Abstract
The Saccharomyces cerevisiae Shu complex, consisting of Shu1, Shu2, Csm2 and Psy3, promotes error-free homologous recombination (HR) by an unknown mechanism. Recent structural analysis of two Shu proteins, Csm2 and Psy3, has revealed that these proteins are Rad51 paralogues and mediate DNA binding of this complex. We show in vitro that the Csm2–Psy3 heterodimer preferentially binds synthetic forked DNA or 3′-DNA overhang substrates resembling structures used during HR in vivo. We find that Csm2 interacts with Rad51 and the Rad51 paralogues, the Rad55–Rad57 heterodimer and that the Shu complex functions in the same epistasis group as Rad55–Rad57. Importantly, Csm2’s interaction with Rad51 is dependent on Rad55, whereas Csm2’s interaction with Rad55 occurs independently of Rad51. Consistent with the Shu complex containing Rad51 paralogues, the methyl methanesulphonate sensitivity of Csm2 is exacerbated at colder temperatures. Furthermore, Csm2 and Psy3 are needed for efficient recruitment of Rad55 to DNA repair foci after DNA damage. Finally, we observe that the Shu complex preferentially promotes Rad51-dependent homologous recombination over Rad51-independent repair. Our data suggest a model in which Csm2–Psy3 recruit the Shu complex to HR substrates, where it interacts with Rad51 through Rad55–Rad57 to stimulate Rad51 filament assembly and stability, promoting error-free repair.
INTRODUCTION
DNA double-strand breaks (DSBs) are cytotoxic lesions whose improper repair can lead to mutations, genomic rearrangements or cell death. Faced with a DSB, eukaryotic cells can differentially use non-homologous end-joining or homologous recombination (HR) to repair the lesion. Misregulation of these pathways is both a hallmark of, and a driving force behind, cancer development. Recent work in the budding yeast Saccharomyces cerevisiae has characterized a novel regulator of HR, the Shu complex, which is also conserved in humans (1–3). Loss of the Shu complex leads to misregulation of HR, resulting in a higher mutation rate and increased genome rearrangements (1–4). Therefore, the Shu complex is likely an important regulator to suppress the chromosomal rearrangements and mutations observed in tumour cells, although the mechanism is largely unknown.
In budding yeast, the primary method of repairing a DSB is through Rad51-mediated HR [reviewed in (5,6)]. After recognition of a DSB by the cell, the 5′-end of the break is resected leading to 3′ single-stranded DNA (ssDNA) overhangs that are coated by the ssDNA-binding complex replication protein A (RPA). RPA on the nucleoprotein filament is displaced by Rad51 in a Rad52-dependent fashion. Formation of the Rad51 nucleoprotein filament is required for the homology search and strand invasion steps of HR. Resolution of the HR intermediates can be achieved through a multistep process leading to either crossover or non-crossover products.
As formation of the Rad51 filament is essential for all recombination events that require strand invasion, Rad51 loading onto the DNA is tightly regulated. Srs2 is a DNA helicase referred to as an ‘anti-recombinase’, as it functions to destabilize Rad51 filaments by translocating along ssDNA mediating Rad51 removal from 3′-DNA overhangs (7,8). Additional proteins promote Rad51 filament formation, for example, Rad52, which displaces RPA to facilitate Rad51 loading (9,10). At the same time, the Rad51 paralogues, the heterodimer Rad55–Rad57, integrate into and stabilize the Rad51 filament, block the progression of Srs2 and allow improved Rad51 nucleation and elongation (11–15). Failure to form Rad51 filaments shifts repair of DSBs away from HR towards Rad51-independent repair pathways, such as single-strand annealing (SSA) (16–18). In SSA, the ends of a break are resected to reveal distal homologous stretches of DNA that base pair to one another, a process that can result in the loss of the intervening genetic sequences.
The Shu complex, comprises Shu1, Shu2, Csm2 and Psy3, was previously identified in a genetic screen to identify mutants that suppress the slow growth phenotype of top3Δ mutants (2). Further analysis revealed that the Shu complex promotes Rad51-dependent HR (19,20). Interestingly, it was found in Schizosaccharomyces pombe that the Shu2 homologue, Sws1, physically interacts with Srs2, and that loss of the Shu complex suppresses the camptothecin-sensitivity of srs2Δ cells (1). More recently, it was demonstrated in S. cerevisiae that the Shu complex suppresses Srs2 recruitment to DSBs (20). This suggests a model whereby the Shu complex promotes Rad51-dependent HR by inhibiting the anti-recombinase Srs2.
Sequence homology between the Shu complex proteins, Shu1 and Psy3, and the human RAD51 paralogues, XRCC2 and RAD51D, respectively, has suggested that this yeast complex is composed of additional Rad51 paralogues (1). Consistent with this hypothesis, recent structural information has revealed that both Psy3 and Csm2 adopt a similar α–β sandwich fold structurally homologous to the ATPase core domain of Rad51 and RecA (21,22). Two independent crystallization studies demonstrate that the Shu complex is able to bind to DNA through the activity of the L2 loops in Psy3 and Csm2 (21,22). Importantly, Shu1 and Shu2 are dispensable for DNA binding in vitro, suggesting that recruitment of the Shu complex to DNA is mediated by Psy3 and Csm2 under endogenous conditions (22). However, both studies analysed the DNA-binding capability of the Shu complex for either ssDNA or dsDNA substrates, both of which are not the DNA structures typically used during HR; therefore, the biological substrates for these proteins are currently unclear. Additionally, it remains unknown whether the Shu complex functions similarly to the other Rad51 paralogues, Rad55 and Rad57, which are incorporated into the Rad51 filament to mediate Rad51 filament nucleation and elongation.
Here, we show that the Shu complex acts during DSB repair, shifting the balance towards error-free DNA repair through gene conversion (GC) and away from other error-prone repair pathways, such as SSA. We report that similar to the other Rad51 paralogues, Csm2 interacts with Rad51, as well as Rad55 and Rad57, and functions epistatically to Rad55–Rad57. Csm2’s interaction with Rad51 is dependent on the presence of Rad55; however, Csm2’s interaction with Rad55 occurs independently of Rad51. Interestingly, we show by fluorescent microscopy that loss of the Shu complex results in impaired Rad55 focus formation, indicating that Csm2 and Psy3 are needed for efficient recruitment of Rad55 to DSB sites. Finally, we find that the loss of the Shu complex alters the balance of HR outcomes away from the Rad51-dependent GC and towards the Rad51-independent SSA pathway. Together our work describes a model whereby the Shu complex, controlled by the DNA-binding activity of the Csm2 and Psy3 heterodimer, interacts with the Rad51 filament to stabilize it.
MATERIALS AND METHODS
Strains, plasmids and media
The strains used in this study are listed in Supplementary Table S1 and are isogenic with W303 and derived from the RAD5+ strains W1588-4C and W5909-1B (23,24), except for PJ69-4A and PJ69-4α strains used during the yeast-2-hybrid experiments (25). Standard protocols were used for crosses, tetrad dissection and yeast transformation (LiOAc method) (26). The media was prepared as described, except with twice the amount of leucine (26).
Co-purification of Csm2 and Psy3
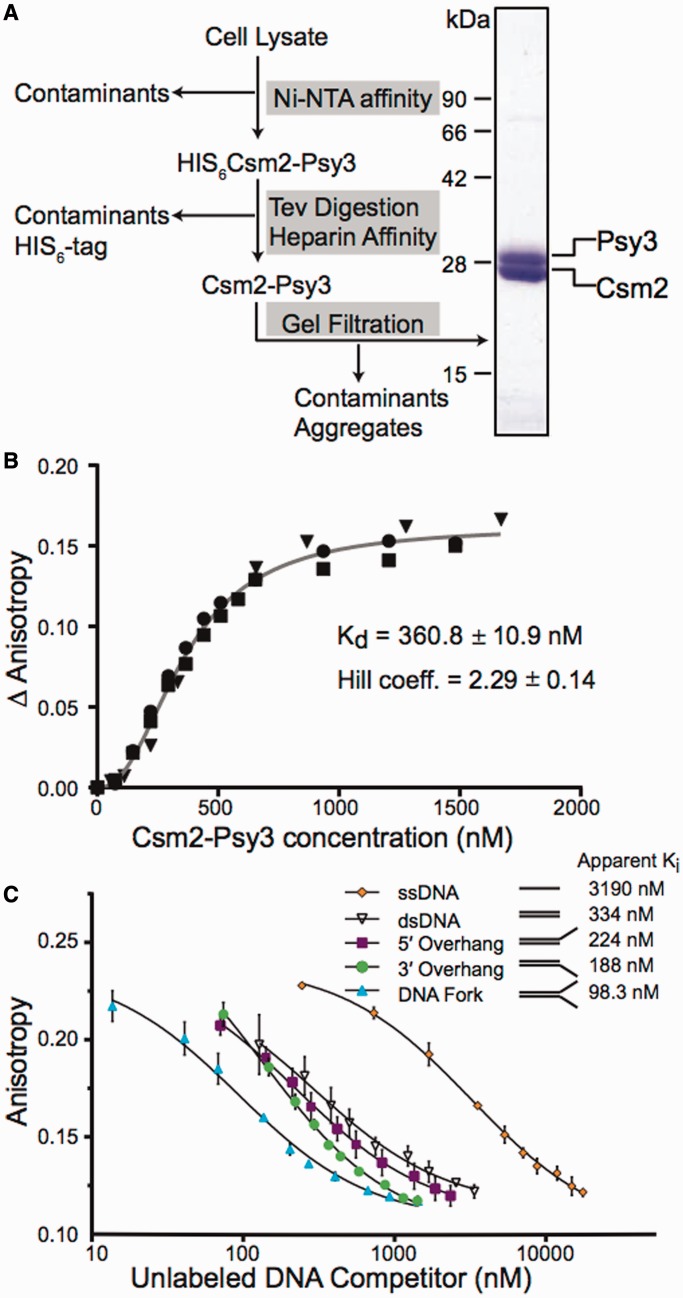
Full-length S. cerevisiae Csm2 and Psy3 were amplified from genomic DNA by polymerase chain reaction and cloned into the bacterial co-expression plasmid pCDF Duet-1 (cloning described in Supplementary Table S1; EMD Millipore). Protein expression was performed in Escherichia coli BL21 (DE3) Codon+(pRIL) via isopropylthio-beta-galatosidase (IPTG) induction. Cells were harvested by centrifugation, lysed in 20 mM Tris (pH 8.0), 300 mM NaCl, 10% glycerol, 5 mM imidazole and 1 mM β-mercaptoethanol and the lysates cleared by centrifugation at 30 000g. Csm2 and Psy3 were co-purified by nickel affinity chromatography (Qiagen) via the His6-tag on Csm2, followed by an overnight digestion with tobacco etch virus (TEV). The Csm2–Psy3 complex was then further purified using HiTrap Heparin HP (GE Healthcare) affinity chromatography and size-exclusion chromatography using a Sephacryl S-200 column (GE Healthcare) with peak fractions eluting as an apparent heterodimer verified by sodium dodecyl sulphate–polyacrylamide gel electrophoresis. The peak fractions were dialysed into a buffer containing 20 mM Tris (pH 8.0), 300 mM NaCl, 8% glycerol and 1 mM dithiothreitol and concentrated to 1.6 mg/ml using a Vivaspin concentrator (Millipore).
Fluorescence anisotropy assays for DNA binding
The basic protocol and mathematical rationale for this technique is outlined in Hey et al. (27). All experiments were performed using a Cary Eclipse Fluorescence Spectrophotometer (Varian) fitted with a peltier thermostatted multicell holder and automated polarizer. Fluorescent anisotropy/polarization measurements were collected with the excitation wavelength of 498 nm (slit-width 5 nm) and the emission wavelength at 520 nm (slit-width 5 nm), with a photomultiplier tube voltage of 780 V. Reactions were carried out at 30°C in a standard reaction buffer (20 mM Tris, pH 8, and 100 mM NaCl) with a total volume of 400 µl. A fluorescein-labelled DNA fork substrate (Table 1) was used in each of the experiments. Purified Csm2–Psy3 was titrated into the reaction volume to the indicated concentration and allowed 7.5 min for equilibration before the anisotropy measurement. Protein was titrated until the anisotropy signal plateaued, indicating saturation of the labelled probe. The Kd and Hill coefficient were calculated in PRISM (GraphPad) using a global non-linear regression from the three Csm2–Psy3 binding isotherms shown in Figure 1B. For the competition curves, fluorescence experiments were carried out with 25 nM fluorescein-labelled DNA fork and 546.6 nM Csm2–Psy3. The unlabelled competitors (Table 1) were added at increasing concentrations and allowed 7.5 min equilibration time before each measurement. Unlabelled DNA probe was added until polarized fluorescence stabilized, indicating saturation of the reaction with unlabelled DNA probe. Experiments were done in triplicate. The apparent Ki for each competitor was calculated in PRISM using a non-linear regression assuming a single-binding site, which is consistent with our electrophoretic mobility shift assays (EMSA) results (Supplementary Figure S1).
Table 1.
Sequences of the DNA substrates used in Figure 1B, Figure 1C, and Supplemental Figure S1
| DNA substrate | Sequence |
|---|---|
| Single stranded | 3′-GACGCTCGAGCTTAAGTGACCTCACTGGAG-5′ |
| dsDNA | 5′-CTGCGAGCTCGAATTCACTGGAGTGACCTC-3′ |
| 3′-GACGCTCGAGCTTAAGTGACCTCACTGGAG-5′ | |
| 5′-overhang | 5′-TCAAAGTCACGACCTAGACACTGCGAGCTCGAATTCACTGGAGTGACCTC-3′ |
| 3′-GACGCTCGAGCTTAAGTGACCTCACTGGAG-5′ | |
| 3′-overhang | 5′-CTGCGAGCTCGAATTCACTGGAGTGACCTC-3′ |
| 3′-AGTTTCAGTGCTGGATCTGTGACGCTCGAGCTTAAGTGACCTCACTGGAG-5′ | |
| Forka | 5′-TTTTTTTTTTTTTTTTTTTTCTTGACAAGCTTGCGCACT-3′ |
| 3′-TTTTTTTTTTTTTTTTCACGGAACTGTTCGAACGCGTGA-5′ |
aThe fluorescein-labelled fork is identical to the fork substrate with a single-fluorescein molecule attached to the 3′-end of the top strand.
Figure 1.
The Csm2 and Psy3 heterodimer preferentially bind forked and 3′-overhang DNA substrates. (A) Csm2–Psy3 was purified using the schematic presented using the conditions outlined in the ‘Materials and Methods’ section. Coomasie stained Csm2–Psy3 heterodimer (2.4 μg) is shown. Csm2 is 25 kDa and Psy3 is 28 kDa. (B) The Csm2–Psy3 heterodimer was assayed for DNA binding. Increasing concentrations of Csm2–Psy3 were added to a reaction mixture containing 25 nM fluorescein-labelled DNA fork in a fluorescence spectrophotometer. The binding isotherm was performed on three separate days, and each data set is shown. These data were fit using a global non-linear regression to obtain the Kd and a Hill coefficient. (C) Complexes of Csm2–Psy3 bound to a fluorescein-labelled fork substrate were pre-assembled as described. Fluorescence anisotropy was then measured with increasing concentrations of the indicated unlabelled competitors. Competition curves were fit using non-linear regression to calculate the apparent Ki. The experiment was done in triplicate, and standard deviations were plotted.
Yeast two-hybrids
The yeast two-hybrid plasmid pGAD was used to express a fusion of GAL4-activation domain, and pGBD was used to express a fusion of the GAL4 DNA-binding domain. The pGBD expressing plasmids (Csm2, Psy3 and empty vector) were transformed into PJ69-4A (25) or PJ69-4A, where the endogenous RAD51 or RAD55 was deleted, and positive colonies were selected on synthetic complete without tryptophane (SC-TRP) medium. The pGAD expressing plasmids (Rad55, Rad57 and Rad51) were transformed in PJ69-4α (25) or PJ69-4α, where the endogenous RAD51 or RAD55 was deleted, and recombinants were selected on SC-LEU medium. PJ69-4A and PJ69-4α haploid yeast cells harbouring their respective plasmids were mated, and diploids were selected on SC-LEU-TRP solid medium. Individual diploid cells were grown to early log phase OD600 0.2, and then 5 µl was spotted onto medium to select for the plasmids (SC-LEU-TRP) or onto medium to select for expression of the reporter HIS3 gene (SC-LEU-TRP-HIS), indicating a yeast two-hybrid interaction. Plates were incubated for 2 days at 30°C and subsequently photographed. Each experiment was done in triplicate.
Serial dilutions
The indicated strains were grown to early log phase, diluted to an OD600 of 0.2 and subsequently 5-fold serially diluted onto rich medium or rich medium exposed to 70 Gy or 0.006% methyl methanesulphonate (MMS) and incubated for 2 days at either 23°C or 30°C.
Fluorescent microscopy
Cells were grown overnight at 30°C in 3-ml cultures of SC with adenine (100 mg/l) and harvested for microscopy as previously described (28). A yellow fluorescent protein (YFP)–Rad55 integrated at its endogenous locus was introduced by mating into wild-type (WT), csm2Δ and psy3Δ cells and was visualized before and after 40 Gy of ionizing radiation (20, 60, 120 and 240 min) using a Nikon TiE inverted live cell system with a 100× oil immersion objective (1.45 numerical aperture) with a Photometrics HQ2 camera and motorized Prior Z-stage. Stacks of 11 0.3-µm sections were captured using the following exposure times: differential interference contrast (60 ms) and YFP–Rad55 (4000 ms). The images were deconvolved using Elements imaging software (Nikon). All images were processed and enhanced identically, and experiments were performed in triplicate with 300–500 total cells analysed.
Mitotic recombination assays
Mitotic recombination rates were calculated from haploid cells with the indicated mutations and the leu2-ΔEcoRI::URA3::leu2-ΔBstEII direct repeat recombination assay as described previously (29). SSA recombinants were measured as Leu+ Ura− colonies, and GC events were measured as Leu+ Ura+ colonies. For each genotype, nine individual colonies were analysed, and the experiment was performed in triplicate. The average mitotic recombination rate and standard deviation was calculated as described by Lea and Coulson (30).
RESULTS
Csm2 and Psy3 preferentially bind forked and 3′-overhang DNA substrates
Recently, the co-structure of two Shu complex members, Csm2 and Psy3, has been solved, and biochemical analysis has revealed that the heterodimer of Csm2 and Psy3 is responsible for binding of the Shu complex to DNA in vitro (21,22). However, the preferred physiological DNA substrates for these proteins are yet to be identified. To understand the function of the Shu complex during HR, we sought to characterize the preferred DNA substrates for these proteins. First, we co-purified Csm2 and Psy3 as a 1:1 heterodimer to homogeneity (Figure 1A). As the Shu complex was previously found to function during post-replicative repair (31), we first examined the ability of a Csm2–Psy3 complex to bind a forked DNA substrate. To accomplish this, Csm2–Psy3 complex was titrated against a fluorescein-labelled forked DNA substrate (Table 1) and binding measured by fluorescence anisotropy (Figure 1B). The binding seemed to be co-operative, and from these data, we calculated an equilibrium dissociation constant, Kd, of 360.8 ± 10.9 nM, with a Hill coefficient of 2.3 (Figure 1B).
To define the best binding substrate for the Csm2–Psy3 heterodimer, we compared the ability of the unlabelled substrates illustrated in Table 1 to compete for binding to a pre-assembled labelled fork by EMSA. In this assay, effective competition is monitored as a decrease in the amount of labelled DNA in complex with Csm2–Psy3 and an increase in the observed unbound free-labelled fork. We find that the forked DNA and, to a lesser extent, the 3′-overhang substrate are the best DNA-binding competitors for Csm2–Psy3 (Supplementary Figure S1). To better quantitate these results, we examined the substrates for their ability to compete for Csm2–Psy3 binding against the fluorescein-labelled fork by fluorescence anisotropy. We added sufficient Csm2–Psy3 protein to obtain ∼50% binding to the fluorescently labelled fork substrate, followed by titration with unlabelled DNA competitors, including ssDNA, double-stranded DNA (dsDNA), 5′-DNA overhang, 3′-DNA overhang or a forked DNA substrate (Table 1 and Figure 1C). Our results show that the forked DNA substrate, and to a lesser extent the 3′ DNA overhang, effectively competes for Csm2–Psy3 binding (Figure 1C). Calculation of apparent Kis for each competitor indicates that forked and 3′-overhang DNA compete the best, consistent with results obtained by EMSA. The 5′-overhang DNA substrate, which does not compete by EMSA, has a small but measurable difference in this assay. In contrast, both the dsDNA and ssDNA substrates are comparatively poor competitors (Figure 1C). Importantly, forked DNA and 3′-overhangs are DNA structures used by the homologous recombination pathway. Together our results show that the Csm2–Psy3 complex can specifically recognize and bind DNA substrates used by the homologous recombination pathway.
Csm2 interacts with Rad51 through the Rad51 paralogues Rad55–Rad57
Two of the human components of the Shu complex, XRCC2 and RAD51D, are RAD51 paralogues, which are proteins that have structural similarity to RAD51 (1,32). Consistent with this finding, the crystal structure of the yeast Shu proteins, Csm2 and Psy3, also reveal structural similarity to Rad51 (21,22). These results strongly suggest that, like the mammalian Shu complex, Csm2 and Psy3 are also Rad51 paralogues. Other Rad51 paralogues in yeast, such as the Rad55–Rad57 heterodimer, are incorporated into the Rad51 filament, thus stabilizing and promoting Rad51 filament formation and elongation (11–14). As Csm2 and Psy3 bind to similar DNA substrates used by Rad51 during HR, we assessed whether they could interact with Rad51 or the other Rad51 paralogues, Rad55 or Rad57, by yeast two-hybrid. We obtained plasmids harbouring fusions of the GAL4 activation domain with RAD51, RAD55 or RAD57 (pGAD) or of the GAL4–DNA–binding domain with PSY3 or CSM2 (pGBD) (Figure 2). Growth of yeast cells transformed with the respective combination of plasmids was assessed on medium lacking leucine and tryptophan (control) or additionally lacking histidine (interaction). Using this assay, we observe that Csm2 interacts with both Rad51 and Rad55, as well as weakly with Rad57 (Figure 2A). In contrast, Psy3 does not interact with Rad51, Rad55 or Rad57 by yeast two-hybrid (Figure 2A). To determine whether Rad51 mediates the interaction between Csm2 and Rad55, we repeated the yeast two-hybrid assay in cells where RAD51 is deleted. In the absence of Rad51, Csm2 and Rad55 still interact by yeast two-hybrid (Figure 2B). In contrast, in a reciprocal experiment where RAD55 is deleted, we no longer detect a yeast two-hybrid interaction between Csm2 and Rad51 (Figure 2B). Therefore, Csm2’s interaction with Rad55 occurs independently of Rad51, but Csm2’s interaction with Rad51 is likely dependent on Rad55. Furthermore, these results are consistent with the model that Csm2 mediates binding of the Shu complex to DNA perhaps through its physical interaction with the Rad51 paralogue Rad55.
Figure 2.
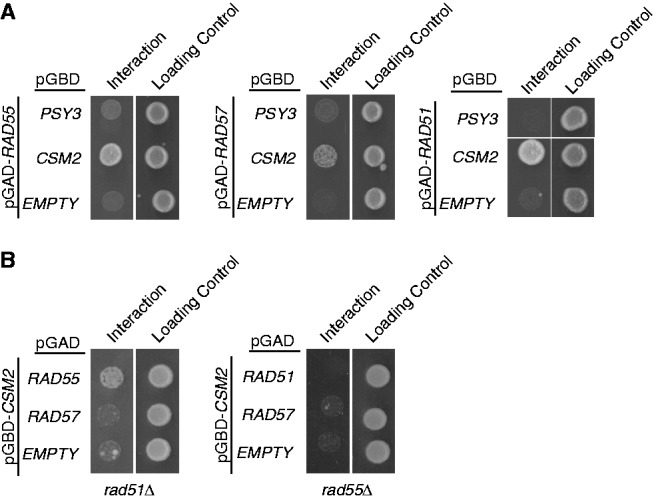
Csm2’s physical interaction by yeast two-hybrid with Rad51 is mediated by Rad55–Rad57. (A) Csm2 or Psy3 cloned into the pGBD plasmid (containing a GAL4-binding domain) was assayed for interaction with Rad55, Rad57 or Rad51, which were individually cloned into the pGAD (containing a GAL4-activating domain), by yeast two-hybrid. Growth on minimal medium lacking histidine indicates a yeast two-hybrid interaction, as HIS3 is the downstream reporter gene activated by interaction between the queried plasmids (interaction). Equal cell plating was determined by growth on minimal media lacking leucine and tryptophan, which selects for the two plasmids (loading control). (B) Csm2 and Rad51 or Csm2 and Rad55 yeast two-hybrid interactions were analysed in strains where RAD55 or RAD51 were, respectively, disrupted as described in (A).
The Shu complex and Rad55–Rad57 function in the same epistasis group
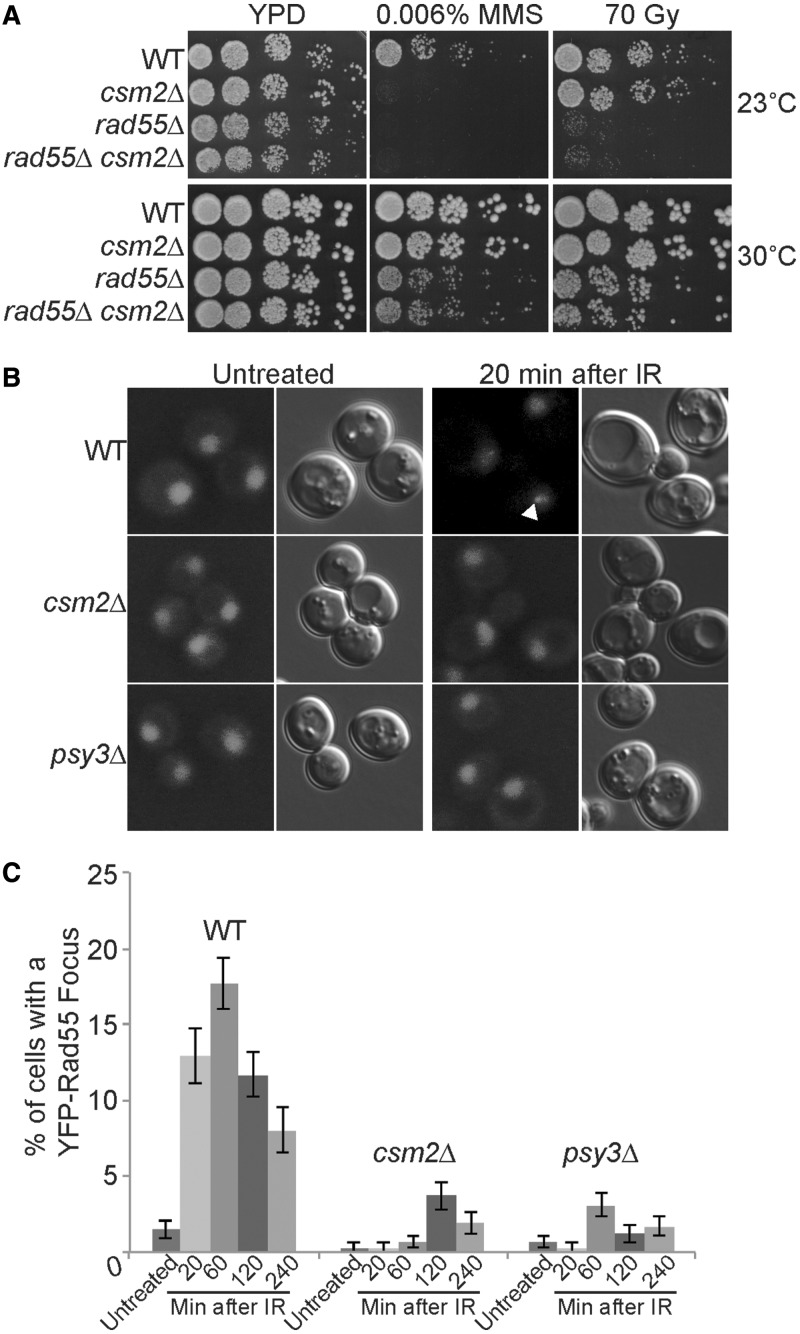
Similar to other Rad51 paralogues, we find that Csm2 interacts with Rad51. Therefore, we asked whether Csm2 functions in the same epistasis group as Rad55–Rad57. To address this question, we compared csm2Δ and rad55Δ single and double mutants for sensitivity to DNA damaging agents, such as MMS (a DNA alkylating agent) or ionizing radiation (IR, which induces DSBs) (Figure 3A). As rad55Δ cells were previously found to be cold sensitive, we analysed these mutants for growth at both 23°C and 30°C (33,34) (Figure 3A). We observe that both rad55Δ single mutant and rad55Δ csm2Δ double mutants are equally sensitive to both MMS and IR treatments when compared with WT or a csm2Δ single mutant, suggesting that Rad55 is epistatic to Csm2 with respect to DNA damage (Figure 3A). These results show that Rad55 and Csm2 likely function in the same epistasis group. Interestingly, csm2Δ cells were only sensitive to 0.006% MMS at lower temperatures (Figure 3A; 23°C panel), suggesting that like Rad55–Rad57, the Shu complex is likely involved in formation or stabilization of a larger complex (33,34).
Figure 3.
Csm2 is in the same epistasis group as Rad55 and regulates Rad55 recruitment to DNA damage sites. (A) WT, csm2Δ, rad55Δ and rad55Δ csm2Δ cells were 5-fold serially diluted onto yeast peptone dextrose (YPD) medium or YPD medium containing 0.006% MMS or exposed to 70 Gy IR and incubated at 23°C or 30°C for 2 days. (B) and (C) YFP-Rad55 expressing strains were analysed for the percentage of cells with a nuclear Rad55 focus before (untreated) or 20, 60, 120, and 240 min after IR (40 Gy). In part (B) images of Rad55 are shown, and a fluorescent Rad55 focus is indicated with a white arrowhead. Each experiment was done in triplicate with 300–500 cells analysed with standard errors plotted.
Csm2 is necessary for efficient recruitment of Rad55 to DNA damage sites
As Csm2 and Rad55 function in the same epistasis group in response to DNA damaging agents, we wondered whether Csm2 or Psy3 would be necessary for Rad55 recruitment to DSB sites. We analysed cells with fluorescently tagged Rad55 (YFP–Rad55) for formation of fluorescent foci, which indicates their redistribution to a DNA damage site, before and after exposure to IR (Figure 3B and C). In WT cells, we observe a Rad55 focus in 1.5% of cells before DNA damage (untreated). Twenty minutes after IR treatment, WT strains exhibit an increase in the percentage of cells with a Rad55 focus to 12.9% peaking after 60 min at 17.9% (Figure 3B and C). Subsequently, fewer Rad55 foci are observed, likely indicating resolution of the IR-induced DNA breaks. In contrast to WT, when CSM2 or PSY3 are deleted, we observe fewer cells with a Rad55 focus at all time points either before or after IR with no >3.7% of these mutants exhibiting a Rad55 focus (Figure 3B and C). Therefore, our data suggest that Csm2 and Psy3 are needed for efficient recruitment of Rad55 to DNA damage sites caused by IR.
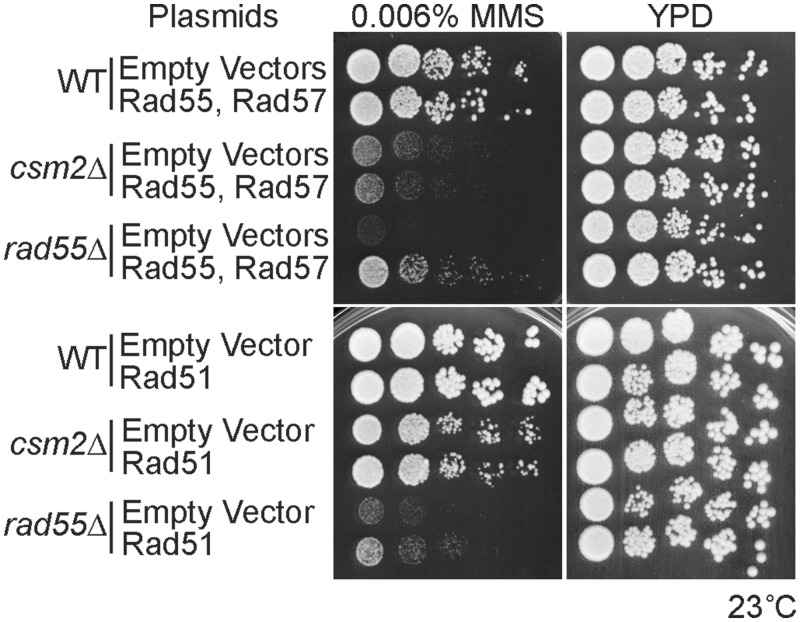
Unlike rad55Δ, the cold sensitivity of csm2Δ cells exposed to MMS is not suppressed by overexpression of Rad55–Rad57 or Rad51
Previously it was reported that the cold sensitivity of rad55Δ cells exposed to IR could be suppressed by overexpressing Rad55, Rad55–Rad57 together or Rad51 using a centromere (CEN) or 2-µ plasmid (15,35,36). Therefore, we asked whether the slow growth of csm2Δ cells exposed to MMS would similarly be suppressed by Rad55–Rad57 or Rad51 overexpression. Akin to what was previously observed with IR, the cold sensitivity of rad55Δ cells exposed to MMS could be partially suppressed by expressing Rad55–Rad57 or Rad51 from a CEN plasmid (Figure 4). In contrast, the slow growth of csm2Δ on MMS exposure was not suppressed by either Rad55–Rad57 co-expression or Rad51 at 23°C (Figure 4). Therefore, it is possible that Csm2 and Rad55 have different roles with respect to their function during repair of MMS-induced lesions. Alternatively, greater expression levels of Rad51 might be needed to observe suppression of this phenotype.
Figure 4.
Overexpression of Rad55–Rad57 or Rad51 does not suppress the MMS sensitivity of csm2Δ cells at 23°C. WT, csm2Δ and rad55Δ cells were either co-transformed with a Rad55 and Rad57 plasmid, a Rad51 plasmid or their respective empty vectors. Cells were grown to early log phase in minimal medium with selection for the plasmids and then 5-fold serially diluted onto YPD or YPD with 0.006% MMS. After 3 or 4 days of growth at 23°C, the plates were photographed.
The Shu complex promotes Rad51-dependent recombination events
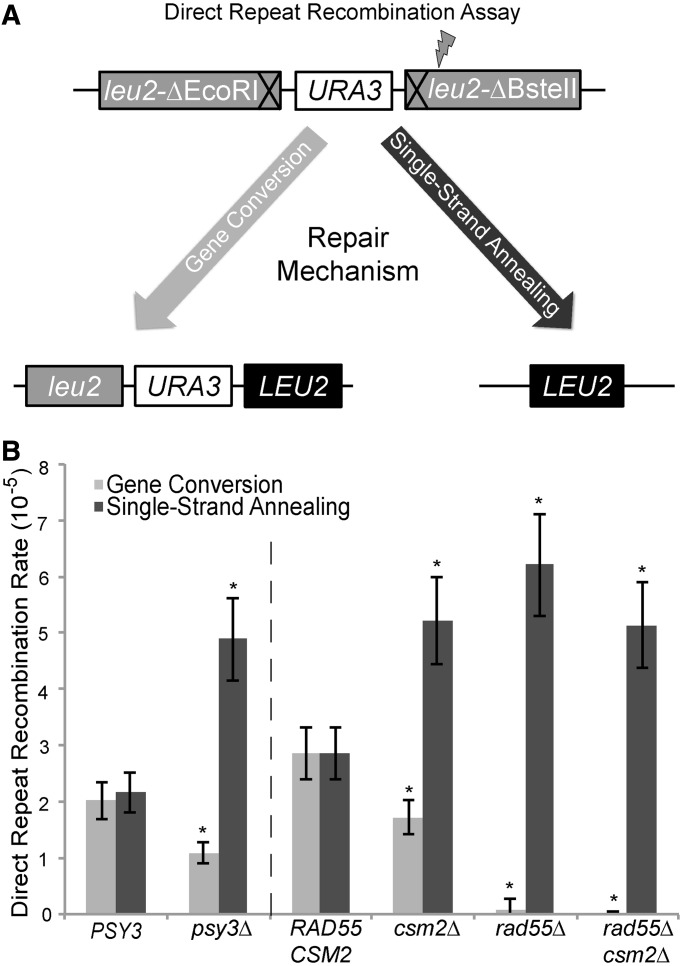
Previously, we hypothesized that the Shu complex promotes Rad51 filament formation, as fewer spontaneous Rad51 fluorescent foci are observed when the Shu complex is disrupted in yeast or mammalian cells (1,20). As Rad51 filaments are essential for HR, we asked whether disruption of the Shu complex might alter repair pathway choice if Rad51 filament formation is limited. To address this question, we used a heteroallelic recombination assay that can distinguish between direct repeat recombination mediated by sister chromatid GC (a Rad51-mediated event) and intrachromosomal SSA (a Rad51-independent event) (Figure 5A). In this assay, a recombination event can generate a functional LEU2 allele between two leu2 heteroalleles. The intervening URA3 marker between the leu2 alleles enables us to differentiate between sister chromatid GC (Leu+ Ura+) and SSA (Leu+ Ura−) recombinants (Figure 5A). Using this assay, WT PSY3 cells exhibit similar rates of GC and SSA (Figure 5B). In contrast, disruption of PSY3 or CSM2 results in significantly more SSA events (P ≤ 0.05 and P ≤ 0.01, respectively) where GC events are modestly, but significantly, reduced (P ≤ 0.001) (Figure 5B). As more SSA recombinants are observed with PSY3 or CSM2 disruption, these results are consistent with the model where inhibiting the Shu complex shifts the repair of spontaneous DSBs towards a Rad51-independent repair process. Alternatively, the Shu complex may suppress SSA.
Figure 5.
Disruption of CSM2 or PSY3 leads to more Rad51-independent recombination events. (A) Strains harbouring a direct repeat recombination assay (leu2-ΔEcoRI::URA3::leu2-ΔBstEII) were used to simultaneously measure rates of direct repeat recombination by GC or intrachromosomal SSA that result in a LEU2+ allele. Generation of a functional LEU2 gene can occur either through a GC event in which the other leu2 allele is used as a template for repair (Rad51-dependent, left side of diagram) resulting in Leu+ Ura+ colonies. Alternatively, repair can also occur by SSA where the intervening regions are resected until a region of homology is exposed and re-ligated (Rad51-independent, right side of diagram) resulting in Leu+ Ura− colonies. (B) The rates of GC and single-strand annealing events in psy3Δ, csm2Δ, rad55Δ or rad55Δ csm2Δ strains were compared with control cells (PSY3 or RAD55 CSM2) where equal rates of GC and SSA events are observed. The dashed line indicates that these strains were analysed independently.
As Rad55 interacts with Csm2 and functions in the same epistasis group in response to DNA damage, we further assessed whether disrupting Rad55 would have a similar effect on recombination rates using this assay. We assayed csm2Δ and rad55Δ single mutants and compared their recombination rates with the double rad55Δ csm2Δ mutant or RAD55 CSM2 cells (Figure 5B). As expected, disruption of RAD55 resulted in few detectable GC events (recombination rate <6.7 × 10−7), and SSA rates were increased to a level similar to a csm2Δ single mutant (Figure 5B). These results show that Rad55 has a more prominent role in mediating GC when compared with disruption of either PSY3 or CSM2. Furthermore, the increased SSA events observed in either rad55Δ or csm2Δ single or double mutants were similar, again suggesting that Rad55 and Csm2 are epistatic.
DISCUSSION
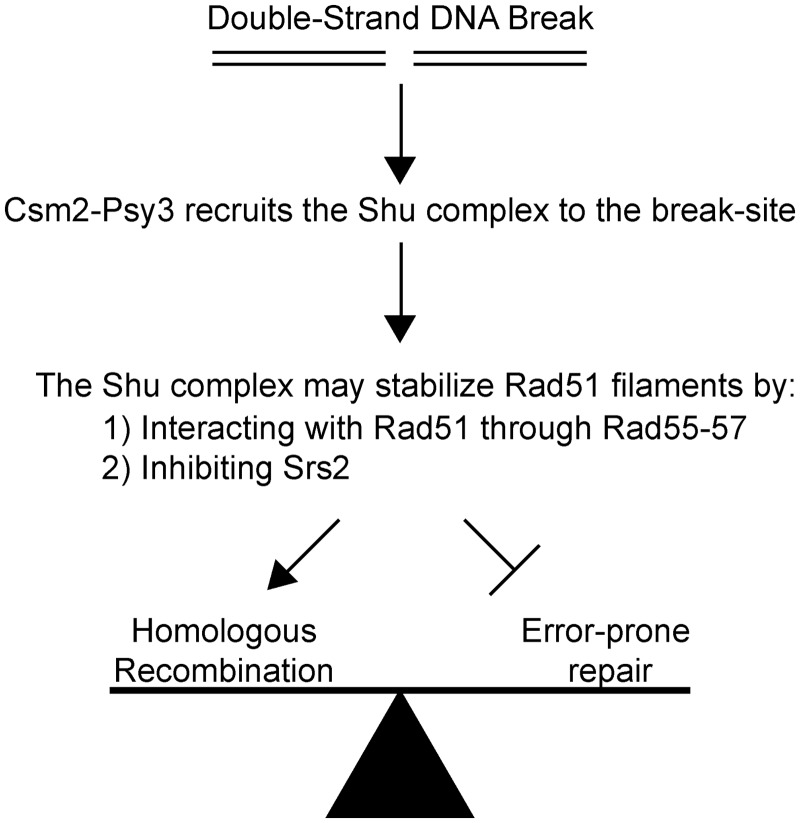
After a DSB occurs, cells can commit to multiple repair pathways to fix the lesion. An important complex in committing the cell to error-free DNA repair is the Shu complex. Recently, the crystal structures of two components of the Shu complex, Csm2 and Psy3, have been solved (21,22). These proteins bind DNA and are structural paralogues of Rad51. With this in mind, we examined the preferred binding substrates of purified Csm2–Psy3 heterodimer and find that these proteins preferentially bind to forked DNA and, to a lesser extent, 3′-overhang DNA substrates (Figure 1 and Supplementary Figure S1). The sigmoid nature of the protein saturation curve of the fluorescein-labelled fork suggests some co-operativity between the Csm2–Psy3 heterodimer, or that dimerization of the heterodimer complex is necessary before DNA binding (Figure 1B). Interestingly, the apparent Kd of Csm2–Psy3 for the forked substrate, 360.8 nM, falls within the predicted concentration of 237–472 nM for the Shu complex in the yeast nucleus (37,38). Importantly, the forked DNA and 3′-overhang structures are used by the HR pathway and are coated by Rad51 nucleoprotein filaments to perform the essential homology search and strand invasion HR steps. Consistent with our findings, the human RAD51B,RAD51C, RAD51D, XRCC2 complex (BCDX2) complex, which contains RAD51 paralogues, also preferentially bind branched DNA (39). As Csm2 and Psy3 exhibit structural similarity to Rad51, we examined whether they interact with Rad51 or the other known Rad51 paralogues in yeast, Rad55–Rad57. Importantly, by yeast two-hybrid, we detected an interaction between Csm2 and Rad51 that is dependent on Rad55 as well as an interaction between Csm2 and Rad55–Rad57 that is independent of Rad51 (Figure 2). Subsequently, we found that Csm2 is epistatic to Rad55, suggesting that they function in the same pathway (Figure 3). Importantly, by fluorescent microscopy, we find that Csm2 and Psy3 are necessary for Rad55 recruitment to DSB sites induced by ionizing radiation (Figure 3). Finally, we discovered, using recombination assays, that both Csm2 and Psy3 are important in Rad51-mediated repair at the expense of error-prone DNA repair mechanisms, such as SSA (Figure 5). Together our results and previous results from the literature (20–22) suggest a working model where the Shu complex, which consists of Rad51 paralogues, is likely recruited to HR substrates through its DNA-binding activity where it can interact with Rad51 through Rad55–Rad57 to mediate Rad51 filament formation and commitment to error-free DNA repair (Figure 6).
Figure 6.
Hypothetical working model of the role of the Shu complex during error-free HR. After a dsDNA break occurs, Csm2–Psy3 heterodimer recruits the Shu complex to the break site, which can either be at a replication fork or at a 3′-DNA overhang. At the break site, the Shu complex could promote Rad51 filament formation and stabilization (i) by interacting with Rad51 through Rad55–Rad57 and/or (ii) by inhibiting Srs2 recruitment to DSB sites. Stabilization of Rad51 filaments promotes DSB repair by an error-free HR pathway while inhibiting other error-prone DNA repair mechanisms, such as SSA.
One of the key steps in HR is the formation of Rad51 filaments. Importantly, there are proteins that mediate Rad51 filament formation, such as Rad52, and its epistasis group of proteins, including the Rad51 paralogues, Rad55–Rad57 (10,13,34,40–42). How do the Rad51 paralogues promote Rad51 nucleofilament formation? Both Rad55 and Rad57 are RecA-like proteins with structural similarity to Rad51 (11,12) and are needed for Rad51-mediated recombination events (13). As disruption of either RAD55 or RAD57 leads to slower and reduced recruitment of Rad51 to DSB sites and dimmer Rad51 foci (43–45), it has been proposed that Rad55–Rad57 nucleate Rad51 filaments where they stabilize Rad51 on the ssDNA end leading to longer Rad51 filament tracks (13,46). In support of this model, the IR sensitivity of rad55Δ or rad57Δ can be suppressed by overexpressing Rad51 (15,35,36). Furthermore, the cold sensitivity of rad55Δ and rad57Δ suggests that they are important for stabilization of larger complexes, such as the Rad51 pre-synaptic filament (33,34).
There are several lines of evidence that suggest that the Shu complex also consists of Rad51 paralogues. First, like the other Rad51 paralogues, the structure of Csm2 and Psy3 shows similarity to Rad51 (21,22). Second, similar to Rad55 and Rad57, the Shu genes are also needed for Rad51 recruitment to DNA damage sites in both yeast and human cells (1,3,20). Additionally, Csm2 interacts with Rad51 through the other Rad51 paralogues (Figure 2), and its disruption leads to decreases in Rad51-mediated DNA repair processes (Figure 5). Finally, Csm2 is in the same epistasis group as Rad55 in response to DNA damaging agents MMS and IR (Figure 3A). Together, we provide further genetic evidence that the Shu complex consists of Rad51 paralogues that is consistent with the structural homology observed by She et al. and Tao et al. (21,22).
In addition to the Rad51 paralogues stabilizing Rad51 on the DNA, there are also factors that mediate Rad51 filament disassembly, such as the DNA helicase Srs2. ATP-bound Rad51 frequently binds ssDNA, although with limited extension. In contrast, adenosine di-phosphate (ADP)-bound Rad51 can be readily disassociated from DNA (5,46,47). Srs2 promotes ATP hydrolysis of DNA bound Rad51 and then subsequently uses its helicase activity to translocate along the DNA filament where it can interact with the next available Rad51–ATP substrate (48–50). In humans, there are multiple proteins that have overlapping functions to Srs2, such as RECQL5, PARI, and RTEL (51–54). Recently, the Rad51 paralogues Rad55–Rad57 have been shown to physically interact with Srs2 in a 1:1 ratio (14). Further biochemical analysis has revealed that Rad55–Rad57 has an additional function in mediating Rad51 filament formation through its interaction with Srs2, where it inhibits Srs2 translocation activity and thus prevents removal of Rad51 from ssDNA substrates (14).
The Shu complex also has a role in regulating Srs2. Shu2 physically interacts with Srs2 in both budding and fission yeast (1,55). Disruption of either SHU1 or SHU2 results in more fluorescently tagged Srs2 recruited into spontaneous DNA repair foci and increased recruitment of Srs2 to inducible DSB sites (20). As we observe fewer Rad55 foci in the absence of CSM2 or PSY3 (Figure 3B and C), one explanation could be that disruption of the Shu complex leads to an increase in Srs2 recruitment to the DSB site where Srs2 may be removing Rad51 and Rad55–Rad57 resulting in fewer foci observed. Therefore, it is possible that the interaction between Rad51 paralogues in promoting Rad51 filament assembly by inhibiting Srs2 may be a shared function. However, it remains unknown whether the Shu complex is incorporated into the Rad51 filament like the other paralogues.
One puzzling observation made here is that Csm2 and Psy3 both promote Rad55 focus formation after IR but are not IR sensitive when disrupted (Figure 3). This is consistent with a reduction in Rad51 foci observed in shu1Δ cells (20). Although fewer Rad55 foci are observed, perhaps enough Rad55 is recruited to these lesions to enable cell viability after IR treatment in the absence of PSY3 or CSM2. Alternatively, there may be a delay in the kinetics of Rad55 focus assembly at DNA repair sites that would not result in a growth defect in IR exposed csm2Δ cells.
How are the roles of the Shu complex different from Rad55–Rad57 during HR? Previously, we proposed a model where the function of the Shu complex was to inhibit Srs2 recruitment to DNA repair sites, thus promoting error-free Rad51-mediated recombination. We find that disruption of either RAD55 or RAD57 leads to a more pronounced defect in HR and increased sensitivity to a broader range of DNA damaging agents. Furthermore, unlike Rad55–Rad57, the Shu genes likely have a more specialized function with respect to HR, perhaps a more dominant role at the replication fork or in response to specific types of DNA lesions. Consistent with this idea, the Shu complex was shown to have a role in post-replicative repair and to influence use of error-free DNA polymerases in response to MMS-induced lesions (2,31). Furthermore, disruption of the Shu complex members leads to sensitivity to MMS specifically but not other DNA damaging agents (i.e. IR, UV, HU and so forth) (1,2,19,31). Unlike the other Rad51 paralogues, csm2Δ cold sensitivity on exposure to MMS is not suppressed by Rad51 overexpression (Figure 4). These results may explain why csm2Δ or psy3Δ cells do not have a more dramatic effect on GC rates like those observed in rad55Δ cells. It is also possible that the Shu complex may suppress other repair mechanisms, such as SSA. Regardless, the interaction between the Shu complex and the other key players in mediating HR (such as Rad51, Rad55, Rad57 and Srs2) underscores the importance of understanding the unique roles these proteins play during Rad51 filament formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 1 and Supplementary Methods.
FUNDING
National Institutes of Health (NIH) [GM088413 to K.A.B., ES019566 to B.V.H.]. Funding for open access charge: NIH [GM088413 to K.A.B.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Rodney Rothstein and Lorraine Symington for providing reagents and insightful discussions for this work. They thank Stefanie Böhm and Cheryl Clauson for careful reading of the manuscript and Ye Peng for help with the initial anistropy experiments.
REFERENCES
- 1.Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shor E, Weinstein J, Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J. Biol. Chem. 2011;286:41758–41766. doi: 10.1074/jbc.M111.271080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl Acad. Sci. USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 8.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277: 31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 10.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 11.Kans JA, Mortimer RK. Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene. 1991;105:139–140. doi: 10.1016/0378-1119(91)90527-i. [DOI] [PubMed] [Google Scholar]
- 12.Lovett ST. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene. 1994;142:103–106. doi: 10.1016/0378-1119(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 13.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald JP, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mankouri HW, Ngo HP, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell. 2011;22:1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She Z, Gao ZQ, Liu Y, Wang WJ, Liu GF, Shtykova EV, Xu JH, Dong YH. Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS Lett. 2012;586:2306–2312. doi: 10.1016/j.febslet.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Tao Y, Li X, Liu Y, Ruan J, Qi S, Niu L, Teng M. Structural analysis of Shu proteins reveals a DNA-binding role essential for resisting damage. J. Biol. Chem. 2012;287:20231–20239. doi: 10.1074/jbc.M111.334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 25.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 27.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry (Mosc.) 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 28.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 31.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol. 2009;73:89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- 32.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Lovett ST, Mortimer RK. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RD, Symington LS. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, Kurumizaka H, Yokoyama S. Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 2004;32:2556–2565. doi: 10.1093/nar/gkh578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 41.Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 43.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 44.Fung CW, Fortin GS, Peterson SE, Symington LS. The rad51-K191R ATPase-defective mutant is impaired for presynaptic filament formation. Mol. Cell. Biol. 2006;26:9544–9554. doi: 10.1128/MCB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J. Biol. Chem. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, Krejci L, Sung P. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009;37:6754–6764. doi: 10.1093/nar/gkp748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karpenshif Y, Bernstein KA. From yeast to mammals: recent advances in genetic control of homologous recombination. DNA Repair. 2012;11:781–788. doi: 10.1016/j.dnarep.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moldovan G-L, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MIR, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 Maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.