Abstract
Cellular genomes are constantly damaged by endogenous and exogenous agents that covalently and structurally modify DNA to produce DNA lesions. Although most lesions are mended by various DNA repair pathways in vivo, a significant number of damage sites persist during genomic replication. Our understanding of the mutagenic outcomes derived from these unrepaired DNA lesions has been hindered by the low throughput of existing sequencing methods. Therefore, we have developed a cost-effective high-throughput short oligonucleotide sequencing assay that uses next-generation DNA sequencing technology for the assessment of the mutagenic profiles of translesion DNA synthesis catalyzed by any error-prone DNA polymerase. The vast amount of sequencing data produced were aligned and quantified by using our novel software. As an example, the high-throughput short oligonucleotide sequencing assay was used to analyze the types and frequencies of mutations upstream, downstream and at a site-specifically placed cis–syn thymidine–thymidine dimer generated individually by three lesion-bypass human Y-family DNA polymerases.
INTRODUCTION
DNA damage often causes replicative DNA polymerases to stall at lesion sites, arresting DNA synthesis, and eventually leading to apoptosis. To rescue stalled replication machinery, cells often switch to a DNA polymerase that is specialized to bypass DNA lesions, a process known as translesion DNA synthesis (TLS). Among the six DNA polymerase families, most of the DNA lesion bypass polymerases phylogenetically belong to the Y-family. The Y-family polymerases lack proof-reading 3′→5′ exonuclease activity and catalyze both error-free and error-prone TLS (1). Notably, 4 of the 16 identified human DNA polymerases, designated as DNA polymerases eta (hPolη), kappa (hPolκ), iota (hPolι) and Rev1 (hRev1), are in the Y-family. Although there is significant overlap in the lesion bypass abilities of these four enzymes, the nucleotide incorporation efficiency and fidelity of each human Y-family polymerase during TLS is likely lesion-specific (2). Therefore, it is possible that each Y-family polymerase has evolved to bypass a specific set of lesions in vivo. However, with the exception of ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPDs) such as cis–syn thymidine-thymidine (TT) dimers, the question of which Y-family polymerase is responsible for the bypass of a specific lesion type remains unanswered.
CPDs are estimated to account for ∼80% of UV-induced mutations within mammalian genomes (3). Among the human Y-family members, hPolη is known to catalyze the mostly error-free bypass of cis–syn TT dimers in vitro (4–6) and in vivo (7–9). Inactivation of hPolη through genetic mutation or deletion leads to the xeroderma pigmentosum variant disease, which predisposes individuals to increased incidence of skin cancer (9–12). In the absence of hPolη, both hPolκ (7) and hPolι (13–15) have been proposed to be responsible for the error-prone TLS of cis–syn TT dimers. Thus, it is biologically important to investigate the mutagenic patterns of TLS of a cis–syn TT dimer catalyzed by these human Y-family enzymes.
Studying the mutagenic consequences of TLS is essential to further our understanding of the mechanisms governing faithful genomic maintenance and cancer induction. However, the current methods for investigating the mutagenic profiles resulting from DNA lesion bypass catalyzed by DNA polymerases are labor-intensive, have low-throughput and often use a shuttle vector system requiring the amplification of lesion bypass products in Escherichia coli before sequencing and analysis (16–21). To overcome these obstacles, we have developed a high-throughput short oligonucleotide sequencing assay (HT-SOSA) that uses next-generation DNA sequencing technology to analyze the mutagenic profile produced as a result of the bypass of a site-specifically placed DNA lesion. This method enables the examination of the types and frequencies of lesion-induced errors generated by specific DNA polymerases. Although next-generation sequencing technology has previously been used to assess the mutagenic properties of carboxymethylated DNA lesions (22), only mutations opposite the lesion site were analyzed. In contrast, our design provides sequencing information for at least 10 template positions upstream and downstream from the DNA lesion site. This sequencing window is essential for the study of DNA damage-induced mutations within a newly replicated strand, as some types of DNA lesions reduce the fidelity of DNA polymerases at template positions adjacent to the lesion site due to a polymerase ‘sensing mechanism’ (17,18,23,24). By directly sequencing polymerase chain reaction (PCR)-amplified in vitro bypass products, our method also eliminates the time-consuming and labor-intensive steps of isolating and amplifying individual shuttle vectors bearing lesion bypass products in E. coli before sequencing a limited number of lesion bypass products. Finally, by using a bar-coding strategy to label DNA products produced by each combination of a DNA substrate and a polymerase, multiple populations of DNA lesion bypass products can be sequenced simultaneously within a single sequencing run. To analyze the mutagenic profiles, we developed specialized software called the ‘Next-Generation Sequencing Position Counter’ to align and quantify millions of DNA sequences. Therefore, in comparison with our first generation of the short oligonucleotide sequencing assay (SOSA) (17,18,21) and other methods in the literature (6,16,19,20), HT-SOSA enables the assessment of the mutagenic consequences of lesion bypass in a cost-effective manner, with exponentially increased sequencing information. Here, as a demonstration of HT-SOSA, we determined the types and frequencies of mutations generated individually by the human Y-family DNA polymerases hPolη, hPolκ and hPolι at template positions upstream, downstream and opposite a site-specifically placed cis–syn TT dimer.
MATERIALS AND METHODS
Materials
Reagents were purchased from the following companies: OptiKinase from USB Corporation, [γ-32P]ATP from MP Biomedicals, dNTPs from GE Healthcare and T4 DNA ligase from Fermentas. Full-length hPolη, truncated hPolκ (residues 9–518) and truncated hPolι (residues 1-420) were expressed and purified as previously described (17).
Synthesis of 21-mer-TT. The oligonucleotide 21-mer-TT (Table 1) containing a site-specifically placed cis–syn TT-dimer photoproduct was synthesized on a 0.2-μmol scale from the cis–syn TT dimer building block DMT-dT-PO(OCE)[c,s]-N3-(pivaloyloxymethyl)-dT-P(OCE)(NiPr2) (25) on an Applied Biosystem DNA synthesizer by standard phosphoramidite chemistry on a controlled pore glass (CPG) solid support. The yield for the dimer coupling step was ∼40%, as observed from trityl monitoring. The oligonucleotide was cleaved from the solid support using concentrated ammonium hydroxide at 55°C in a sealed tube overnight. The sample was dried in a Savant Speedvac, dissolved in ddH2O and filtered. The oligonucleotide was purified by reverse-phase high performance liquid chromatography (HPLC). The HPLC purified 21-mer-TT oligo was then confirmed to be correct by Matrix-assisted laser desorption ionisation time of flight (MALDI-TOF) mass spectrometry: (M+H)+ 6477.3 calculated, 6478.1 found.
Table 1.
Undamaged and damaged DNA oligomers containing a cis–syn TT dimera
| 31-mer | 3′-CCTGCTGTCCTGCCGTAGTCGTTACAACTGG-5′ |
| 21-mer-TT | 5′- GTTGAGTTACAGCTAGGTTAC-3′ |
| 25-mer | 3′- CTCCGCACGACACGCTCGCCTATCC-5′ |
| 77-mer-TT | 3′-CCTGCTGTCCTGCCGTAGTCGTTACAACTGGGTTGAGTTACAGCTAGGTTACCTCCGCACGACACGCTCGCCTATCC-5′ |
| 77-mer-ctl | 3′-CCTGCTGTCCTGCCGTAGTCGTTACAACTGGGTTGAGTTACAGCTAGGTTACCTCCGCACGACACGCTCGCCTATCC-5′ |
| 17-mer | 5′-CGGCATCAGCAATGTTG-3′ |
aTT represent the site of the cis–syn TT dimer.
DNA substrates
All DNA oligomers (Table 1 and Supplementary Table S1), except for 21-mer-TT, were purchased from Integrated DNA Technologies. All oligomers in Table 1 were gel-purified by using denaturing polyacrylamide gel electrophoresis (PAGE). Due to the difficulty in chemically synthesizing long DNA oligos containing a cis–syn TT dimer, the damaged DNA template 77-mer-TT was generated by ligation of the 21-mer-TT with a 31-mer and 25-mer DNA oligo by using standard protocols (Supplementary Figure S1). Briefly, the 31-mer, 21-mer-TT, (5′-32P)-labeled 25-mer and a guide 51-mer were mixed in a 1:1:1:1 molar ratio, heated to 70°C for 5 min and allowed to cool slowly. The annealed 77-mer-TT was ligated by T4 DNA ligase at 16°C for 24 h. Unligated DNA products were separated from the ligated 77-mer-TT product by using denaturing PAGE. The 5′-32P-labeled 17-mer was annealed to either 77-mer-TT or 77-mer-ctl in a 1:1 molar ratio by heating solutions for 5 min to 70°C and 95°C, respectively, followed by slow cooling to 25°C over several hours.
High-throughput short oligonucleotide sequencing assay
To generate the lesion bypass products, either radiolabeled 17-mer/77-mer-ctl or 17-mer/77-mer-TT (30 nM) was briefly preincubated at 37°C with an enzyme (120 nM), and subsequently mixed with all four dNTPs (200 nM each) in reaction buffer S (50 mM HEPES, pH 7.5 at 37°C, 5 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 7% glycerol and 0.1 mg/ml BSA). The reactions were incubated at 37°C for 1 hour for hPolη and hPolκ, and 4 hours for hPolι. As the SOSA primer 17-mer annealed 11 bases downstream of the 3′-end of the 77-mer-TT and 77-mer-ctl templates, full-length DNA products were effectively separated from the template by using denaturing PAGE.
Generation of sequencing libraries and DNA sequencing
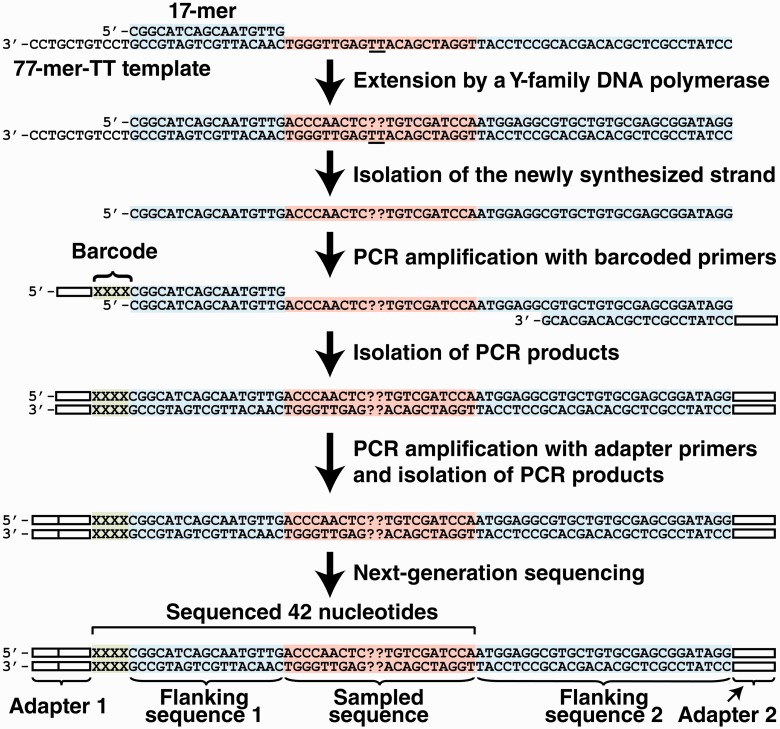
To generate the sequencing libraries of the lesion bypass products, each purified SOSA bypass product was PCR-amplified by using one of six primers containing a unique four-nucleotide barcode sequence and the HT-SOSA reverse primer (Supplementary Table S1). All PCRs were performed by using the following protocol: 95°C for 60 s and 15 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 4 min. The PCR products were subsequently gel-purified by using the QIAquick Gel Extraction Kit (Qiagen). To add the remaining adapter sequences necessary for next-generation sequencing, the purified PCR products were PCR-amplified with Illumina PCR primers 1 and 2 (Supplementary Table S1) by using the following protocol: 95°C for 60 s and 15 cycles of 95°C for 30 s, 63°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 4 min. The resulting PCR products were then gel-purified as described previously. The purity and concentration of the PCR-amplified DNA products with full-length adapter sequences were determined by using a bioanalyzer (Agilent Technologies). The DNA products were mixed in equal molar ratios (2.9 fmol each). The sequencing library solution was subsequently mixed with an equal amount of a sequence library derived from the genome of bacteriophage ΦX. The final sequencing library solution was then subjected to next-generation sequencing by using a GAII Genome Analyzer (Illumina). This method is summarized in Scheme 1.
Scheme 1.
Lesion bypass products were initially generated by extension of a control or damaged primer/template pair by an individual polymerase. The newly synthesized strands were then isolated by denaturing PAGE and subsequently amplified by two rounds of PCR amplification with primers containing a four nucleotide barcode sequence, and the adapter sequences necessary for next-generation sequencing. The adapter sequences are shown as white bars. The four nucleotide barcode is shown as ‘XXXX’ in green. The sampled sequence and the flanking sequences are shown in red and blue, respectively. The sequencing primer used for next-generation sequencing anneals to the PCR products within Adapter 1 and the 42 nucleotides of sequencing information obtained are indicated by a bracket. The position of the cis-syn TT dimer within the damaged template is underlined and the nucleotide incorporations opposite from the lesion are shown as ‘??’.
Analysis of DNA sequences
Initially, all of the raw sequence reads that matched the ΦX genome were removed. Sequence reads that contained one or more base calls that were not identified with >99.9% accuracy (Phred quality score of <30) were subsequently removed by using the NGS QC Toolkit (26). The remaining sequence reads were sorted into groups corresponding to the six unique barcode sequences used for analysis (i.e. Iota-control, Iota-damage, Kappa-control, Kappa-damage, Eta-control and Eta-damage) and stored as individual Sequence Alignment/Map (SAM) files. After sorting, erroneous sequences were further removed by eliminating all sequence reads that did not perfectly match the reference sequences within the first six nucleotides, which consist of the four-nucleotide barcode sequence and two adjacent nucleotides (Supplementary Figures S4–S6, Positions −31 to −26). Each SAM file of sequence reads was then analyzed by using a novel computer program called ‘The Next-Generation Sequencing Position Counter’. This program, written in Java, aligned each query sequence within the SAM file to a reference sequence (Supplementary Figure S3) by using a Needleman–Wunsch algorithm (citation: http://code.google.com/p/gal2009/) and produced an audit file containing an annotated ‘best fit’ alignment for each sequence (Supplementary Figure S7). The program simultaneously tallied the total number of matches, mismatches (i.e. substitutions), insertions and deletions at each template position of the query sequence relative to the reference sequence by using the ‘best fit’ alignments. In cases where the Needleman–Wunsch algorithm reported multiple ‘best fit’ alignments of the quarry and reference sequences with equivalent alignment scores, one alignment was selected for analysis at random to avoid alignment biases within our data sets. The sequence analysis for each SAM file was calculated and summarized in an output text file. Additionally, the details pertaining to how the program aligned and tabulated the mutations for each individual sequence read were also output to a separate audit file (Supplementary Figure S7). The Next-Generation Position Base Counter software is available for download at: http://bisr.osumc.edu/temp/NGSPositionCounter.tar.gz and https://chemistry.osu.edu/∼suo.3/index.html.
The base comparisons at each template position along the sequence read were scored as follows. Each base in the query sequence read was compared with the corresponding base in the reference sequence. If the two bases were identical, the comparison was scored as a match. If the two bases were different, the comparison was scored as a mismatch. Insertions or deletions were indicated with a dash. If the dash was reported in the reference read, the comparison was scored as an insertion in the query sequence. If the dash was reported in the query sequence, then the comparison was scored as a deletion. For insertions, the numbering for the reference read was adjusted from the spot of the insertion (see Supplementary Figure S7). Source codes are available on request.
RESULTS
The design of HT-SOSA to assess the mutagenic consequences of translesion synthesis
To investigate the frequencies and patterns of mutations induced by the TLS of a cis–syn TT dimer catalyzed by the human Y-family DNA polymerases by using a high-throughput approach, we designed HT-SOSA, as described in Scheme 1. First, a ‘77-mer-TT’ template containing a single cis–syn TT dimer site was generated by standard DNA ligation techniques (Table 1, Supplementary Figure S1). Subsequently, a 5′-radiolabeled 17-mer primer was annealed 11 nucleotides from the 3′-end of the 77-mer-TT template (17-mer/77-mer-TT) and extended separately by purified hPolη, hPolκ and hPolι to generate full-length lesion bypass products. For comparison, DNA products were also synthesized by using a control DNA substrate (17-mer/77-mer-ctl) containing a pair of undamaged template dTs in place of the cis–syn TT dimer (Table 1). As previously observed (17), hRev1 failed to produce full-length products with either the damaged or control DNA substrates (data not shown) and was not included in our analysis. This result was expected, as hRev1 is specialized to function as a dCMP transferase to preferentially incorporate dCTP opposite various lesion sites, as well as undamaged template bases (27,28). The full-length DNA products synthesized by hPolη, hPolκ or hPolι were separated from the longer DNA template through denaturing PAGE. Next-generation DNA sequencing libraries were then created by PCR amplification of the isolated bypass products with primers bearing both a unique four-nucleotide barcode to identify each DNA product and the adapter sequences necessary for next-generation sequencing (Supplementary Table S1, Supplementary Figure S2). Although the adapter and barcode sequences may be added within a single round of PCR, we chose to sequentially build these sequences by two rounds of PCR to reduce the sizes of the PCR primers. Before sequencing, the six sequencing libraries were mixed in equal molar ratios and subsequently combined with an equal amount of a DNA sequencing library derived from the bacteriophage ΦX genome to increase sequence diversity. Such sequence diversity is a necessity, as cluster detection algorithms for next-generation sequencing are often optimized for a balanced representation of all four nucleotides. After next-generation sequencing, the raw sequence reads that either aligned to the ΦX genome (1.4 × 107, 38% of total) or contained one or more base calls that were not assigned with ≥99.9% accuracy (6.6 × 106, 18% of total) were removed. The remaining sequences were then sorted according to the four-nucleotide barcodes (Supplementary Figure S3), and sequence reads that contained an error within the first six nucleotides or sequences that lacked a barcode altogether (8.4 × 105, 2% of total) were also removed. The sorted sequences (1.5 × 107, 42% of total) were subsequently aligned, and the mutation frequencies at each template position were calculated by using our novel ‘Next-Generation Sequencing Position Counter’ software. This custom software produced an annotated alignment of each sequencing read with a reference sequence (Supplementary Figure S7), allowing for the analysis of multiple mutations that occurred on the same template, and also tabulated the total number of observed base substitution, insertion and deletion mutations as a function of template position. As an internal control for errors introduced by PCR amplification and next-generation sequencing, we determined the error rates within flanking sequence 1 (Positions −25 to −11), which were initially derived from the 17-mer primer (Scheme 1). The average base substitution, insertion and deletion frequencies within this control region were calculated to be 5.5 × 10−4, 9.4 × 10−5 and 5.6 × 10−4 per base, respectively. The average relative error at each template position within this control region, calculated as the total number of mutations (insertions, deletions and substitutions) divided by the total number of dNTP incorporation events, was found to be 0.12 ± 0.02%. This background error rate is lower than the average error rates (0.1–1%) reported for the Illumina next-generation sequencing platform (29), partially due to the fact that our DNA template was derived from purified synthetic oligos rather than cellular DNA, which is often modified or damaged. Although this background error rate precludes the use of HT-SOSA for the examination of extremely rare mutation events, such as those produced by high-fidelity DNA polymerases, the total error rate of each error-prone Y-family polymerase investigated was found to be >145-fold above the background error rate opposite the lesion site and >10-fold above the background error rate at nearly every other template position analyzed (Supplementary Figures S4–S6). Thus, we concluded that HT-SOSA is a viable method for the investigation of the mutagenic profiles induced by TLS catalyzed by various Y-family polymerases.
Mutagenic profiles induced by TLS of a cis–syn TT dimer catalyzed by three human Y-family DNA polymerases
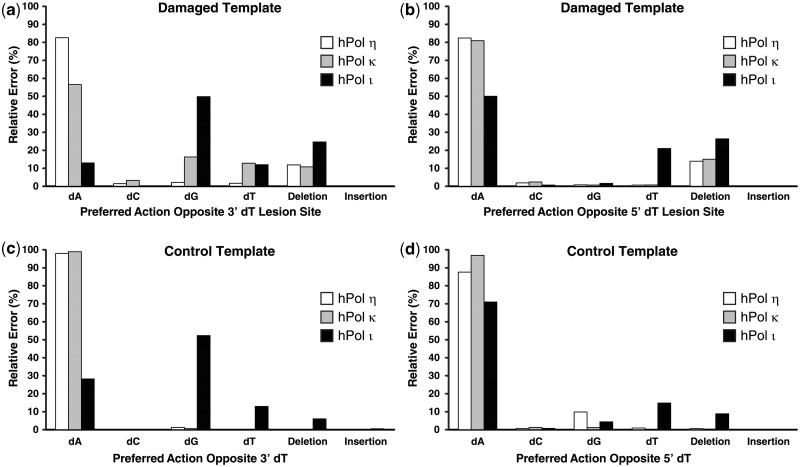
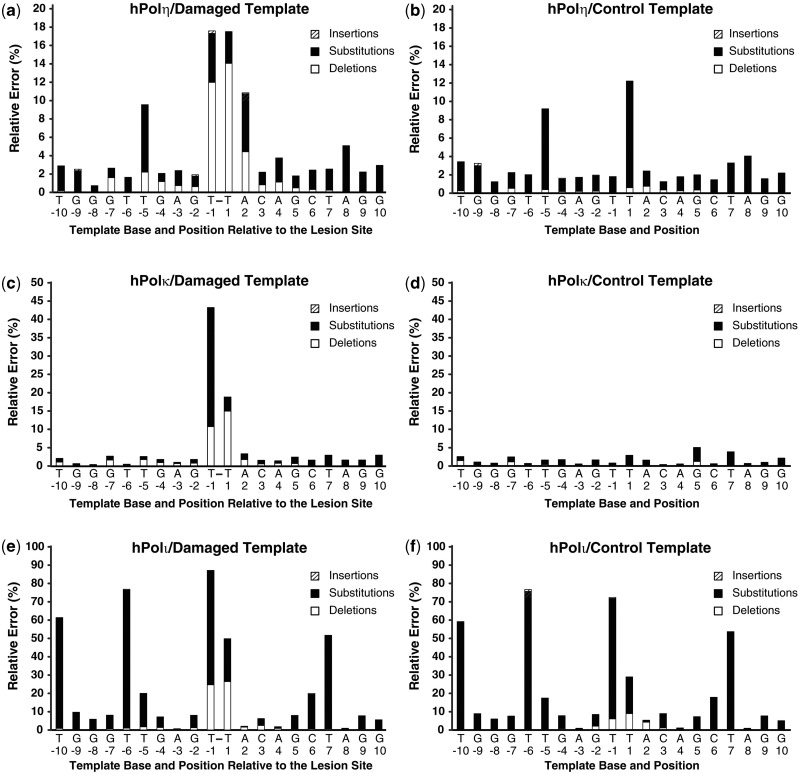
hPolη. Human Polη correctly incorporated dATP opposite the 3′-dT or 5′-dT of the cis–syn TT dimer in 82.5 and 82.6% of the 1.88 × 106 sequences analyzed, respectively (Figure 1a and b), with an average error frequency of 17.5%. Notably, hPolη correctly incorporated dATP opposite the 3′-dT or 5′-dT of the control template 77-mer-ctl within 98.2 and 87.8% of the sequences analyzed, respectively (Figure1c and d), with an average error frequency of 7.0%. Therefore, hPolη was only 2.5-fold more error-prone while incorporating dNTPs opposite the cis–syn TT dimer than opposite undamaged DNA. However, the base deletion frequency of hPolη at template Positions −1 and +1 increased 37-fold in the presence of the cis–syn TT dimer, with the majority of these deletions arising as double-base deletions (Figure 1 and Table 2). A similar increase in the base deletion error rate of hPolη opposite a cis–syn TT dimer has also been demonstrated by using ‘gap filling’ assays (6). We concluded that the cis–syn TT dimer altered both the types and the average frequencies of errors generated by hPolη. To quantitatively compare the error frequencies of hPolη at the damaged site with other template positions, we plotted the relative errors produced by hPolη as a function of template position (Figure 2a and b). Most notably, the relative error frequency of hPolη at Position +2 increased 4.5-fold in the presence of the cis–syn TT dimer, indicating the fidelity of hPolη was also reduced during the first extension step after TLS. Furthermore, the average deletion error frequency at template positions upstream and downstream of the lesion site increased 3.4- and 3.5-fold, respectively, in the presence of the cis–syn TT dimer (Supplementary Table S2). This increase in the base deletion frequency of hPolη was centered at template Positions −5 and +4, suggesting that the double-base lesion influenced the fidelity of hPolη most when the enzyme was approximately one-half helical turn upstream or downstream of the lesion site. The base substitution rate for hPolη replicating the undamaged DNA template was calculated to be 2.8 × 10−2 by using HT-SOSA. This value is comparable with the dNTP misincorporation fidelity of 5.6 × 10−2, as measured by steady-state kinetic assays (30), and 2.0 × 10−3 to 2.1 × 10−2, as measured by pre-steady-state kinetic assays (31), validating HT-SOSA as an effective method to determine the error rates of lesion bypass.
Figure 1.
Comparison of the preferred actions of human Y-family DNA polymerases opposite a cis–syn TT-dimer. The relative frequency of nucleotide incorporations opposite the (a) 3′-dT or (b) 5′-dT of the cis–syn TT-dimer is indicated. For comparison, relative frequency of nucleotide incorporations opposite the corresponding (c) 3′-dT or (d) 5′-dT of the control template is indicated.
Table 2.
Number of sequences that contain deletion mutations opposite the cis-syn TT dimer site and the corresponding percent of total sequences analyzed
| Sequencea | hPolη |
hPolκ |
hPolι |
|||
|---|---|---|---|---|---|---|
| Control template | Damaged template | Control template | Damaged template | Control template | Damaged template | |
| TCAATG | 2 106 445 (80.7%) | 1 257 192 (66.8%) | 3 388 119 (92.5%) | 1 407 584 (49.2%) | 318 649 (16.3%) | 168 671 (7.1%) |
| –-ATG | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (<0.1) | 0 (0.0%) |
| T–ATG | 2 (<0.1%) | 6 (<0.1%) | 2 (<0.1%) | 15 (<0.1%) | 57 (<0.1%) | 1014 (<0.1%) |
| T–-TG | 165 (<0.1%) | 1887 (0.1%) | 19 (<0.1%) | 4201 (0.1%) | 33 460 (1.7%) | 16 950 (0.7%) |
| TC-ATGb | 7434 (0.3%) | 27 201 (1.4%) | 11 492 (0.3%) | 122 986 (4.2%) | 25 694 (1.3%) | 51 550 (2.2%) |
| TC–TG | 933 (<0.1%) | 214 826 (11.4%) | 359 (<0.1%) | 294 250 (10.3%) | 58 960 (3.0%) | 551 921 (23.1%) |
| TC–-G | 461 (<0.1%) | 5783 (0.3%) | 26 (<0.1%) | 3924 (0.1%) | 8325 (0.4%) | 5653 (0.2%) |
| TCA–G | 6363 (0.2%) | 14 063 (0.7%) | 1043 (<0.1%) | 10 821 (0.4%) | 63 975 (3.3%) | 21 486 (0.9%) |
| TCA–- | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| T––G | 683 (<0.1%) | 684 (<0.1%) | 38 (<0.1%) | 1182 (<0.1%) | 44 (<0.1%) | 193 (<0.1%) |
aIncorporations opposite from the cis–syn TT dimer are in bold. The sequence TCAATG represents a sequence with no substitution, deletion or insertion mutations from Position −3 to +3. Sequences are depicted 5′–3′.
bIncludes all single-base deletions that aligned opposite either the 5′-dT or 3′-dT of the control or damaged templates, as these two events cannot be resolved.
Figure 2.
Histogram of the relative percent error as a function of template position. The relative number of base insertions (striped bar), substitutions (black bar) and deletions (white bar) as a percentage of the total dNTP incorporations is shown at each template position. The indicated template position is relative to the cis–syn TT dimer site within the 77-mer-TT template. The template bases are indicated and the cis–syn TT dimer is represented as T-T. Lesion bypass analysis for (a) hPolη, (c) hPolκ and (e) hPolι with the cis–syn TT dimer-containing DNA substrate is shown. The relative error as a function of template position with the control DNA substrate containing a pair of undamaged template dTs was also analyzed for (b) hPolη, (d) hPolκ and (f) hPolι.
hPolκ. We observed that hPolκ correctly incorporated dATP opposite the 3′-dT and 5′-dT of the cis–syn TT dimer in 56.6 and 81.2% of the sequences analyzed, respectively (Figure 1a and b). Therefore, hPolκ was more error-prone than hPolη when bypassing the cis–syn TT dimer. In contrast to hPolη, the total error frequency of hPolκ at Position +2 increased only 2-fold in the presence of the lesion (Figure 2c and d). This finding is consistent with the hypothesized role of hPolκ as the enzyme to extend lesion bypass products during TLS (32,33). Opposite the 3′dT of the cis–syn TT dimer, hPolκ preferentially misincorporated dGTP (16.3%), dTTP (12.9%) or dCTP (3.3%) over generating a base insertion (0.10%) or deletion (10.8%) mutation (Figure 1a). However, whenever hPolκ generated a deletion mutation opposite the cis–syn TT dimer, a double-base deletion was favored over a single-base deletion (Table 2). Comparing the error rate of hPolκ at template positions upstream and downstream of the double-base lesion site, we found that the base deletion error rate of hPolκ increased 1.8- and 1.9-fold, respectively, in the presence of the lesion (Figure 2 and Supplementary Table S2). Therefore, like hPolη, the lesion influenced the error rate of hPolκ before entering and after exiting the polymerase active site. The total base substitution rate for hPolκ replicating the undamaged DNA template was calculated to be 1.3 × 10−2 by HT-SOSA. This value is similar to hPolκ misincorporation fidelity of 1.4 × 10−2, determined by steady-state kinetic assays (34), and 3.5 × 10−3 to 2.9 × 10−2, established by pre-steady-state kinetic assays (31).
hPolι. HT-SOSA analysis indicated that hPolι correctly incorporated dATP opposite the 3′-dT and 5′-dT of the cis–syn TT dimer in 13.0 and 50.2% of the sequences analyzed, respectively (Figure 1a and b). Thus, hPolι was more error-prone than either hPolη or hPolκ for dNTP incorporations opposite the cis–syn TT dimer. Although hPolι generated a significant number of base deletions and substitutions opposite the template dTs of the control template, the cis–syn TT dimer increased the total average error frequency of hPolι at Positions −1 and +1 from 50.5 to 68.0% (Figures 1 and 2). This increase in relative error frequency was almost entirely due to a 7.7-fold increase in double-base deletions generated opposite the cis–syn TT dimer (Table 2). Thus, the cis–syn TT dimer altered both the type and the average frequency of errors generated by hPolι. Overall, hPolι produced more errors than hPolη and hPolκ at nearly every template position. Consistent with previous studies demonstrating the higher fidelity of hPolι for nucleotide incorporation opposite template purines than template pyrimidines (35–37), we found the fidelity of hPolι was highest for dNTP incorporations opposite template base dA, followed by dG, dC and dT (Figure 2). Surprisingly, hPolι preferred to misincorporate dGTP opposite template dTs at every template position (−10, −6, −5, −1 and +7), except for Position +1, where hPolι preferred to incorporate dTTP (Figure 1). Furthermore, the fidelity of hPolι increased opposite the 5′-dT relative to the 3′-dT of dT pairs, including the cis–syn TT dimer (Figure 2e and f, compare Positions −6 with −5, and −1 with +1). Therefore, we hypothesize that the fidelity of hPolι for dNTP incorporations opposite the cis–syn TT dimer may be dependent on the sequence context of the lesion. The base substitution error rate for hPolι replicating the undamaged template was calculated to be 1.8 × 10−1 by HT-SOSA, which is comparable with the dNTP incorporation fidelity of 1.0 × 10−1, measured by steady-state kinetic assays (38), and 9.3 × 10−3 to 1.1 × 10−1, as measured by single-turnover kinetic assays (31).
DISCUSSION
We have developed HT-SOSA, a high-throughput approach, to analyze the types and frequencies of mutations (deletions, additions and substitutions) produced as a result of DNA lesion bypass catalyzed by individual DNA polymerases (Scheme 1). Although HT-SOSA provides an unparalleled level of statistically robust information, the analysis of these vast data sets was problematic with the currently available computational approaches. We found that the local sequence alignment methods typically used to align the relatively short sequences generated by next-generation sequencing to much larger reference genomes poorly aligned the 42-bp HT-SOSA sequences with an equal-length reference sequence (Supplementary Figure S3). Therefore, we developed the software ‘Next-Generation Sequencing Position Counter’. This software uses a Needleman–Wunsch global sequence alignment algorithm to align the sequence of each lesion bypass product to an error-free reference sequence, and scores the types and frequencies of mutations generated by each DNA polymerase during TLS of a specific lesion. Therefore, lesion-induced mutations up to 10 template positions upstream and downstream from the damaged site can be investigated by using this strategy. Furthermore, by producing an annotated alignment for each sequence, this program facilitates the high-throughput analysis of sequences containing multiple mutations (Table 2, Supplementary Table S3, and Supplementary Figure S7).
To demonstrate the quantitative assessment of mutations induced by a site-specifically placed DNA lesion by using HT-SOSA, we analyzed the mutagenic profile generated during TLS of a cis–syn TT dimer catalyzed by hPolη, hPolκ and hPolι. We concluded that hPolη was the least error-prone Y-family enzyme for dNTP incorporation opposite the cis–syn TT dimer, whereas hPolκ was less error-prone than hPolη for dNTP incorporation opposite Position +2 of the damaged template (Figures 1 and 2). These findings are consistent with the established role of hPolη for the bypass of cis–syn TT dimers in vivo (7–9) and the hypothesized role of hPolκ for the extension of TLS products (32,33,39). Indeed the specialization of hPolη for bypassing CPDs such as cis–syn TT dimers is supported by X-ray crystal structural studies (40) demonstrating that the spacious and flexible active site of hPolη can readily accommodate an intra-strand dinucleotide lesion. Additionally, hPolη acts as a ‘molecular splint’ to straighten the template strand bearing a cis–syn TT dimer. The cis–syn TT dimer typically induces significant distortions to local DNA structure, including a 30° bend in the DNA helix (41). However, hPolη stabilizes the lesion-containing template strand in the B-form conformation through extensive interactions between the enzyme and the template strand (40).
We observed that the dNTP incorporation accuracy of hPolι was sequence-dependent, with the fidelity of hPolι being the highest for dNTP incorporations opposite dA, followed by dG, dC and dT (Figure 2). This template sequence-bias of dNTP incorporation fidelity for hPolι has been predicted by kinetic assays (37). For example, hPolι has been shown kinetically to incorporate dGTP more efficiently than dATP when opposite dT (36). Consistently, structural studies demonstrate that the templating nucleotide is switched from the cis to syn conformation at the active site of hPolι, leading to Hoogsteen base pairing, rather than Watson–Crick base pairing, between an incoming dNTP and the template base (35).
The base substitution error rates of hPolη, hPolκ and hPolι calculated by HT-SOSA with undamaged DNA templates closely match the dNTP incorporation fidelities predicted by steady-state and pre-steady-state kinetic assays (see RESULTS). Thus, the base substitution error rates calculated by HT-SOSA are corroborated by established kinetic methods. However, for the determination of polymerase fidelity opposite a lesion, it should be noted that HT-SOSA provides a more comprehensive analysis of the mutagenic profile of DNA damaged-induced mutations than kinetic assays because kinetic assays assume each dNTP misincorporation is a base substitution event, whereas HT-SOSA analysis accounts for all mutagenic events, i.e. base substitutions, deletions and insertions. Furthermore, unlike kinetic assays that calculate the polymerase fidelity based on the dNTP incorporation efficiency opposite individual template positions, HT-SOSA allows for the analysis of multi-base mutations within a single full-length product (Supplementary Table S3).
The majority of UV-induced mutations detected within cells lacking hPolη are T→C transitions and T→A transversions (14,20,42–44), indicating that the error-prone DNA polymerase(s) that substitutes for hPolη principally misincorporates dGTP or dTTP opposite CPDs. Interestingly, we found that hPolι generated the most mutations opposite both template positions of the double-base lesion, and preferentially misincorporated dGTP and dTTP opposite the 3′-dT and dTTP opposite the 5′-dT of the cis–syn TT dimer, rather than correctly incorporating two dATPs (Figure 1). Thus, our HT-SOSA data are most consistent with the proposed lesion bypass model wherein hPolι (13–15) is responsible for the error-prone TLS of cis–syn TT dimers in the absence of hPolη. However, given that hPolκ also misincorporated dGTP and dTTP opposite the 3′-dT of the cis–syn TT dimer (Figure 1a), we cannot completely rule out the possibility that hPolκ also plays a role in the error-prone bypass of cis–syn TT dimers. Once hPolι or hPolκ bypasses cis–syn TT dimers in the absence of hPolη, the mostly error-free extension of lesion bypass products may be carried out by either hPolκ (see previously) or a B-family enzyme, human DNA polymerase ζ (7,32,45).
Given that the next-generation sequencing technology used by HT-SOSA has consistently and frequently improved with respect to the length, number and accuracy of obtained sequences since its inception, we predict that our method will become far more powerful and versatile in the near future. We have demonstrated that this approach is a valuable tool for defining the mutagenic profile of DNA lesion bypass by individual polymerases in vitro. However, HT-SOSA can be readily adapted to assess the mutagenic profile induced by DNA lesions within cell culture by using established methods (46) to construct damaged DNA templates for cell transfection. Furthermore, owing to the capability to combine multiple samples in a single sequencing reaction and the subsequent sorting of the sequences by using unique barcodes, we expect that HT-SOSA will serve as the basis for larger and more complex applications, while remaining cost-effective. For example, by using a four-nucleotide barcode, up to 256 individual sequence libraries may be sequenced within a single reaction and subsequently sorted before analysis. Such a scheme allows for the simultaneous assessment of the mutagenic profiles induced by TLS catalyzed by a polymerase in combination with one or more auxiliary proteins, wild-type or mutated enzymes, or in the presence of exogenous agents, such as carcinogens. Finally, the general approach that we have described here is not limited to the study of TLS by error-prone DNA polymerases, but can be modified for the study of a number of cellular DNA repair pathways, such as base excision repair or non-homologous end joining.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–7.
FUNDING
National Science Foundation [MCB-0960961 to Z.S.]; National Institutes of Health [GM079403 and ES009127 to Z.S.]; National Institutes of Health (NIH) [CA40463 to J.S.T.]; National Science Foundation REU Training [DBI-1062144 to J.K.H.] and an American Heart Association Predoctoral Fellowship [GRT00014861 to S.M.S.]. Funding for open access charge: NIH [GM079403 to Z.S.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jason Fowler and Sean Newmister for the purification of hRev1 and hPolκ, respectively. They also thank Laura Sanman for making the DNA template 77-mer-TT. In addition, they thank Pearly Yan and Benjamin Rodriguez for their support in the design of the Illumina sequencing approach.
REFERENCES
- 1.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 2.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 5.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc. Natl Acad. Sci. USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 7.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl Acad. Sci. USA. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 9.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 10.Yamada A, Masutani C, Iwai S, Hanaoka F. Complementation of defective translesion synthesis and UV light sensitivity in xeroderma pigmentosum variant cells by human and mouse DNA polymerase eta. Nucleic Acids Res. 2000;28:2473–2480. doi: 10.1093/nar/28.13.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuasa M, Masutani C, Eki T, Hanaoka F. Genomic structure, chromosomal localization and identification of mutations in the xeroderma pigmentosum variant (XPV) gene. Oncogene. 2000;19:4721–4728. doi: 10.1038/sj.onc.1203842. [DOI] [PubMed] [Google Scholar]
- 12.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 13.Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota. EMBO J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 15.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol. Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang H, Taylor JS. Serial analysis of mutation spectra (SAMS): a new approach for the determination of mutation spectra of site-specific DNA damage and their sequence dependence. Nucleic Acids Res. 2008;36:6004–6012. doi: 10.1093/nar/gkn595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherrer SM, Fiala KA, Fowler JD, Newmister SA, Pryor JM, Suo Z. Quantitative analysis of the efficiency and mutagenic spectra of abasic lesion bypass catalyzed by human Y-family DNA polymerases. Nucleic Acids Res. 2011;39:609–622. doi: 10.1093/nar/gkq719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 19.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 20.Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair (Amst) 2008;7:1636–1646. doi: 10.1016/j.dnarep.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherrer SM, Taggart DJ, Pack LR, Malik CK, Basu AK, Suo Z. Quantitative analysis of the mutagenic potential of 1-aminopyrene-DNA adduct bypass catalyzed by Y-family DNA polymerases. Mutat Res. 2012;737:25–33. doi: 10.1016/j.mrfmmm.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan B, Wang J, Cao H, Sun R, Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 24.Gruz P, Shimizu M, Pisani FM, De Felice M, Kanke Y, Nohmi T. Processing of DNA lesions by archaeal DNA polymerases from Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:4024–4030. doi: 10.1093/nar/gkg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kshetry AK. 2008. Design and synthesis of modified deoxynucleotides, DNA, and PNA for biological studies, accession number 3316638, Ph.D. Thesis. Washington University in St. Louis, United States – Missouri. 233 pages. [Google Scholar]
- 26.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- 29.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 31.Sherrer SM, Sanman LE, Xia CX, Bolin ER, Malik CK, Efthimiopoulos G, Basu AK, Suo Z. Kinetic analysis of the bypass of a bulky DNA lesion catalyzed by human Y-family DNA polymerases. Chem. Res. Toxicol. 2012;25:730–740. doi: 10.1021/tx200531y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol. Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Prakash L, Prakash S. Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase iota. Proc. Natl Acad. Sci. USA. 2005;102:10466–10471. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasquez-Del Carpio R, Silverstein TD, Lone S, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Role of human DNA polymerase kappa in extension opposite from a cis-syn thymine dimer. J. Mol. Biol. 2011;408:252–261. doi: 10.1016/j.jmb.2011.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl Acad. Sci. USA. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YC, Maher VM, Mitchell DL, McCormick JJ. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGregor WG, Wei D, Maher VM, McCormick JJ. Abnormal, error-prone bypass of photoproducts by xeroderma pigmentosum variant cell extracts results in extreme strand bias for the kinds of mutations induced by UV light. Mol. Cell Biol. 1999;19:147–154. doi: 10.1128/mcb.19.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc. Natl Acad. Sci. USA. 2009;106:11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 46.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.