequence inserted in the opposite orientation so that it could be targeted by the sense or passenger strand of TS6 siRNA. RKO cells were plated in 24-well plates at a density of 6 × 104 cells/well. On the following day, ssiRNAs (0.1 nM) and luciferase vectors (400 ng) were co-transfected into cells using LF2000. A renilla-expressing luciferase plasmid (pRL-SV40) (10 ng) was included in each transfection and used to normalize transfection conditions. After 24 h, cells were washed with PBS, lysed in 1× Passive Lysis buffer and firefly and renilla luciferase levels were determined by the Dual-Luciferase™ Reporter Assay system (Promega). The ratio of firefly/renilla luciferase activity obtained from transfection of the pGL3 plasmids in the absence of ssiRNA was normalized to 100%. Each experiment was performed in duplicate. Firefly/renilla ratios represent the mean ± S.D. from at least four separate experiments.
Immunoblot analysis
Protein concentrations of cell lysates were determined using the DC Protein Assay (Bio-Rad; Hercules, CA). Equal amounts of protein (30 µg) from each cell lysate were resolved on sodium dodecyl sulphate–polyacrylamide gel electrophoresis using the method of Laemmli (19) and transferred onto 0.45 µm nitrocellulose membranes (Bio-Rad). Membranes were blocked and incubated overnight with primary antibodies at 4°C. The following antibodies were used in the experiments: anti-TS (Clone 4H4B1; Invitrogen), anti-TS (#5449; Cell Signaling; San Francisco, CA), anti-p21 Waf1/Cip1 (#2947; Cell Signaling), anti-p53 (#sc-126; Santa Cruz Biotechnology; Santa Cruz, CA), anti-PARP (#9542; Cell Signaling), anti-Fas (#4233; Cell Signaling), anti-phospho-histone H2A.X(Ser139) (#2577; Cell Signaling), anti-histone H2A.X (#7631; Cell Signaling), anti-phospho-ATM (pS1981; EPITOMICS Inc.; Burlingame, CA), anti-ATM (#A1106; Sigma) and anti-α-tubulin (EMD Biosciences; San Diego, CA). After multiple TBST washes (1× Tris-buffered saline, 0.1%Tween-20), membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) for 1 h at room temperature. Proteins were detected by the enhanced chemiluminescence method (SuperSignal West Pico substrate; Pierce; Rockford, IL). Quantitation of signal intensities was performed by densitometry on a Xerox scanner using NIH IMAGEJ software.
Odyssey system immunoblot analysis
Immunoblots were prepared as described earlier in the text. After incubation with anti-TS polyclonal antibody, membranes were blocked with the Odyssey blocking buffer and incubated with IRDye 800CW goat anti-rabbit antibody (LI-COR Biosciences; Lincoln, NE, USA) according to the manufacturer’s instructions. The fluorescent signal was detected and quantified using the Odyssey infrared imaging system (LI-COR).
Real time quantitative polymerase chain reaction analysis
Complementary DNA was synthesized from 1.0 µg total RNA using the iScriptTM Reverse Transcription Supermix for real-time quantitative polymerase chain reaction (RT-qPCR) (Bio-Rad; Hercules, CA). PCR was performed in triplicate using the SsoFastTM Probes Supermix (Bio-Rad) in a final reaction volume of 10 µl with gene-specific primer/probe sets and a standard thermal cycling procedure (40 cycles) on a Bio-Rad CFX96TM Real-Time PCR System. The mRNA levels of TS and 18S RNA were assessed using the TaqMan Gene Expression real-time PCR assays (Applied Biosystems; assay ID: Hs00426586_m1 and Hs03928990_g1, respectively). Results were expressed as the threshold cycle (Ct). The relative quantification of the target transcripts was determined by the comparative Ct method (ΔΔCt) according to the manufacturer’s protocol. The 2−ΔΔCt method was used to analyse the relative changes in gene expression. Control experiments without reverse transcription were performed to confirm that the total RNA was not contaminated with genomic DNA.
siRNA melting temperature analysis
A Beckman Coulter Du800 spectrophotometer was used to collect all ultraviolet absorption data (260 nm). siRNA was diluted to 5 µM in DEPC-treated water cooled to 20°C for 15 min before beginning the melting experiments. Ultraviolet absorption data were taken every 1°C, from 20°C to 85°C, with a ramp rate of 0.7°C/min. The melting temperature (Tm) of the siRNA was determined by taking the first derivative of the melting curve. All experiments were performed in triplicate.
Flow cytometry
RKO cells were plated in T-25 cm2 flasks at a density of 2.5 × 105/flask. Cells were transfected on the following day with siRNA/LF2000 complexes for 6 h. After an additional 48 and 72 h, cells were harvested, washed twice with PBS, resuspended in 1× binding buffer at a concentration of 1 × 106 cells/ml, stained with FITC Annexin V Apoptosis Detection Kit according to the manufacturer’s protocol (BD Biosciences; San Jose, CA) and analysed on the BD Accuri C6 Flow Cytometer (BD Accuri Cytometers Inc.; Ann Arbor, MI) at the UPCI Flow Cytometry Facility.
Statistical analysis
All results are expressed as the mean ± S.D. and represent data from at least three independent experiments. Student’s t-tests (two-tailed) were used to analyse differences between two groups, and P < 0.05 was considered statistically significant.
RESULTS
We had previously shown that the activity of siRNAs with extended complementary 3′-end overhangs [termed ‘sticky’ siRNAs or ssiRNAs (20)] was at least equivalent or perhaps slightly more effective than standard siRNAs with respect to target mRNA suppression (17). However, in these studies, we determined that the extended dT overhangs on the ssiRNA were degraded, resulting in rescue from the cytotoxic effects of a TS-targeted ssiRNA. Although this issue was eventually resolved by replacement of dT with dU, we hypothesized that this process of siRNA degradation might be used to release nucleoside moieties to further enhance the cytotoxicity associated with the siRNA. With this in mind, we designed and synthesized multiple TS-targeted ssiRNAs in which FdU analogues replaced uridine residues at various locations on the ssiRNA molecule (Table 1).
In our initial series of experiments, we determined whether the presence of FdU could alter the gene-silencing effects of the TS-targeted ssiRNA. The unmodified ssiRNA, TS6-dU/dA (5 dU on the 3′-end of the sense strand; 5 dA on the 3′-end of the antisense strand), at a concentration of 1 nM suppressed expression of TS protein by ∼80% (Figure 1A, lane 2) (quantified in Table 2). An ssiRNA (TS6-1xF(4)-dU/dA) with incorporation of a single FdU residue at the fourth position in the sense strand significantly inhibited expression of TS protein (Figure 1A, lane 3). A long exposure of the blot revealed a slower migrating band at ∼38 kDa. This band corresponds to the presence of the covalent inhibitory ternary complex (ITC), which is made up of TS protein, the reduced folate 5,10-methylenetetrahydrofolate and the fluoropyrimidine metabolite FdUMP (12). The presence of this complex suggests that the ssiRNA was degraded resulting in the intracellular release of FdU, which would then be metabolized to the FdUMP metabolite. Similar effects were observed for an ssiRNA containing a single FdU residue at the 21st position on the sense strand (data not shown). Additional ssiRNA molecules were designed in which all of the 3′-end overhang dU nucleotides were replaced with FdU. Transfection of TS6-FdU/dA ssiRNA resulted in a similar inhibitory effect on TS protein expression (Figure 1A, lane 4) when compared with the unmodified ssiRNA.
Figure 1.
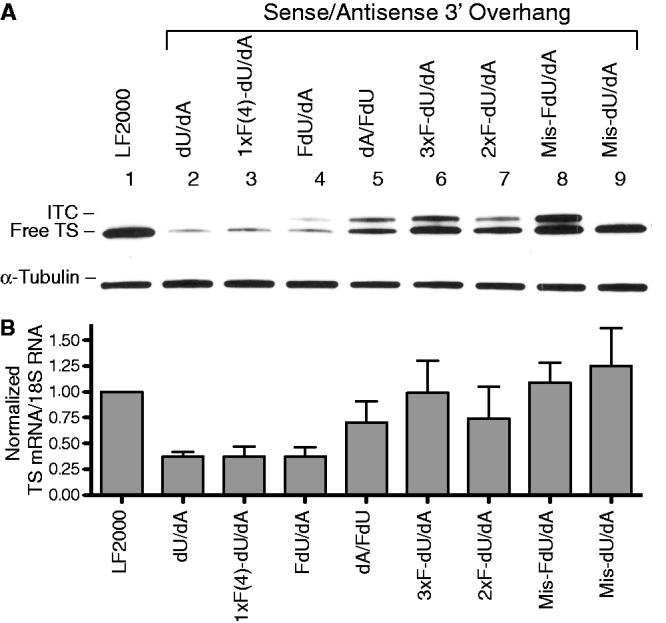
Effect of FdU incorporation in ssiRNA on TS protein expression. RKO cells were transfected with TS6 ssiRNAs (1 nM) containing various 3′-overhangs. After 6 h, the medium was replaced. After an additional 48 h, cells were harvested and processed for immunoblot and qPCR analysis as described in the ‘Materials and Methods’ section. (A) Representative blot from four experiments. (B) All qPCR values represent the mean ± S.D. from at least three separate experiments performed in triplicate. The normalized amount of TS mRNA from mock-transfected cells (LF2000) was set to 1.0, and all other values compared against that value. ITC, inhibitory ternary complex.
Table 2.
Quantitation of free versus ITC-bound TS protein
| ssiRNA | Free TS (%) | ITC-bound TS (%) |
|---|---|---|
| LF2000 | 100 | |
| TS6-dU/dA | 22.6 ± 7.5 | |
| TS6-1xF(4)-dU/dA | 27.3 ± 8.3 | 4.4 ± 2.6 |
| TS6-FdU/dA | 29.8 ± 12.0 | 22.0 ± 11.8 |
| TS6-dA/FdU | 48.0 ± 18.6 | 30.6 ± 15.6 |
| TS6-3xF-dU/dA | 73.6 ± 14.7 | 26.2 ± 10.7 |
| TS6-2xF-dU/dA | 54.1 ± 14.1 | 15.4 ± 9.5 |
| Mis-FdU/dA | 89.5 ± 26.2 | 45.6 ± 21.1 |
| Mis-dU/dA | 90.1 ± 4.0 |
TS protein levels in immunoblots were quantified using an Odyssey LICOR scanner. Values represent the mean ± S.D. from six separate experiments. All values are normalized to the amount of free TS protein in mock-transfected cells (LF2000) set to 100%.
To gain further insight into the process of siRNA catabolism, experiments were performed to determine whether FdU analogues were released from degraded ssiRNAs as nucleosides or nucleotides. A FdU nucleoside would require phosphorylation by TK to yield the 5-FU metabolite FdUMP. As seen in Figure 2A, siRNA knockdown of TK prevented formation of the ITC after transfection of TS6-FdU/dA (lane 4 versus lane 2), suggesting that ssiRNAs are initially degraded into FdU nucleosides and that TK is required for synthesis of FdUMP. Thus, ssiRNAs are degraded into nucleosides that require further metabolism to yield nucleotides.
Figure 2.
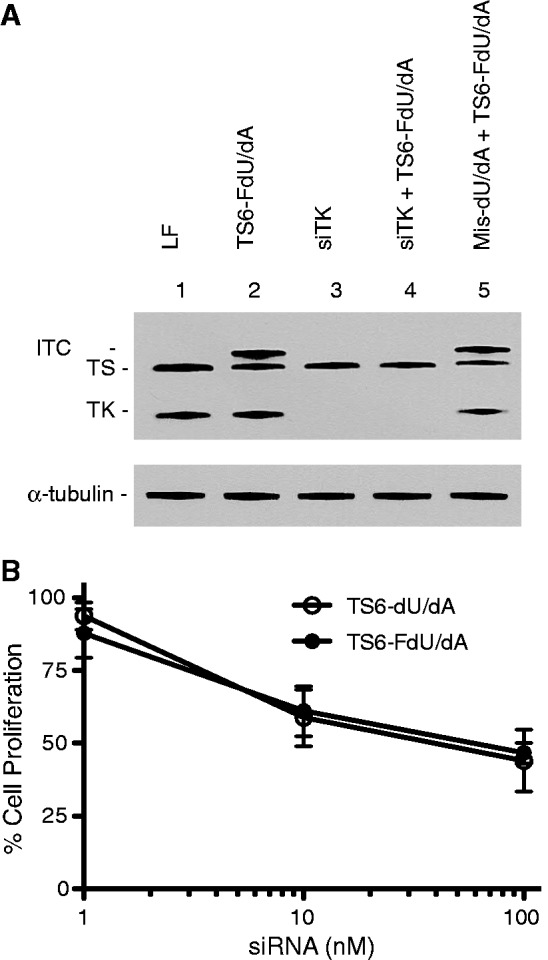
Effect of TK knockdown on the cytotoxicity of the FdU-modified ssiRNA. (A) RKO cells were transfected with 1 nM TK siRNA (lanes 3 and 4) or Mis-dU/dA (lane 5). After 6 h, the medium was replaced with fresh medium. On the following day, cells were transfected with TS6-FdU/dA ssiRNA (1 nM) for 6 h (lanes 2, 4 and 5) after which time, cells were harvested for immunoblot analysis. (B) GC3/TK- cells were transfected with various concentrations of TS6-dU/dA or TS6-FdU/dA ssiRNA. After 6 h, the medium was replaced, and after an additional 5 days, cell proliferation was determined with the WST-1 assay.
It has been well-established that once siRNAs are incorporated into the RNA-induced silencing complex (RISC), the sense or non-targeting strand is degraded, whereas the antisense strand remains bound in the RISC and guides mRNA cleavage (21). As our previous work showed that pre-treatment of TS siRNA, as opposed to simultaneous addition, enhanced cytotoxicity of TS inhibitors (11), we postulated that modification of the antisense strand with FdU residues might delay release of FdU, allowing time for TS protein reduction by the siRNA. However, FdU modification of the 3′-end of antisense strand reduced the ability of the ssiRNA (TS6-dA/FdU) to suppress TS protein levels (Figure 1, lane 5). We then designed ssiRNAs with internal modifications replacing U with FdU at positions 10, 16 and 18, respectively, in the antisense strand. This ssiRNA (TS6-3xF-dU/dA) was even less effective at targeting TS (Figure 1, lane 6). Previous studies have shown that replacement of NTP for dNTP at the RISC cleavage site (position 10) has little impact on silencing (4). Thus, it appears that the fluorine atom on FdU somehow interfered with silencing. Transfection of the ssiRNA with FdU on positions 16 and 18 resulted in slight improvement in target knockdown (Figure 1, lane 7); however, this modified ssiRNA was still less effective than the unmodified ssiRNA molecule (Figure 1, lane 2). This finding suggests that placement of FdU within the antisense strand interferes with RNAi efficiency.
To separate the effects of siRNA knockdown function from that of nucleoside release, we synthesized a control, non-targeted ssiRNA containing five FdU residues on the sense strand (Mis-FdU/dA). This ssiRNA had absolutely no inhibitory effect on expression of TS protein (Figure 1A, lane 8) or TS mRNA (Figure 1B). Total levels of TS protein (free TS + ITC) were greater than what was observed in LF2000-treated cells, a finding that supports the negative autoregulatory model for TS translational regulation that our laboratory has previously identified (22). In this model, we proposed binding of TS inhibitor compounds to TS protein abrogates the normal interaction between TS protein and its own mRNA, thereby leading to increased mRNA translation and subsequent synthesis of new TS protein. Finally, the ability of each of these ssiRNAs to inhibit the expression of TS mRNA levels correlated with TS protein levels (Figure 1B).
We next investigated whether FdU modifications might be altering the specific RNA strand that was actively incorporated into RISC. The respective 20-nt target site of the antisense and sense strands were cloned into separate luciferase-expressing plasmids. These plasmids were transfected into RKO cells with different ssiRNAs, and luciferase activity was determined 24 h later. As seen in Figure 3, a standard TS6 siRNA with two uridine overhangs suppressed both antisense- and sense-strand guided targets. The inhibitory effect by the sense strand is likely a result of the lack of a thermodynamic difference between the two ends of the TS6 siRNA, resulting in an inability of RISC to select only one of the RNA strands (23). After transfection of TS6-dU/dA, antisense strand targeting was maintained, whereas sense strand targeting was significantly reduced. This finding suggests that the 3′-end dU modification interferes with incorporation of the sense strand into RISC and provides an explanation for the slightly greater knockdown of TS previously observed with this ssiRNA (17). Transfection of the modified ssiRNA TS6-FdU/dA resulted in similar strand selection as with TS6-dU/dA. However, transfection of TS6-dA/FdU resulted in 2-fold less targeting by the antisense strand and enhanced sense strand targeting, suggesting that the FdU modification on the 3′-end is able to block RNA strand incorporation into RISC.
Figure 3.
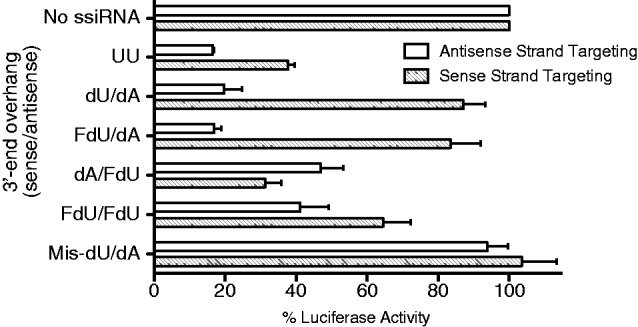
Effect of 3′-end modification on ssiRNA strand selection. RKO cells were transfected with luciferase vectors [including the TS6 target site for either the antisense (open bars) or sense strand (hatched bars)] in combination with ssiRNAs containing different 3′-end overhangs. After 24 h, cells were harvested, and luciferase activity was determined as described in the ‘Materials and Methods’ section. Firefly/renilla luciferase activity from vector-alone transfection was normalized to 100%. All values represent the mean ± S.D. from at least four separate experiments performed in duplicate.
To further investigate the influence of 3′-end dU nucleotides on RNAi efficiency, additional ssiRNAs were designed and transfected into RKO cells. When the 3′-end overhang nucleotides were reversed from dU/dA to dA/dU, the ssiRNA was less able to suppress TS (Figure 4A, lane 2 versus lane 3). This effect was also seen at the mRNA level (Figure 4B) and in the luciferase reporter assay (Figure 4C) where the antisense targeting activity was reduced and the sense targeting activity was increased. Placing dU on both 3′-ends resulted in an ssiRNA (TS6-dU/dU) in which both strands had reduced targeting ability. Transfection of TS6-dA/dA ssiRNA resulted in efficient silencing by the antisense strand (knockdown of TS protein and mRNA) but suppressed the sense luciferase target (similar to UU overhang in Figure 3). This result suggests that dA overhangs do not influence strand selection by RISC. Incorporation of ribonucleotides for the overhangs, as opposed to deoxynucleotides, the TS6-U/A ssiRNA effectively targeted TS protein and mRNA but demonstrated slightly more sense strand targeting than the deoxy version. These results suggest that either dU or FdU can block ssiRNA strand selection by RISC.
Figure 4.
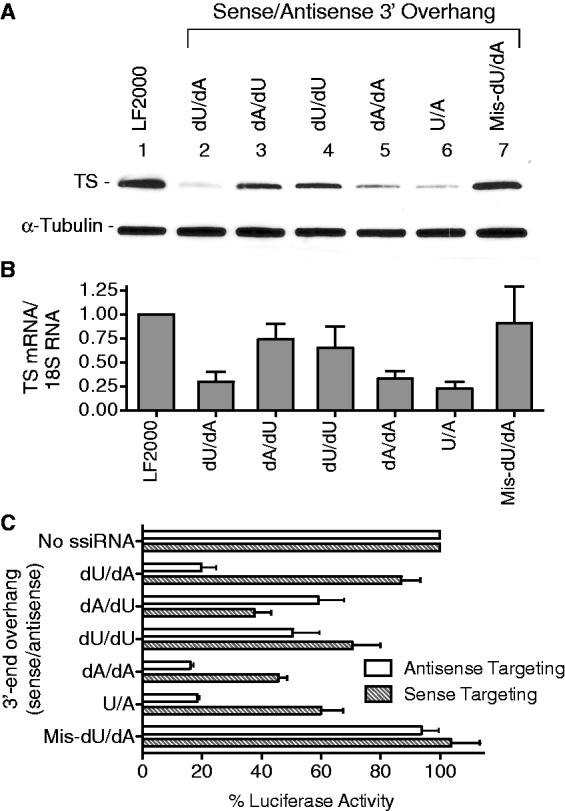
Effect of 3′-end dU incorporation on ssiRNA strand selection. RKO cells were transfected with various TS6 ssiRNAs (1 nM) for 6 h. After an additional 48 h, cells were harvested for immunoblot (A) and qPCR (B) analysis. (C) Luciferase vectors (containing the siRNA target site for either the sense or antisense strand) were transfected into RKO cells in combination with different ssiRNAs. After 24 h, cells were harvested, and luciferase activity was determined as described in the ‘Materials and Methods’ section. Luciferase activity from vector-alone transfection was normalized to 100%. All values represent the mean ± S.D. from at least four separate experiments performed in duplicate.
The nucleotide composition of double-stranded RNAs has been shown to influence their thermodynamic stability (23,24). To determine whether the stability of these modified ssiRNAs influenced their RNAi ability, the Tm of the ssiRNAs was determined. Compared with a standard siRNA containing a 2 nt overhang (TS6-UU), the extended overhang TS6-dU/dA ssiRNA had a significantly lower Tm (Table 3). However, this lower Tm did not improve its RNAi ability, as both TS6-UU and TS6-dU/dA suppressed their intended target sequence equally (Figure 3). Addition of one or five FdU residues in the sense strand of the ssiRNA (TS6-1xF(4)-dU/dA or TS6-FdU/dA) had no influence on Tm compared with TS6-dU/dA (Table 3). Interestingly, incorporation of FdU in the antisense strand overhang of the ssiRNA (TS6-dA/FdU) significantly increased the Tm, which may, in part, explain the ssiRNAs limited ability to suppress TS mRNA levels. Incorporation of three FdU within the antisense strand of the ssiRNA (TS6-3xF-dU/dA) had no significant effect on Tm, whereas incorporating two FdU residues increased Tm of the ssiRNA (TS6-2xF-dU/dA). However, TS6-2xF-dU/dA ssiRNA suppressed TS expression to a greater extent than TS6-3xF-dU/dA (Table 2). Thus, the thermodynamic stability of these FdU-modified ssiRNAs does not seem to play a major role in their RNAi activity.
Table 3.
siRNA thermal stability
| siRNA | Tm (°C) |
|---|---|
| TS6-UU | 55.6 ± 2.3* |
| TS6-dU/dA | 47.5 ± 0.9 |
| TS6-1xF(4)-dU/dA | 49.7 ± 1.1 |
| TS6-FdU/dA | 49.6 ± 5.8 |
| TS6-dA/FdU | 55.2 ± 0.9* |
| TS6-3xF-dU/dA | 49.8 ± 1.5 |
| TS6-2xF-dU/dA | 54.7 ± 1.6* |
| TS6-dA/dU | 51.7 ± 1.2* |
| TS6-dA/dA | 46.4 ± 5.2 |
The melting temperature (Tm) of dissociation was calculated by first order-derivative method. Data represent the mean ± SD from three separate determinations.
*P < 0.02 compared with TS6-dU/dA.
To determine whether FdU modifications enhance cytotoxicity of these ssiRNAs, cell proliferation and clonogenic assays were performed. TS6-dU/dA had modest cytotoxic activity but displayed a greater effect on clonal growth with IC50 values of 81.92 and 1.77 nM, respectively (Table 4). Incorporation of one FdU residue into the ssiRNA (TS6-1xF(4)-dU/dA) at the fourth position enhanced the IC50 by nearly 30-fold for the cell growth assay and 5-fold for the clonogenic assay. Similar effects were observed for an ssiRNA containing a single FdU residue at the 21st position on the sense strand (data not shown). Transfection of TS6-FdU/dA, which contains five FdU residues, resulted in further improvement in inhibition of cell growth and clonal growth with IC50 values of 0.81 and 0.17 nM, respectively. The ssiRNA TS-dA/FdU was significantly less effective than the FdU-sense-modified ssiRNA in both assays (P = 0.01 and P = 0.005, respectively). This finding is in agreement with our western blots and qPCR analyses, which show reduced effects on gene silencing by this particular ssiRNA. Modification of both 3′-ends with FdU (TS6-FdU/FdU) resulted in similar cytotoxic effects as TS6-FdU/dA. However, in the clonogenic assay, these additional FdU residues reduced efficacy by 2-fold as compared with TS6-FdU/dA (P = 0.01). Thus, there may be a limit as to the total number of FdU residues that can enhance the overall cytotoxicity of the TS-targeted siRNA. Of note, the cytotoxic effects of these modified ssiRNAs were completely reversed by the addition of thymidine (10 µM; data not shown), suggesting that analogue incorporation did not alter the TS6 ssiRNA specificity for its target mRNA.
Table 4.
Effect of ssiRNA sequence on cell growth and clonal growth
| IC50 (nM) |
||
|---|---|---|
| ssiRNA | WST-1 | Clonogenic |
| TS6-dU/dA | 81.92 ± 37.15 | 1.77 ± 1.02 |
| TS6-1xF(4)-dU/dA | 2.85 ± 0.83 | 0.34 ± 0.08 |
| TS6-FdU/dA | 0.81 ± 0.29 | 0.17 ± 0.06 |
| TS6-dA/FdU | 1.27 ± 0.32 | 0.40 ± 0.20 |
| TS6-FdU/FdU | 0.77 ± 0.12 | 0.43 ± 0.16 |
| Mis-FdU/dA | 2.88 ± 0.88 | 1.75 ± 0.46 |
| FdU (TS inhibitor) | 6.93 ± 2.08 | 7.42 ± 1.33 |
IC50 values denote the concentration that inhibits 50% of cell or clonal growth. Values represent the mean ± S.D. from at least three separate experiments.
As an important control, the unmodified TS6 mismatched ssiRNA had no effect on cell proliferation or clonogenic growth (data not shown). Transfection of a different control ssiRNA (Mis-FdU/dA) allowed us to begin to isolate the effect of TS knockdown from FdU release. The IC50 for this control ssiRNA was 2.88 nM in the WST-1 assay, which was similar to that observed with TS6-1xF-dU/dA, an active TS-targeted ssiRNA with a single FdU. Comparison between the non-targeted mismatch-FdU/dA and the targeted TS6-FdU/dA reveals that ssiRNA-mediated TS knockdown enhanced the IC50 by 3.5-fold in the growth assay. The effect is further enhanced in the clonogenic assay with a 10-fold lower IC50 value (0.17 versus 1.75 nM). To provide further evidence that the enhanced cytotoxicity was dependent on FdU release with subsequent metabolism to FdUMP, RKO cells were transfected with a siRNA targeting TK followed by subsequent transfection of TS6-FdU/dA. Unfortunately, these experiments were inconclusive, as the TK siRNA was cytotoxic by itself. Thus, TK-deficient GC3/TK- cells were used to compare the cytotoxicity of TS6-dU/dA and TS6-FdU/dA. Cell proliferation assays revealed that IC50 values of these two ssiRNAs were identical in these cells (51.4 ± 27.8 and 45.9 ± 18.6 nM, respectively) (Figure 2B), confirming that FdU analogues are released from the ssiRNA as nucleosides and that TK is required for subsequent activation of FdU and enhancement of cytotoxicity.
To investigate the associated cellular signalling pathways that are induced by these ssiRNAs, the expression of certain key proteins was analysed. Immediately after transfection of TS6-FdU/dA or Mis-FdU/dA (0 h), formation of the ITC is observed along with reduced levels of free TS, suggesting that the ssiRNA was rapidly degraded releasing FdU (Figure 5). The reduction in free TS at this early time point was not due to RNAi knockdown, as TS protein levels were minimally affected after TS6-dU/dA transfection (left panel; pre versus 0 h). After 24 h, TS protein levels were greatly decreased except after transfection of Mis-FdU/dA. It has been well-established that p53 is one of the first cellular proteins to respond to genotoxic stress (25). p53 levels are rapidly induced only after transfection of TS6-FdU/dA. The expression of the p53 responsive gene, p21, was subsequently induced. One of the hallmarks of ‘thymineless death’ owing to TS inhibition is activation of the death receptor pathway (26). The receptor for this pathway, Fas, was upregulated after the FdU-modified TS-targeted ssiRNA, but not by the other ssiRNAs. It has been reported that Fas activation triggers apoptosis (26). As seen in Figure 5, cleaved PARP, a marker of apoptosis, was elevated only after TS6-FdU/dA transfection. Furthermore, two biomarkers of DNA damage, γH2AX and p-ATM, were strongly induced by TS6-FdU/dA with little-to-no effect in response to treatment with TS6-dU/dA or Mis-FdU/dA.
Figure 5.
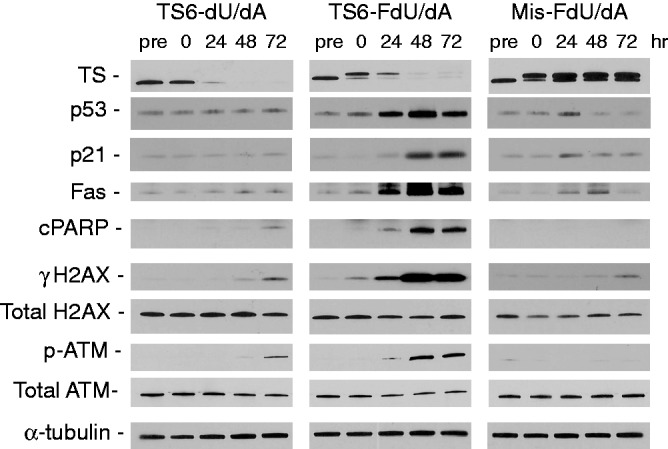
Time-dependent effects of FdU-containing ssiRNAs on protein expression. RKO cells were transfected with various TS6 ssiRNAs (10 nM). After 6 h, the medium was replaced, and cells were harvested for immunoblot blot analysis at the following times: before transfection (pre) and 0, 24, 48 and 72 h after transfection. Images shown are representative blots from at least four different experiments.
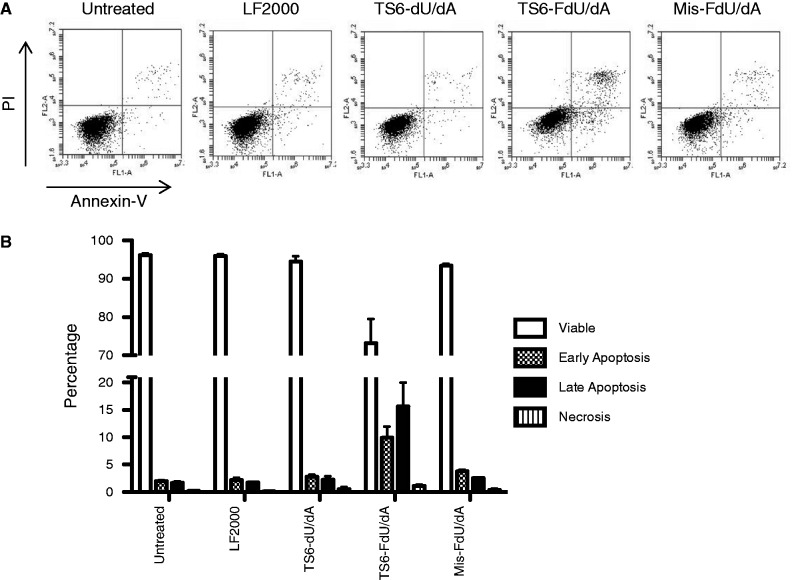
Finally, we performed a series of experiments to more directly investigate the effects of the modified ssiRNA on apoptosis. Although smaller increases in apoptosis were observed after 48 h (data not shown), transfection of TS6-FdU/dA resulted in a significant increase in both early and late apoptotic cells after 72 h (Figure 6). Transfection of the unmodified or mismatched ssiRNA had essentially no effect on inducing apoptosis. These results suggest that the combination of TS targeting with siRNA and TS enzyme inhibition from FdU released from degraded siRNA enhanced the ability of this modified siRNA to induce DNA damage and the process of apoptosis.
Figure 6.
Induction of apoptosis after transfection of FdU-containing ssiRNAs. RKO cells were transfected with various TS6 ssiRNAs (10 nM). After 6 h, the medium was replaced with fresh medium. After 72 h, cells were harvested, fixed, stained and analysed by flow cytometry as described in the ‘Materials and Methods’ section. (A) Representative cytometry images from one experiment. Lower left quadrant represents viable cells; lower right quadrant, early apoptotic cells; upper right quadrant, late apoptotic cells; upper left quadrant, necrotic cells. (B) Percentage values represent the mean ± S.D. from three separate experiments.
DISCUSSION
The potential therapeutic application of siRNAs is slowing being realized, as issues relating to siRNA stability, target selection, selective delivery and off-target effects have been mostly resolved with improved nanoparticles, advanced target selection algorithms and the use of chemically modified nucleotides (7,27). One issue that remains poorly defined is the effect of such modified nucleotides on cellular metabolism once the siRNA is eventually degraded within the cell. It is unknown whether these nucleotides are eliminated from the cell or whether they might be directly incorporated into cellular RNA or DNA with resultant effects on synthesis and function.
In earlier studies, we had shown that traditional siRNAs and ssiRNAs with 3′-end dT overhangs are rapidly degraded after transfection with intracellular release of dT and other nucleosides (17). The release of these nucleosides was sufficient to rescue from the growth inhibitory effects of a TS-targeted siRNA. To exploit this process of siRNA degradation, we hypothesized that a siRNA molecule could be designed with nucleoside analogues that would enhance its overall cytotoxic effects. In this regard, we have shown that the incorporation of FdU moieties into the ssiRNA greatly enhanced its cytotoxic effects. The addition of one FdU into the ssiRNA backbone reduced the IC50 value by 27-fold using WST-1 assay and by 5-fold using the clonogenicity assay (Table 4). Incorporation of 5 FdU on the 3′-end of the sense strand further enhanced siRNA toxicity (TS6-FdU/dA). Modification of both strands with FdU did not provide any additional enhancement of either cytotoxicity or clonogenicity. In the clonal assay, the unmodified TS-targeted ssiRNA and the untargeted control, FdU-modified ssiRNA yielded similar IC50 values, suggesting that both approaches (siRNA knockdown and nucleoside analogue release) were effective at growth inhibition. However, combining both approaches into a single ssiRNA enhanced growth inhibitory effects by 10-fold providing further support for our initial hypothesis. Previously, we demonstrated that resistant colon cancer cells could be re-sensitized to TS inhibitor compounds using our TS-targeted siRNA (11). Taken together, these results support the potential advantage of such a combined approach.
The design and synthesis of modified siRNAs with nucleoside analogues appears promising. The rationale for considering the incorporation of various nucleoside analogues comes from the fact that they have been used widely for antiviral and anticancer treatment (9,10). In particular, it is conceivable that ssiRNAs or siRNAs that incorporate HIV nucleoside analogues, such as stavudine and emtricitabine, or anticancer agents, such as gemcitabine, cytarabine, fludarabine and decitabine, could be easily synthesized. The incorporation of such modifications in ssi/siRNAs might then give rise to molecules with enhanced cytotoxicity. For instance, a synergistic effect might be observed with a gemcitabine-containing siRNA that suppresses the key DNA synthesis enzyme, ribonucleotide reductase (RRM1). Two groups have demonstrated that siRNAs directed against RRM1 can enhance gemcitabine toxicity in multiple cancer cell lines (28,29). Currently, modified nucleotides are incorporated into siRNAs or antisense oligonucleotides to enhance RNA stability against nucleases and to increase their binding affinity for their target mRNA (4). There have been several reports in which two or more nucleoside analogues are joined together to form ‘prodrug’-like molecules that depend on intracellular degradation for cytotoxicity (30,31). More recently, it was shown that the incorporation of a 5′-triphosphate on a viral-specific siRNA was able to enhance potency owing to an innate immune response activated by the 5′-triphosphate (32).
In conclusion, we have designed a novel dual-acting siRNA molecule, which to our knowledge, is the first of its class to have been developed. This siRNA has the usual gene-silencing effects typically seen with siRNAs. However, within the backbone of the siRNA, we have incorporated the fluoropyrimidine nucleoside FdU, which when released on siRNA degradation, can be metabolized via successive phosphorylation steps to nucleotide metabolites that then have biological activity and cytotoxicity. Of importance, this dual-acting siRNA displays enhanced cytotoxic and apoptotic effects when compared with the more traditionally designed TS-targeted siRNA. With the rapid development of nanoparticle delivery technologies, such modified siRNAs may find clinical value to be used as monotherapy or in combination with other chemotherapy agents to treat a wide range of human cancers. Moreover, this strategy of dual-acting siRNAs represents a potentially novel drug development approach for the treatment of other human diseases.
FUNDING
Veterans Administration Merit Award (to J.C.S.); National Natural Science Foundation of China [#30901823 to S.Y.W.]; University of Pittsburgh Cancer Institute; UPCI Clinical Pharmacology Analytical Facility; The Translational Research Core; The Cytometry Facility [P30-CA047904, in part]. Funding for open access charge: University of Pittsburgh Cancer Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank their laboratory members and members of the UPCI Merrill Egorin Writing Group for their critical review of this manuscript. They also thank Chris Stuart for his assistance with siRNA melting temperature analysis.
REFERENCES
- 1.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeVincenzo J, Lambkin-Williams R, Wilkinson T, Cehelsky J, Nochur S, Walsh E, Meyers R, Gollob J, Vaishnaw A. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl Acad. Sci. USA. 2010;107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Cihlar T, Ray AS. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 2010;85:39–58. doi: 10.1016/j.antiviral.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Sampath D, Rao VA, Plunkett W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene. 2003;22:9063–9074. doi: 10.1038/sj.onc.1207229. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz JC, Chen TM, Chu E. Small interfering double-stranded RNAs as therapeutic molecules to restore chemosensitivity to thymidylate synthase inhibitor compounds. Cancer Res. 2004;64:1431–1435. doi: 10.1158/0008-5472.can-03-1203. [DOI] [PubMed] [Google Scholar]
- 12.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Ann. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 13.Danenberg PV. Thymidylate synthetase - a target enzyme in cancer chemotherapy. Biochim. Biophys. Acta. 1977;473:73–92. doi: 10.1016/0304-419x(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 14.Shoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- 15.Smith SG, Lehman NL, Moran RG. Cytotoxicity of antifolate inhibitors of thymidylate and purine synthesis to WiDr colonic carcinoma cells. Cancer Res. 1993;53:5697–5706. [PubMed] [Google Scholar]
- 16.Yin MB, Guimaraes MA, Zhang ZG, Arredondo MA, Rustum YM. Time dependence of DNA lesions and growth inhibition by ICI D1694, a new quinazoline antifolate thymidylate synthase inhibitor. Cancer Res. 1992;52:5900–5905. [PubMed] [Google Scholar]
- 17.Schmitz JC, Chu E. Effect of small interfering RNA 3′-end overhangs on chemosensitivity to thymidylate synthase inhibitors. Silence. 2011;2:1. doi: 10.1186/1758-907X-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houghton PJ, Houghton JA, Germain G, Torrance PM. Development and characterization of a human colon adenocarcinoma xenograft deficient in thymidine salvage. Cancer Res. 1987;47:2117–2122. [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc. Natl Acad. Sci. USA. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 22.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl Acad. Sci. USA. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.O’Toole AS, Miller S, Haines N, Zink MC, Serra MJ. Comprehensive thermodynamic analysis of 3′ double-nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006;34:3338–3344. doi: 10.1093/nar/gkl428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 26.Houghton JA, Harwood FG, Tillman DM. Thymineless death in colon carcinoma cells is mediated via fas signaling. Proc. Natl Acad. Sci. USA. 1997;94:8144–8149. doi: 10.1073/pnas.94.15.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 28.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 29.Funamizu N, Kamata Y, Misawa T, Uwagawa T, Lacy CR, Yanaga K, Manome Y. Hydroxyurea decreases gemcitabine resistance in pancreatic carcinoma cells with highly expressed ribonucleotide reductase. Pancreas. 2012;41:107–113. doi: 10.1097/MPA.0b013e318224b5fb. [DOI] [PubMed] [Google Scholar]
- 30.Bijnsdorp IV, Schwendener RA, Schott H, Fichtner I, Smid K, Laan AC, Schott S, Losekoot N, Honeywell RJ, Peters GJ. Cellular pharmacology of multi- and duplex drugs consisting of ethynylcytidine and 5-fluoro-2'-deoxyuridine. Invest. New Drugs. 2011;29:248–257. doi: 10.1007/s10637-009-9353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gmeiner WH, Skradis A, Pon RT, Liu J. Cytotoxicity and in-vivo tolerance of FdUMP[10]: a novel pro-drug of the TS inhibitory nucleotide FdUMP. Nucleosides Nucleotides. 1999;18:1729–1730. doi: 10.1080/07328319908044836. [DOI] [PubMed] [Google Scholar]
- 32.Ahn J, Ko A, Jun EJ, Won M, Kim YK, Ju ES, Jeon ES, Lee H. Antiviral effects of siRNA simultaneously inducing RNA interference and type 1 interferon in coxsackievirus myocarditis. Antimicrob. Agents Chemother. 2012;56:3516–3523. doi: 10.1128/AAC.06050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]