Abstract
Salmonella Typhi and Typhimurium diverged only ∼50 000 years ago, yet have very different host ranges and pathogenicity. Despite the availability of multiple whole-genome sequences, the genetic differences that have driven these changes in phenotype are only beginning to be understood. In this study, we use transposon-directed insertion-site sequencing to probe differences in gene requirements for competitive growth in rich media between these two closely related serovars. We identify a conserved core of 281 genes that are required for growth in both serovars, 228 of which are essential in Escherichia coli. We are able to identify active prophage elements through the requirement for their repressors. We also find distinct differences in requirements for genes involved in cell surface structure biogenesis and iron utilization. Finally, we demonstrate that transposon-directed insertion-site sequencing is not only applicable to the protein-coding content of the cell but also has sufficient resolution to generate hypotheses regarding the functions of non-coding RNAs (ncRNAs) as well. We are able to assign probable functions to a number of cis-regulatory ncRNA elements, as well as to infer likely differences in trans-acting ncRNA regulatory networks.
INTRODUCTION
Salmonella enterica subspecies enterica serovars Typhi (S. Typhi) and Typhimurium (S. Typhimurium) are important human pathogens with distinctly different lifestyles. S. Typhi is host-restricted to humans and causes typhoid fever. This potentially fatal systemic illness affects at least 21 million people annually, primarily in developing countries (1–3) and is capable of colonizing the gall bladder creating asymptomatic carriers; such individuals are the primary source of this human restricted infection, exemplified by the case of ‘Typhoid Mary’ (4). S. Typhimurium, conversely, is a generalist, infecting a wide range of mammals and birds in addition to being a leading cause of foodborne gastroenteritis in human populations. Control of S. Typhimurium infection in livestock destined for the human food chain is of great economic importance, particularly in swine and cattle (5,6). Additionally, S. Typhimurium causes an invasive disease in mice, which has been used extensively as a model for pathogenicity in general and human typhoid fever specifically (7).
Despite this long history of investigation, the genomic factors that contribute to these differences in lifestyle remain unclear. More than 85% of predicted coding sequences are conserved between the two serovars in sequenced genomes of multiple strains (8–11). The horizontal acquisition of both plasmids and pathogenicity islands during the evolution of the salmonellae is believed to have impacted upon their disease potential. A 100 kb plasmid, encoding the spv (Salmonella plasmid virulence) genes, is found in some S. Typhimurium strains and contributes significantly towards systemic infection in animal models (12,13). S. Typhi is known to have harboured IncHI1 plasmids conferring antibiotic resistance since the 1970’s (14), and there is evidence that these strains present a higher bacterial load in the blood during human infection (15). Similar plasmids have been isolated from S. Typhimurium (16–18). Salmonella pathogenicity islands (SPI)-1 and -2 are common to both serovars and are required for invasion of epithelial cells [reviewed in (19)] and survival inside macrophages respectively (20,21). S. Typhi additionally incorporates SPI-7 and SPI-10, which contain the Vi surface antigen and a number of other putative virulence factors (22–24).
Acquisition of virulence determinants is not the sole explanation for the differing disease phenotypes displayed in humans by S. Typhimurium and S. Typhi; genome degradation is an important feature of the S. Typhi genome, in common with other host-restricted serovars such as S. Paratyphi A (humans) and S. Gallinarum (chickens). In each of these serovars, pseudogenes account for 4–7% of the genome (9,25–27). Loss of function has occurred in a number of S. Typhi genes that have been shown to encode intestinal colonization and persistence determinants in S. Typhimurium (28). Numerous sugar transport and degradation pathways have also been interrupted (9) but remain intact in S. Typhimurium, potentially underlying the restricted host niche occupied by S. Typhi.
In addition to its history as a model organism for pathogenicity, S. Typhimurium has recently served as a model organism for the elucidation of non-coding RNA (ncRNA) function (29). These include cis-acting switches, such as RNA-based temperature and magnesium ion sensors (30,31), together with a host of predicted metabolite-sensing riboswitches. Additionally, a large number of trans-acting small RNAs (sRNAs) have been identified within the S. Typhimurium genome (32), some with known roles in virulence (33). These sRNAs generally control a regulon of mRNA transcripts through an antisense binding mechanism mediated by the protein Hfq in response to stress. The functions of these molecules have generally been explored in either S. Typhimurium or Escherichia coli, and it is unknown how stable these functions and regulons are over evolutionary time (34).
Transposon mutagenesis has previously been used to assess the requirement of particular genes for cellular viability. The advent of next-generation sequencing has allowed simultaneous identification of all transposon insertion sites within libraries of up to 1 million independent mutants (35–38), enabling us to answer the basic question of which genes are required for in vitro growth with extremely fine resolution. By using transposon mutant libraries of this density, which in S. Typhi represents on average >80 unique insertions per gene (35), shorter regions of the genome can be interrogated, including ncRNAs (38). In addition, once these libraries exist, they can be screened through various selective conditions to further reveal which functions are required for growth/survival.
Using Illumina-based transposon-directed insertion-site sequencing [TraDIS (35)] with large mutant libraries of both S. Typhimurium and S. Typhi, we investigated whether these Salmonellae require the same protein-coding and ncRNA gene sets for competitive growth under laboratory conditions, and whether there are differences that reflect intrinsic differences in the pathogenic niches these bacteria inhabit.
MATERIALS AND METHODS
Strains
S. Typhimurium strain SL3261 was used to generate the large transposon mutant library, and contains a deletion relative to the parent strain SL1344. The 2166 bp deletion ranges from 153 bp within aroA (normally 1284 bp) to the last 42 bp of cmk, forming two pseudogenes and deleting the intervening gene SL0916 completely. For comparison, we used our previously generated S. Typhi Ty2 transposon library (35).
Annotation
For S. Typhimurium strain SL3261, we used feature annotations drawn from the SL1344 genome (EMBL-Bank accession FQ312003.1), ignoring the deleted aroA, ycaL and cmk genes. We re-analysed our S. Typhi Ty2 transposon library with features drawn from an updated genome annotation (EMBL-Bank accession AE014613.1). We supplemented the EMBL-Bank annotations with ncRNA annotations drawn from Rfam 10.1 (39), Sittka et al. (40), Chinni et al. (41), Raghavan et al. (42) and Kröger et al. (32). Selected protein-coding gene annotations were supplemented using the HMMER webserver (43) and Pfam (44).
Creation of S. Typhimurium transposon mutant library
S. Typhimurium was mutagenized using a Tn5-derived transposon as described previously (35). Briefly, the transposon was combined with the EZ-Tn5 transposase (Epicenter, Madison, USA) and electroporated into S. Typhimurium. Transformants were selected by plating on LB agar containing 15 μg/ml kanamycin and harvested directly from the plates following overnight incubation. A typical electroporation experiment generated a batch of between 50 000 and 150 000 individual mutants. Ten batches were pooled together to create a mutant library comprising ∼930 000 transposon mutants.
DNA manipulations and sequencing
Genomic DNA was extracted from the library pool samples using tip-100 g columns and the genomic DNA buffer set from Qiagen (Crawley, UK). DNA was prepared for nucleotide sequencing as described previously (35). Before sequencing, a 22-cycle PCR was performed as previously described (35). Sequencing took place on a single end Illumina flowcell using an Illumina GAII sequencer, for 36 cycles of sequencing, using a custom sequencing primer and 2× Hybridization Buffer (35). The custom primer was designed such that the first 10 bp of each read was transposon sequence.
Sequence analysis
The Illumina FASTQ sequence files were parsed for 100% identity to the 5′ 10 bp of the transposon (TAAGAGACAG). Sequence reads, which matched, were stripped of the transposon tag and subsequently mapped to the S. Typhimurium SL1344 or S. Typhi Ty2 chromosomes using Maq version maq-0.6.8 (45). Approximately 12 million sequence reads were generated from the sequencing run, which used two lanes on the Illumina flowcell. Precise insertion sites were determined using the output from the Maq mapview command, which gives the first nucleotide position to which each read mapped. The number and frequency of insertions mapping to each nucleotide in the appropriate genome was then determined.
Statistical analysis of required genes
The number of insertion sites for any gene is dependent on its length; therefore, the values were made comparable by dividing the number of insertion sites by the gene length, giving an ‘insertion index’ for each gene. As before (35), the distribution of insertion indices was bimodal, corresponding to the required (mode at 0) and non-required models. We fitted gamma distributions for the two modes using the R MASS library (http://www.r-project.org). Log2-likelihood ratios (LLR) were calculated between the required and non-required models, and we called a gene required if it had an LLR of less than −2, indicating it was at least four times more likely according to the required model than the non-required model. ‘Non-required’ genes were assigned for an LLR of >2. Genes falling between the two thresholds were considered ‘ambiguous’ for the purpose of this analysis. This procedure lead to genes being called as required in S. Typhimurium when their insertion index was <0.020 and ambiguous between 0.020 and 0.027. The equivalent cut-offs for the S. Typhi library are 0.0147 and 0.0186, respectively.
We calculated a P-value for the observed number of insertion sites per gene using a Poisson approximation with rate R = N/G where N is the number of unique insert sites (549 086) and G is the number of bases in the genome (4 878 012). The P-value for at least X consecutive bases without an insert site is e(-RX), giving a 5% cut-off at 27 bp and a 1% cut-off at 41 bp.
For every gene g with ng,A reads observed in S. Typhi and ng,B reads observed in S. Typhimurium, we calculated the log2 fold change ratio Sg,A,B = log2 [(ng,A + 100)/(ng,B + 100)]. The correction of 100 reads smoothes out the high scores for genes with very low numbers of observed reads. We fitted a normal model to the mode +/− 2 sample standard deviations of the distribution of SA,B and calculated P-values for each gene according to the fit. We considered genes with a P-value of ≤0.05 under the normal model to be uniquely required by one serovar.
RESULTS AND DISCUSSION
TraDIS assay of every Salmonella Typhimurium protein-coding gene
Approximately 930 000 mutants of S. Typhimurium were generated using a Tn5-derived transposon. In all, 549 086 unique insertion sites were recovered from the mutant library using short-read sequencing with transposon-specific primers. This is a substantially higher density than the 371 775 insertions recovered from S. Typhi previously (35). The S. Typhimurium library contains an average of one insertion every 9 bp or >100 unique inserts per gene (Figure 1). The large number of unique insertion sites allowed every gene to be assayed; assuming random insertion across the genome, a region of 41 bp without an insertion was statistically significant (P < 0.01). As previously noted in S. Typhi, the distribution of length-normalized insertions per gene is bimodal (see Supplementary Figure S1), with one mode at 0. We interpret genes falling in to the distribution around this mode as being required for competitive growth within a mixed population under laboratory conditions (hereafter ‘required’). Of these, 57 contained no insertions whatsoever and were mostly involved in core cellular processes (see Table 1, Supplementary Data Set).
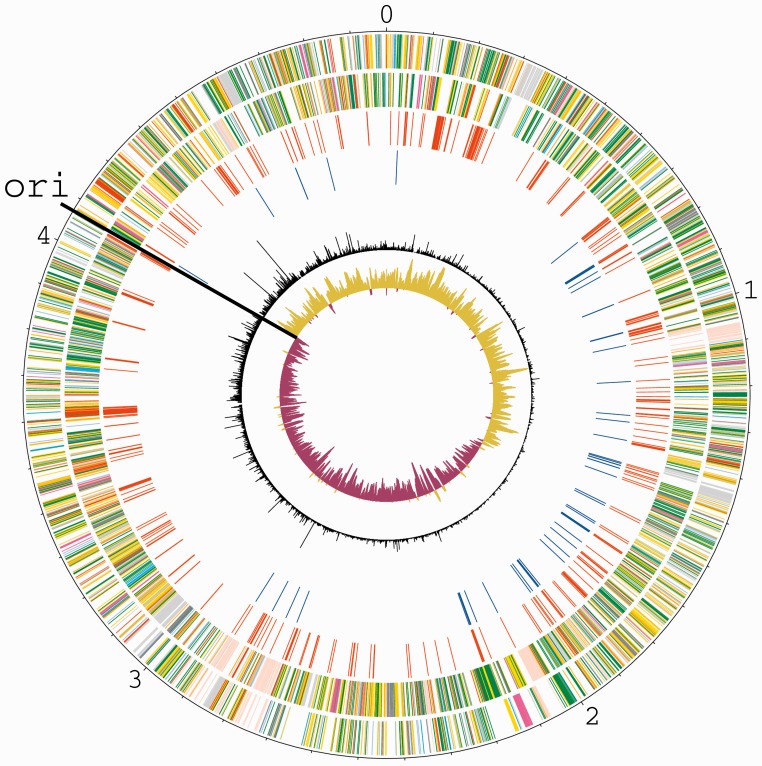
Figure 1.
Genome-wide transposon mutagenesis of S. Typhimurium. Circular plot showing gene content, distribution of required genes and insertion density along the S. Typhimurium chromosome. The outer scale is marked in megabases. Circular tracks range from 1 (outer track) to 6 (inner track). Track 1, all forward-strand genes (colour-coded according to function: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, membranes/surface structures; cyan, degradation of macromolecules; purple, degradation of small molecules; yellow, central/intermediary metabolism; light blue, regulators; pink, phage/IS elements; orange, conserved hypothetical; pale green, unknown function; brown, pseudogenes.); track 2, all reverse-strand genes (colour-coded as for forward-strand genes); track 3, S. Typhimurium required genes (red); track 4, 56 genes required by S. Typhimurium, but not by S. Typhi (dark blue, see also Table 1); track 5, transposon insertion density; track 6, GC bias [(G − C)/(G + C)]), khaki indicates values >1; purple <1.
Table 1.
Core gene functions in S. Typhimurium
| Biological process | Sub-process | Required genes | Non-required genes |
|---|---|---|---|
| Cell division | ftsALKQWYZ, minE, mukB, SL2391 | ftsHJNX*, minCD, sdiA, cedA, sulA | |
| DNA replication | Polymerases I, II and III | dnaENQX, holAB | polAB, holCDE |
| Supercoiling | gyrAB, parCE | ||
| Primosome-associated | dnaBCGT, priA, ssb | priB*C, rep | |
| Transcription | RNA polymerase | rpoABC | |
| Sigma, elongation, anti- and termination factors | nusBG, rpoDH, rho | nusA, rpoENS | |
| Translation | tRNA-synthetases | alaS, argS, asnS, aspS, cysS, glnS, gltX, glyQS, hisS, ileS, leuS, lysS, metG, pheST, proS, serS, thrS, tyrS, valS, | trpS, trpS2 |
| Ribosome components | rplBCDEFJKLMNOPQRSTUVWXY, rpmABCDHI, rpsABCDEFGHIJKLMNPQST | rplAI, rpmEE2, rpmFGJJ2, rpsOR*U*V | |
| Initiation, elongation and peptide chain release factors | fusA, infABC, prfAB, tsf, yrdC | efp, prfCH, selB, tuf | |
| Biosynthetic pathways | |||
| Peptidoglycan | murABCDEFGI, | ddl, ddlA | |
| Fatty acids | accABCD, fabABDGHIZ | – |
Protein-coding genes providing fundamental biological functions in S. Typhimurium. Genes in bold are required in S. Typhi (LLR between required and non-required models less than −2; see ‘Materials and Methods’ section). Asterisk indicates genes ambiguous in S. Typhimurium, having a LLR between −2 and 2.
There was a bias in the frequency of transposon insertion towards the origin of replication. This likely occurred as the bacteria were in exponential growth phase immediately before transformation with the transposon. In this phase of growth, multiple replication forks would have been initiated, meaning genes closer to the origin were in greater copy number and hence more likely to be a target for insertion. We also observed a bias for transposon insertions in A + T rich regions, as was previously observed in the construction of an S. Typhi mutant library (35). However, the insertion density achieved is sufficient to discriminate between required and non-required genes easily. As was first seen in S. Typhi (35), we observed transposon insertions into genes upstream of required genes in the same operon, suggesting that most insertions do not have polar effects leading to the inactivation of downstream genes.
Analysis of the S. Typhimurium mutant library allowed us to identify 353 coding sequences required for growth under laboratory conditions, and 4112 non-required coding sequences (see Supplementary Data Set). We were unable to assign 65 genes to either the required or non-required category. Sixty of these genes, which we will refer to as ‘ambiguous’, had LLRs between −2 and 2. The final five unassigned genes had lengths <60 bases, and they were removed from the analysis. All other genes contained enough insertions or were of sufficient length to generate credible LLR scores. Thus, every gene was assayed, and we were able to draw conclusions for 98.7% of the coding genome in a single sequencing run (Figure 1).
Cross-species comparison of genes required for growth
Gene essentiality has previously been assayed in Salmonella using insertion-duplication mutagenesis (46). Knuth et al. estimated 490 genes are essential to growth in clonal populations, though 36 of these have subsequently been successfully deleted (47). Although TraDIS assays gene requirements after a brief period of competitive growth on rich media, we identify a smaller required set than Knuth et al. of ∼350 genes in each serovar, closer to current estimates of ∼300 essential genes in E. coli (48).
To demonstrate that TraDIS does identify genes known to have strong effects on growth, as well as to test our predictive power for determining gene essentiality, we compared our required gene sets in S. Typhimurium and S. Typhi to essential genes determined by systematic single-gene knockouts in the E. coli K-12 Keio collection (48). We identified orthologous genes in the three data sets by best reciprocal FASTA hits exhibiting >30% sequence identity for the amino acid sequences. Required orthologous genes identified in this manner share a significantly higher average percentage sequence identity with their E. coli counterparts than expected for a random set of orthologues, at ∼94% identity as compared with ∼87% for all orthologous genes. In 100 000 randomly chosen gene sets of the same size as our required set, we did not find a single set where the average shared identity exceeded 90%, indicating that required genes identified by TraDIS are more highly conserved at the nucleotide level than other orthologous protein-coding sequences.
Baba et al. (48) have defined an essentiality score for each gene in E. coli based on evidence from four experimental techniques for determining gene essentiality: targeted knockouts using λ-red mediated homologous recombination (48), genetic footprinting (49,50), large-scale chromosomal deletions (51) and transposon mutagenesis (52). Scores range from −4 to 3, with negative scores indicating evidence for non-essentiality and positive scores indicating evidence for essentiality. Comparing the overlap between essential gene sets in E. coli, S. Typhi and S. Typhimurium, we find a set of 228 E. coli genes, which have a Keio essentiality score of at least 0.5 (i.e. there is evidence for gene essentiality; see Figure 2) that have TraDIS-predicted required orthologues in both S. Typhi and S. Typhimurium, constituting ∼85% of E. coli genes with evidence for essentiality indicating that gene requirements are largely conserved between these genera. Including orthologous genes that are only predicted to be essential by TraDIS in S. Typhi or S. Typhimurium raises this figure to nearly 93%. The majority of shared required genes between all three bacteria are responsible for fundamental cell processes, including cell division, transcription and translation. A number of key metabolic pathways are also represented, such as fatty acid and peptidoglycan biosynthesis (Table 1). A recent study in the alphaproteobacteria Caulobacter crescentus reported 210 shared essential genes with E. coli, despite C. crescentus sharing less than a third as many orthologous genes with E. coli as Salmonella serovars (38). This suggests the existence of a shared core of ∼200 essential proteobacterial genes, with the comparatively rapid turnover of 150–250 ‘non-core’ lineage-specific essential genes.
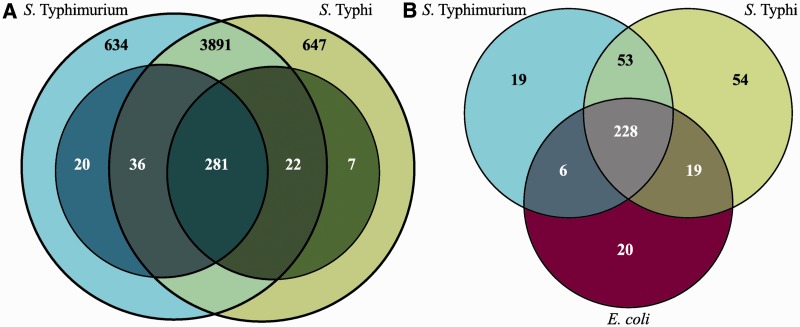
Figure 2.
Comparison of required genes. Venn diagrams showing (A) the overlap of all genes (outer circles, light colours) and required genes (inner circles, dark colours) between S. Typhimurium and S. Typhi (excluding genes required in one serovar only, which do not have significantly different read-counts). Black numbers refer to all genes, white numbers to required genes. (B) the overlap of all required genes between S. Typhimurium (blue), S. Typhi (green) and E. coli (purple). White numbers refer to genes with Keio essentiality scores ≥0.5.
If we make the simplistic assumption that gene essentiality should be conserved between E. coli and Salmonella, we can use the overlap of our predictions with the Keio essential genes to provide an estimate of our TraDIS libraries’ accuracy for predicting that a gene will be required in a clonal population. Of the 2632 orthologous E. coli genes, which have a Keio essentiality score of less than −0.5 (i.e. there is evidence for gene non-essentiality), only 33 are predicted to be required by TraDIS in both Salmonella serovars. S. Typhi contains the largest number of genes predicted by TraDIS to be required with E. coli orthologues with negative Keio essentiality scores. However, even if we assume these are all incorrect predictions of gene essentiality, this still gives a gene-wise false positive rate (FPR) of ∼2.7% (81 of 2981 orthologues) and a positive predictive value (PPV) of ∼75% (247 with essentiality scores ≥0.5 of 328 predictions with some Keio essentiality score). Under these same criteria, the S. Typhimurium data set has a lower gene-wise FPR of ∼1.6% (51 of 3122 orthologues) and a higher PPV of ∼82% (234 of 285 predictions as before), as we would expect given the library’s higher insertion density. In reality, these FPRs and PPVs are only estimates; genes that are not essential in E. coli may become essential in the different genomic context of Salmonella serovars and vice versa, particularly in the case of S. Typhi where wide-spread pseudogene formation has eliminated potentially redundant pathways (26,27). Additionally, TraDIS will naturally over-predict essentiality in comparison with targeted knockouts, as our library creation protocol necessarily contains a short period of competitive growth between mutants during the recovery from electro-transformation and selection. As a consequence, genes that cause major growth defects, but not necessarily a complete lack of viability in clonal populations, may be reported as ‘required’.
Serovar-specific genes required for growth
Many of the required genes present in only one serovar encoded phage repressors, for instance the cI proteins of Fels-2/SopE and ST35 (see Supplementary Tables S2 and S3). Repressors maintain the lysogenic state of prophage, preventing transcription of early lytic genes (53). Transposon insertions into these genes will relieve this repression and trigger the lytic cycle, resulting in cell death, and consequently mutants are not represented in the sequenced library. This again broadens the definition of ‘required’ genes; such repressors may not be required for cellular viability in the traditional sense, but once present in these particular genomes, their maintenance is required for continued viability as long as the rest of the phage remains intact.
Serovars Typhimurium and Typhi both contain eight apparent large phage-derived genomic regions (54,55). We were able to identify required repressors in all the intact lambdoid, P2-like and P22-like prophage in both genomes, including Gifsy-1, Gifsy-2 and Fels-2/SopE (see Supplementary Tables S2 and S3). With the exception of the SLP203 P22-like prophage in S. Typhimurium, all of these repressors lack the peptidase domain of the classical lambda repressor gene cI. This implies that the default anti-repression mechanism of Salmonella prophage may be more similar to a trans-acting mechanism recently discovered in Gifsy phage (56) than to the phage lambda repressor’s RecA-induced self-cleavage mechanism. We are also able to confirm that most phage remnants and fusions contained no active repressors, with the exception of the SLP281 degenerate P2-like prophage in S. Typhimurium. This degenerate prophage contains both intact replication and integration genes, but appears to lack tail and head proteins, suggesting it may depend on another phage for production of viral particles. Both genomes also encode P4-like satellite prophage, which rely on ‘helper’ phage for lytic functions and use a complex antisense-RNA based regulation mechanism for decision pathways regarding cell fate (57) using structural homologs of the IsrK (58) and C4 ncRNAs (59), known as seqA and CI RNA in the P4 literature, respectively. Although the mechanism of P4 lysogenic maintenance is not known, the IsrK-like ncRNAs of two potentially active P4-like prophage in S. Typhi are required under TraDIS. This sequence element has previously been shown to be essential for the establishment of the P4 lysogenic state (60), and we predict based on our observations that it may be necessary for lysogenic maintenance as well. The fact that some lambdoid prophage in S. Typhimurium encode non-coding genes structurally similar to the IsrK-C4 immunity system of P4 raises the possibility that these systems may be acting as a defense mechanism of sorts, protecting the prophage from predatory satellite phage capable of co-opting its lytic genes.
In addition to repressors, 4 prophage cargo genes in S. Typhimurium and one in S. Typhi are required (See Tables 2 and 3; Supplementary Tables S2 and S3). The S. Typhimurium prophage cargo genes encode a PhoPQ-regulated protein, a protein predicted to be involved in natural transformation, an endodeoxyribonuclease and a hypothetical protein. The S. Typhi prophage cargo gene encodes a protein containing the DNA-binding HIRAN domain (62) believed to be involved in the repair of damaged DNA. These warrant further investigation, as they are genes that have been recently acquired and become necessary for survival in rich media.
Table 2.
Genes uniquely required in S. Typhimurium
| Ty inserts | Ty reads | SL inserts | SL reads | SL ID | SL gene length | Ty ID | Ty gene length | Name | Function |
|---|---|---|---|---|---|---|---|---|---|
| No orthologue in S. Typhi | |||||||||
| 18 | 123 | SL0742 | 1269 | putative cation transporter | |||||
| 9 | 80 | SL0830 | 516 | conserved hypothetical protein | |||||
| 4 | 21 | SL0831 | 855 | putative electron transfer flavoprotein (beta subunit) | |||||
| 0 | 0 | SL0950 | 323 | predicted bacteriophage protein, potential phage repressor Gifsy-2 | |||||
| 11 | 75 | SL1179 | 789 | envF | lipoprotein | ||||
| 3 | 18 | SL1480 | 249 | antitoxin Phd_YefM, type II toxin-antitoxin system | |||||
| 4 | 32 | SL1527 | 264 | ydcX | putative inner membrane protein | ||||
| 1 | 3 | SL1560 | 717 | putative membrane protein | |||||
| 7 | 50 | SL1601 | 859 | putative transcriptional regulator (pseudogene) | |||||
| 4 | 36 | SL1799 | 201 | bacteriophage encoded pagK (phoPQ-activated protein) | |||||
| 5 | 22 | SL1830A | 434 | conserved hypothetical protein (pseudogene) | |||||
| 3 | 27 | SL1967 | 677 | predicted bacteriophage protein, potential phage repressor SLP203 | |||||
| 1 | 15 | SL2045A | 63 | yoeI | short ORF | ||||
| 17 | 107 | SL2066 | 900 | rfbJ | CDP-abequose synthase | ||||
| 3 | 34 | SL2549 | 209 | endodeoxyribonuclease | |||||
| 4 | 149 | SL2593 | 449 | putative DNA-binding protein, potential phage repressor Gifsy-1 SLP272 | |||||
| 3 | 7 | SL2633 | 846 | putative repressor protein, phage SLP281 | |||||
| 21 | SL2695 | 978 | smf | putative competence protein | |||||
| 5 | 39 | SL4132 | 291 | hypothetical protein | |||||
| 5 | 45 | SL4354A | 303 | conserved hypothetical protein | |||||
| Present in S. Typhi but required only in S. Typhimurium* | |||||||||
| 36 | 474 | 5 | 26 | SL0032 | 441 | t0033 | 306 | putative transcriptional regulator | |
| 71 | 349 | 11 | 48 | SL0623 | 642 | t2232 | 576 | lipB | lipoate-protein ligase B |
| 151 | 3546 | 10 | 64 | SL0702 | 897 | t2156 | 894 | putative glycosyl transferase | |
| 194 | 3007 | 9 | 61 | SL0703 | 1134 | t2155 | 1134 | galactosyltransferase | |
| 231 | 3499 | 15 | 67 | SL0706 | 1779 | t2152 | 1780 | putative glycosyltransferase, cell wall biogenesis | |
| 84 | 1041 | 2 | 4 | SL0707 | 834 | t2151 | 834 | putative glycosyltransferase, cell wall biogenesis | |
| 49 | 367 | 14 | 70 | SL0722 | 1569 | t2136 | 1569 | cydA | cytochrome d ubiquinol oxidase subunit I |
| 74 | 1613 | 5 | 22 | SL1069 | 693 | t1789 | 693 | putative secreted protein | |
| 20 | 199 | 1 | 1 | SL1203 | 150 | t1146 | 156 | hypothetical protein | |
| 20 | 290 | 1 | 5 | SL1264 | 315 | t1209 | 315 | putative membrane protein | |
| 84 | 384 | 6 | 26 | SL1327 | 402 | t1261 | 384 | spiC | putative pathogenicity island 2 secreted effector protein |
| 66 | 769 | 5 | 35 | SL1331 | 270 | t1265 | 327 | sseA | T3SS chaperone |
| 36 | 307 | 2 | 5 | SL1341 | 228 | t1275 | 228 | ssaH | putative pathogenicity island protein |
| 47 | 407 | 1 | 3 | SL1342 | 249 | t1276 | 249 | ssaI | putative pathogenicity island protein |
| 144 | 3197 | 5 | 14 | SL1343 | 750 | t1277 | 750 | ssaJ | putative pathogenicity island lipoprotein |
| 63 | 847 | 5 | 26 | SL1354 | 267 | t1288 | 267 | ssaS | putative type III secretion protein |
| 73 | 762 | 4 | 44 | SL1355 | 780 | t1289 | 780 | ssaT | putative type III secretion protein |
| 30 | 226 | 12 | 48 | SL1386 | 693 | t1322 | 693 | rnfE/ydgQ | Electron transport complex protein rnfE |
| 265 | 3337 | 29 | 165 | SL1473 | 1557 | t1463 | 1557 | pqaA | PhoPQ-activated protein |
| 85 | 765 | 6 | 35 | SL1532 | 951 | t1511 | 951 | sifB | putative virulence effector protein |
| 22 | 156 | 16 | 174 | SL1561 | 1227 | t1534a | 141 | sseJ | Salmonella translocated effector protein (SseJ) |
| 119 | 1639 | 10 | 44 | SL1563 | 762 | t1536 | 762 | putative periplasmic amino acid-binding protein | |
| 107 | 2440 | 5 | 44 | SL1564 | 648 | t1537 | 648 | putative ABC amino acid transporter permease | |
| 183 | 1646 | 20 | 118 | SL1628 | 1355 | t1612 | 1364 | hypothetical protein | |
| 23 | 177 | 1 | 5 | SL1659 | 183 | t1640 | 183 | yciG | conserved hypothetical protein |
| 78 | 617 | 16 | 104 | SL1684 | 1014 | t1664 | 1014 | Hnr | putative regulatory protein |
| 37 | 277 | 4 | 25 | SL1785 | 396 | t1022 | 396 | conserved hypothetical protein | |
| 166 | 2823 | 9 | 27 | SL1793 | 915 | t1016 | 915 | pagO | inner membrane protein |
| 28 | 311 | 3 | 22 | SL1794 | 159 | t1015 | 159 | putative inner membrane protein | |
| 23 | 155 | 1 | 4 | SL1823 | 972 | t0988 | 972 | msbB | lipid A acyltransferase |
| 60 | 402 | 11 | 58 | SL2064 | 1002 | t0786 | 1002 | rfbV | putative glycosyl transferase |
| 87 | 524 | 7 | 59 | SL2065 | 1293 | t0785 | 1299 | rfbX | putative O-antigen transporter |
| 66 | 559 | 13 | 74 | SL2069 | 774 | t0780 | 774 | rfbF | glucose-1-phosphate cytidylyltransferase |
| 41 | 204 | 5 | 14 | SL3828 | 1830 | t3658 | 1830 | glmS | glucosamine-fructose-6-phosphate aminotransferase |
| 27 | 288 | 5 | 23 | SL4250 | 288 | t4220 | 288 | putative GerE family regulatory protein | |
| 148 | 2633 | 16 | 89 | SL4251 | 876 | t4221 | 876 | araC family regulatory protein | |
Genes determined to be uniquely required in S. Typhimurium. SL, S. Typhimurium; Ty, S. Typhi; inserts refer to the number of unique insertion sites within a gene; reads refer to the number of sequence reads over all insertions sites within a gene. Shaded rows indicate genes shown to be H-NS repressed in (61).
*P-value (associated with log2 read ratio) < 0.05.
asseJ is a pseudogene in S. Typhi.
Table 3.
Genes uniquely required in S. Typhi
| SL inserts | SL reads | Ty inserts | Ty reads | Ty ID | Ty gene length | SL ID | SL gene length | Name | Function |
|---|---|---|---|---|---|---|---|---|---|
| No orthologue in S. Tm | |||||||||
| 1 | 5 | t1332 | 132 | malY | pseudogene | ||||
| 2 | 32 | t1920 | 405 | possible repressor protein, prophage 10/Gifsy-2 | |||||
| 2 | 12 | t3157 | 165 | conserved hypothetical protein | |||||
| 2 | 12 | t3166 | 228 | spurious ORF annotation overlapping the RnaseP/M1 RNA | |||||
| 6 | 196 | t3402 | 570 | cI | repressor protein, cs 73 prophage | ||||
| 4 | 58 | t3415 | 741 | HIRAN-domain family gene, potential DNA repair | |||||
| 1 | 6 | t4531 | 150 | hypothetical secreted protein | |||||
| Present in S. Typhimurium but required only in S. Typhi* | |||||||||
| 199 | 1792 | 18 | 59 | t0095 | 1287 | SL0093 | 1287 | surA | survival protein SurA precursor |
| 45 | 498 | 3 | 22 | t0123 | 459 | SL0119 | 459 | yabB/mraZ | conserved hypothetical protein |
| 120 | 589 | 11 | 32 | t0203 | 1281 | SL0203 | 1281 | hemL | glutamate-1-semialdehyde 2,1-aminomutase |
| 123 | 982 | 2 | 25 | t0224 | 1353 | SL0224 | 1353 | yaeL/rseP | Zinc metallopeptidase |
| 67 | 452 | 1 | 14 | t0270 | 576 | SL2604 | 576 | rpoE | RNA polymerase sigma-E factor |
| 140 | 760 | 0 | 0 | t0587 | 2286 | SL2246 | 2286 | nrdA | ribonucleoside-diphosphate reductase 1 alpha chain |
| 113 | 641 | 15 | 42 | t2140 | 2802 | SL0718 | 2802 | sucA | 2-oxoglutarate dehydrogenase E1 component |
| 116 | 753 | 13 | 36 | t2177 | 1641 | SL0680 | 1641 | pgm | phosphoglucomutase |
| 80 | 542 | 9 | 15 | t2276 | 1008 | SL0580 | 1008 | fepD | ferric enterobactin transport protein FepD |
| 93 | 591 | 2 | 2 | t2277 | 990 | SL0579 | 990 | fepG | ferric enterobactin transport protein FepG |
| 64 | 508 | 5 | 6 | t2278 | 795 | SL0578 | 795 | fepC | ferric enterobactin transport ATP-binding protein FepC |
| 201 | 1129 | 12 | 116 | t2410 | 2355 | SL0444 | 2355 | lon | Lon protease |
| 95 | 518 | 8 | 20 | t2730 | 1062 | SL2809 | 1062 | recAa | recA protein |
| 135 | 719 | 16 | 39 | t2996 | 1992 | SL3052 | 1947 | tktA | transketolase |
| 76 | 358 | 3 | 9 | t3120 | 1434 | SL3173 | 1434 | rfaE | ADP-heptose synthase |
| 213 | 1976 | 6 | 50 | t3265 | 1071 | SL3321 | 1071 | degS | serine protease |
| 43 | 448 | 3 | 10 | t3326 | 606 | SL3925 | 606 | yigP | conserved hypothetical protein |
| 124 | 571 | 17 | 36 | t3384 | 2025 | SL3872 | 2025 | rep | ATP-dependent DNA helicase |
| 175 | 1208 | 6 | 21 | t3621 | 2787 | SL3947 | 2787 | polA | DNA polymerase I |
| 117 | 797 | 9 | 13 | t3808 | 1047 | SL3677 | 1047 | waaF | ADP-heptose-LPS heptosyltransferase II |
| 176 | 1628 | 14 | 59 | t4153 | 1080 | SL4183 | 1080 | alr | alanine racemase |
| 140 | 1127 | 10 | 38 | t4411 | 951 | SL4294 | 951 | miaA | tRNA delta-2-isopentenylpyrophosphate transferase |
Genes determined to be uniquely required in S. Typhi. SL, S. Typhimurium; Ty, S. Typhi; inserts refer to the number of unique insertion sites within a gene; reads refer to the number of sequence reads over all insertions sites within a gene.
*P-value (associated with log2 read ratio) < 0.05.
aThe assignment of recA as a required gene has been described previously (35), but briefly is believed to be due to the presence of the priC pseudogene in Typhi.
To compare differences between requirements for orthologous genes in both serovars, we calculated log-fold read ratios to eliminate genes, which were classified differently in S. Typhi and S. Typhimurium but did not have significantly different read densities (see ‘Materials and Methods’ section.) Even after this correction, 36 S. Typhimurium genes had a significantly lower frequency of transposon insertion compared with the equivalent genes in S. Typhi (P < 0.05), including four encoding hypothetical proteins (Table 2). This indicates that these gene products play a vital role in S. Typhimurium, but not in S. Typhi when grown under laboratory conditions.
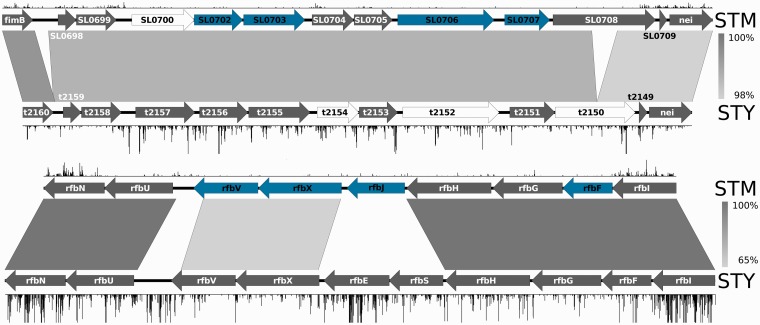
A major difference between the two serovars is in the requirement for genes involved in cell wall biosynthesis (see Figure 3). A set of four genes (SL0702, SL0703, SL0706 and SL0707) in an operonic structure putatively involved in cell wall biogenesis is required in S. Typhimurium, but not in S. Typhi. The protein encoded by SL0706 is a pseudogene in S. Typhi (Ty2 unique ID: t2152) owing to a 1 bp deletion at codon 62 that causes a frameshift (Figure 4a). This operon contains an additional two pseudogenes in S. Typhi (t2154 and t2150), as well as a single different pseudogene (SL0700) in S. Typhimurium, indicating that this difference in gene requirements reflects the evolutionary adaptation of these serovars to their respective niches. Similarly, four genes (rfbV, rfbX, rfbJ and rfbF) within an O-antigen biosynthetic operon are required by S. Typhimurium, but not S. Typhi. There appears to have been a shuffling of O-antigen biosynthetic genes since the divergence between the two serovars, and rfbJ, encoding a CDP-abequose synthase, has been lost from S. Typhi altogether. These broader requirements for cell wall-associated biosynthetic and transporter genes suggest that surface structure biogenesis is of greater importance in S. Typhimurium.
Figure 3.
Comparison of cell surface operon structure and requirements. Diagram illustrating cell surface operons with different requirement patterns in S. Typhimurium and S. Typhi. The top figure is of an uncharacterized operon putatively involved in cell wall biogenesis, whereas the bottom figure shows a portion of the rfb operon involved in O-antigen biosynthesis. Plots along the top and bottom of each figure show insertions in S. Typhimurium and S. Typhi, respectively, with read depth on the y-axis with a maximum cut-off of 100 reads. Genes in blue are required in S. Typhimurium, genes in white are pseudogenes; others are in grey. Grey rectangles represent BLAST hits between orthologous genes, with percentage nucleotide identity coloured on the scale to the right of each figure.
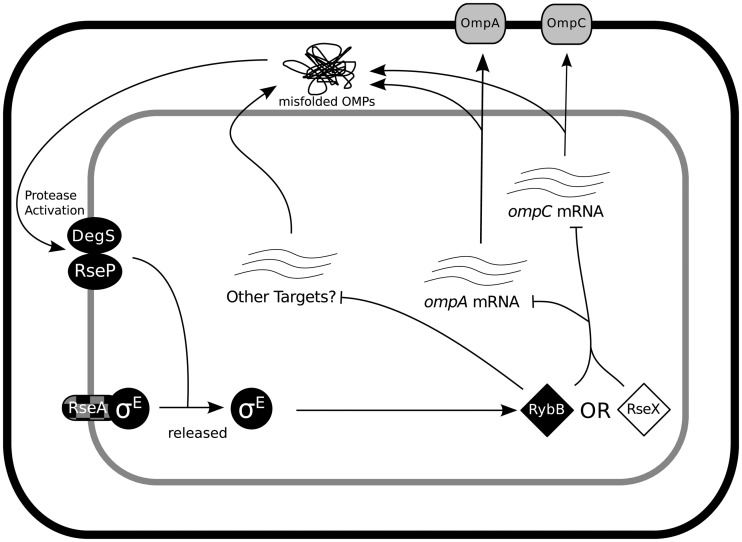
Figure 4.
Proposed differences in sRNA utilization. Diagram illustrating inferred required sRNA regulatory networks under TraDIS. Molecules required in S. Typhi are in black and in S. Typhimurium are in white. RseA, in black/grey check, is ambiguous in S. Typhi. Non-required molecules are in grey. Diamonds indicate sRNAs, circles regulatory proteins, ovals proteases, oblong shapes are membrane-anchored proteins, and rounded squares are outer membrane porins.
We also identified seven genes from the shared pathogenicity island SPI-2 that appear to contain few or no transposon insertions only in S. Typhimurium under laboratory conditions. These genes (spiC, sseA and ssaHIJT) are thought to encode components of the SPI-2 type III secretion system apparatus (T3SS) (63). In addition, the effector genes sseJ and sifB, whose products are secreted through the SPI-2-encoded T3SS (64,65), also fell into the ‘required’ category in S. Typhimurium alone. All of these genes display high A + T nucleotide sequence and have been previously shown (in S. Typhimurium) to be strongly bound by the nucleoid-associated protein H-NS, encoded by hns (61,66). Therefore, rather than being ‘required’, it is instead possible that access for the transposon was sufficiently restricted that very few insertions occurred at these sites. In further support of this hypothesis, a comparison of the binding pattern of H-NS detected in studies using S. Typhimurium LT2 with the TraDIS results from the SPI-2 locus indicated that high regions of H-NS enrichment correlated well with both the ssa genes described here and with sseJ (61,66) (see Supplementary Figure S1). An earlier study also suggests that high-density DNA-binding proteins can block Mu, Tn5 and Tn10 insertion (67); however, a genome-wide study of the effects of H-NS binding on transposition would be necessary to confirm this effect.
Indeed, the generation of null S. Typhimurium mutants in sseJ and sifB, as well as many others generated at the SPI-2 locus suggest that these genes are not truly a requirement for growth in this serovar (65,68–70). Although this is a reminder that the interpretation of gene requirement needs to be made with care, the effect of H-NS on transposon insertion is not genome-wide. If this were the case, there would be an under-representation of transposon mutants in high A + T regions (known for H-NS binding), which is not what we observed. In total, only 21 required genes fall into the ‘hns-repressed’ category described in Navarre et al. (61) (see Table 2; Supplementary Table S1); the remainder (almost 400) contained sufficient transposon insertions to conclude they were non-required. In addition, all SPI-1 genes that encode another T3SS and are of high A + T content were also found to be non-required. This phenomenon was not observed in S. Typhi, possibly because the strain used harbours the pHCM1 plasmid, which encodes the H-NS-like protein sfh and has been shown to affect H-NS binding (71,72).
Twenty-two S. Typhi genes had a significantly lower frequency of transposon insertion compared with orthologues in S. Typhimurium (P < 0.05), indicating that they are required only in S. Typhi for growth under laboratory conditions (Table 3), including the fepBDGC operon. This indicates a requirement for ferric [Fe(III)] rather than ferrous [Fe(II)] iron. This can be explained by the presence of Fe(III) in the bloodstream, where S. Typhi can be found during typhoid fever (15). These genes function to recover the ferric chelator enterobactin from the periplasm, acting with a number of proteins known to aid the passage of this siderophore through the outer membrane (73). It has long been noted that aroA mutants of S. Typhi, deficient in their ability to synthesize enterobactin, exhibit severe growth defects on complex media, whereas similar mutants of S. Typhimurium grow normally under the same conditions (74), though the mechanism has not been clear. Our results suggest that this difference in growth of aroA mutants is caused by a requirement for iron uptake through the fep system in S. Typhi. During host adaptation, S. Typhi has accumulated pseudogenes in many iron transport and response systems (27), presumably because they are not necessary for survival in the niche S. Typhi occupies in the human host, which may have led to this dependence on fep genes. In contrast, S. Typhimurium generally causes intestinal rather than systemic infection and is able to use a wider range of iron sources, including Fe(II), a soluble form of iron present under anaerobic conditions such as those found in the intestine (75).
TraDIS provides resolution sufficient to evaluate ncRNA contributions to fitness
Under a Poisson approximation to the transposon insertion process, a region of 41 (in S. Typhimurium) or 60 bases (in S. Typhi) has only a 1% probability of not containing an insertion by chance. NcRNAs tend to be considerably shorter than their protein-coding counterparts, but this gives us sufficient resolution to assay most of the non-coding complement of the Salmonella genome. As a proof of principle, we performed an analysis of the best-understood class of small ncRNAs, the tRNAs. Francis Crick hypothesized that a single tRNA could recognize more than one codon through wobble recognition (76), where a non-canonical G-U base pair is formed between the first (wobble) position of the anticodon and the third nucleotide in the codon. As a result, some codons are covered by multiple tRNAs, whereas others are covered non-redundantly by a single tRNA. We expect that singleton wobble-capable tRNAs, i.e. wobble tRNAs which recognize a codon uniquely, will be required. In addition, we inferred the requirement for other tRNAs through the non-redundant coverage of their codons and used this to benchmark our ability to use TraDIS to reliably interrogate short genomic intervals.
The S. Typhi and S. Typhimurium genomes encode 78 and 85 (plus one pseudogene) tRNAs, respectively, with 40 anticodons, as identified by tRNAscan-SE (77). In S. Typhi, 10 of 11 singleton wobble tRNAs are predicted to be required or ambiguous, compared with 16 tRNAs below the ambiguous LLR cut-off overall (significant enrichment at the 0.05 level, two-tailed Fisher’s exact test P-value: 6.4e-08). Similarly in S. Typhimurium, 9 of 11 singleton wobble tRNAs are required or ambiguous compared with 15 required or ambiguous tRNAs overall, again showing a significant enrichment of required tRNAs in this subset (Fisher’s exact test P-value: 5.2e-07). The one singleton wobble tRNA, which is consistently not required in both serovars is the tRNA-Pro(GGG), which occurs within a four-member codon family. It has previously been shown in S. Typhimurium that tRNA-Pro(UGG) can read all four proline codons in vivo owing to a cmo5U34 modification to the anticodon, obviating the need for a functional tRNA-Pro(GGG) (78) and making this tRNA non-required. The other non-required singleton wobble tRNA in S. Typhimurium, tRNA-Leu(GAG), is similarly a member of a four-member codon family. We predict tRNA-Leu(TAG) may also be capable of recognizing all four leucine codons in this serovar; such a leucine ‘four-way wobble’ has been previously inferred in at least one other bacterial species (79,80).
Of the six required non-wobble tRNAs in each serovar, four are shared. These include two non-wobble singleton tRNAs covering codons uniquely, as well as a tRNA with the ATG anticodon, which is post-transcriptionally modified by the required protein mesJ/tilS to recognize the isoleucine codon ATA (80). An additional two required tRNAs in both serovars, one shared and one with a differing anticodon, contain Gln anticodons and are part of a polycistronic tRNA operon containing other required tRNAs. This operon is conserved in E. coli with the exception of an additional tRNA-Gln at the 3′ end that has been lost in the Salmonella lineage. It is possible that transposon insertions early in the operon may interfere with processing of the polycistronic transcript in to mature tRNAs. Finally, we do not observe insertions in a tRNA-Met and a tRNA-Val in S. Typhi and S. Typhimurium, respectively.
Using this analysis of the tRNAs, we estimate a worst-case PPV for these short molecules (∼76 bases) at 81%, in line with our previous estimates for conserved protein-coding genes, and a FPR of <4%, higher than for protein-coding genes but still well within the typical tolerance of high-throughput experiments. This assumes that the ‘required’ operonic tRNA-Glns and the serovar-specific tRNA-Met and tRNA-Val are all false positives; it is not clear that this is in fact the case.
Surveying the shared required ncRNA content of both serovars (see Table 4), we find that the RNA components of the signal recognition particle (SRP) and RNaseP, two universally conserved ncRNAs, are required as expected. The SRP is an essential component of the cellular secretion machinery, whereas RnaseP is necessary for the maturation of tRNAs. We also find a number of required known and potential cis-regulatory molecules associated with genes required for growth under laboratory conditions in both serovars. The RFN riboswitch controls ribB, a 3,4-dihydroxy-2-butanone 4-phosphate synthase involved in riboflavin biosynthesis, in response to flavin mononucleotide concentrations (83). Additionally, we are able to assign putative functions to a number of previously uncharacterized required non-coding transcripts through their 5′ association with required genes. SroE, a 90 nt molecule discovered in an early sRNA screen (84), is consistently located at the 5′ end of the required hisS gene across its phylogenic distribution in the Enterobacteriaceae. Given this consistent association and the function of HisS as a histidyl-tRNA synthetase, we hypothesize that this region may act in a manner similar to a T-box leader, inducing or repressing expression in response to tRNA-His levels. The thrU leader sequence, recently discovered in a deep-sequencing screen of E. coli (42), appears to regulate a polycistronic operon of required singleton wobble tRNAs. Three additional required cis-regulatory elements, t44, S15 and StyR-8, are associated with required ribosomal proteins, highlighting the central role ncRNA elements play in regulating fundamental cellular processes.
Table 4.
Candidate required ncRNAs greater than 60 nt in length, excluding rRNA and tRNA
| Element name | Rfam accession | Function | Hfq-binding | Downstream protein-coding gene(s) | Downstream gene required | References |
|---|---|---|---|---|---|---|
| Required or ambiguous in both S. Typhi and S. Typhimurium | ||||||
| SRP | RF00169 | RNA component of the signal recognition particle | (81) | |||
| RNase P | RF00010 | RNA component of RNase P | ybaZ | N | (82) | |
| RFN | RF00050 | FMN-sensing riboswitch controlling the ribB gene | ribB | Y | (83) | |
| SroE | RF00371 | Putative cis-regulatory element controlling the hisS gene | hisS | Y | (84) | |
| ThrU Leader | NA | Putative cis-regulatory element controlling the ThrU tRNA operon | (42) | |||
| t44 | RF00127 | Cis-regulatory element controlling the ribosomal rpsB gene | rpsB | Y | (85-87) | |
| S15b | RF00114 | Translational regulator of the ribosomal S15 protein | rpsO | Y | (88) | |
| StyR-8 | NA | Putative cis-regulatory element controlling the ribosomal rpmB gene | rpmB | Y | (41) | |
| MicA | RF00078 | sRNA involved in cellular response to extracytoplasmic stress | Y | luxS | N | (29) |
| DsrAb | RF00014 | sRNA regulator of H-NS | Y | mngB | N | (89) |
| STnc10 | NA | Putative sRNA | nhaA | N | (40) | |
| STnc60b | NA | Putative sRNA | (40) | |||
| STnc840 | NA | Verified sRNA derived from 3' UTR of the flgL gene | Y | (90) | ||
| IS0420a,b | NA | Putative ncRNA | rmf | N | (42,91) | |
| RGO0b | NA | Putative sRNA identified in E. coli | (42) | |||
| Required or ambiguous in S. Typhimurium only | ||||||
| rne5 | RF00040 | RNase E autoregulatory 5' element | rne | Y | (92) | |
| RydC | RF00505 | sRNA regulator of the yejABEF ABC transporter | Y | (93) | ||
| RydB | RF00118 | Putative ncRNA | (94) | |||
| STnc510 | NA | Putative sRNA | pagD/pagC | Y/N | (40) | |
| STnc460b | NA | Putative sRNA | (40) | |||
| STnc170 | NA | Putative sRNA | SL1458 | N | (40) | |
| STnc130 | NA | Putative sRNA | dmsA | N | (40) | |
| RseX | RF01401 | sRNA regulator of OmpA and OmpC | Y | (95) | ||
| IsrJ | RF01393 | sRNA regulator of SPI-1 effector protein secretion | (40,58) | |||
| IsrI | RF01392 | Island-encoded Hfq-binding sRNA | Y | SL1028 | Y | (40,58,90) |
| Required or ambiguous in S. Typhi only | ||||||
| RybB | RF00110 | sRNA involved in cellular response to extracytoplasmic stress | Y | (29) | ||
| tk5a | NA | Putative ncRNA | (42,96) | |||
| STnc750 | NA | Verified sRNA | Y | SpeB | N | (32,90) |
| StyR-44a | RF01830 | Putative multicopy (2/6 copies required in S. Typhi) ncRNA associated with ribosomal RNA operon | 23S rRNA | N | (41) | |
| tp2 | NA | Putative ncRNA | aceE | N | (42,96) | |
| RdlD | RF01813 | RdlD RNA anti-toxin, 1/2 copies required in S. Typhi | (97) | |||
| STnc120a | NA | Putative sRNA | (40) | |||
| tp28a | NA | Putative ncRNA | fur | N | (42,96) | |
| Phe Leadera | RF01859 | Phenylalanine peptide leader sequence associated with the required PheST operon | PheS | Y | (98) | |
| RimP Leader | RF01770 | Putative cis-regulator of the rimP-nusA-infB operon | rimP | Y | (99) | |
| GlmY | RF00128 | Trans-acting regulator of the GlmS gene | (100) | |||
Known and putative non-coding elements classified as required or ambiguous in this screen. Required ncRNAs have an LLR between required and non-required models of less than −2; see ‘Materials and Methods’ section.
a,bncRNAs that are amibiguous (LLR between −2 and 2) in S. Typhi(a) or in S. Typhimurium(b). Hfq-binding annotations are taken from (90). The downstream protein-coding genes columns report annotated CDS or ribosomal RNA start sites within 100 bases of each candidate required non-coding element on either strand and whether these downstream sequences are also classified as required.
The sRNAs required for competitive growth
Inferring functions for potential trans-acting ncRNA molecules, such as anti-sense binding sRNAs, from requirement patterns alone is more difficult than for cis-acting elements, as we cannot rely on adjacent genes to provide any information. It is also important to keep in mind that TraDIS assays requirements after a brief competition within a large library of mutants on permissive media. This may be particularly important when surveying the bacterial sRNAs, which are known to participate in responses to stress (29).
This is demonstrated by two sRNAs involved in the σE-mediated extracytoplamic stress response, RybB and RseX, both of which can be successfully knocked out in S. Typhimurium (101). In S. Typhi, rpoE is required, as it also is in E. coli (48,102). However, in S. Typhimurium, rpoE tolerates a heavy insertion load, implying that σE mutants are not disadvantaged in competitive growth. In S. Typhimurium, the sRNA RseX is required. Overexpression of RseX has previously been shown to compensate for σE essentiality in E. coli by degrading ompA and ompC transcripts (95). This suggests that RseX may also be short-circuiting the σE stress response network in S. Typhimurium (Figure 4). To our knowledge, this is the first evidence of a native (i.e. not experimentally induced) activity of RseX.
S. Typhi on the other hand requires σE along with its activating proteases RseP and DegS and anchoring protein RseA, as well as the σE-dependent sRNA RybB, which also regulates OmpA and OmpC in S. Typhimurium, along with a host of other OMPs (103). It is unclear why the σE response is required in S. Typhi, but not S. Typhimurium, though it may partially be due to the major differences in the cell wall and outer membrane between the two serovars. In addition, there are significant differences in the OMP content of the S. Typhi and S. Typhimurium membranes that may be driving alternative mechanisms for coping with membrane stress. For instance, S. Typhi completely lacks OmpD, a major component of the S. Typhimurium outer membrane (104) and a known target of RybB (29).
Two additional sRNAs involved in stress response are also required by both S. Typhi and S. Typhimurium. The first, MicA, is known to regulate ompA and the lamB porin-coding gene in S. Typhimurium (105), contributing to the extracytoplasmic stress response. The second, DsrA, has been shown to negatively regulate the nucleoid-forming protein H-NS and enhance translation of the stationary-phase alternative sigma factor σS in E. coli (89), though its regulation of σS does not appear to be conserved in S. Typhimurium (106). Both have been previously deleted in S. Typhimurium and thus are not essential. H-NS knockouts have previously been shown to have severe growth defects in S. Typhimurium that can be rescued by compensatory mutations in either the phoPQ two-component system or rpoS, implying that the lack of H-NS is allowing normally silenced detrimental regions to be transcribed (61). As MicA has recently been shown to negatively regulate phoPQ expression in E. coli (107), it is tempting to speculate that MicA may be moderating the effects of DsrA-induced H-NS repression; however, it is currently unclear whether sRNA regulons are sufficiently conserved between E. coli and S. enterica to justify this hypothesis.
CONCLUSION
The extremely high resolution of TraDIS has allowed us to assay gene requirements in two very closely related Salmonellae with different host ranges. We found, under laboratory conditions, that 58 genes present in both serovars were required in only one, suggesting that identical gene products do not necessarily have the same phenotypic effects in the two different serovar backgrounds. Many of these genes occur in genomic regions or metabolic systems, which contain pseudogenes and/or have undergone reorganization since the divergence of S. Typhi and S. Typhimurium, demonstrating the complementarity of TraDIS and phylogenetic analysis. These changes may, in part, explain differences observed in the pathogenicity and host specificity of these two serovars. In particular, S. Typhimurium showed a requirement for cell surface structure biosynthesis genes; this may be partially explained by the fact that S. Typhi expresses the Vi-antigen, which masks the cell surface, though these genes are not required for survival in our assay. S. Typhi on the other hand has a requirement for iron uptake through the fep system, which enables ferric enterobactin transport. This dependence on enterobactin suggests that S. Typhi is highly adapted to the iron-scarce environments it encounters during systemic infections. Furthermore, this appears to represent a single point of failure in the S. Typhi iron utilization pathways and may present an attractive target for narrow-spectrum antibiotics.
Of the ∼4500 protein-coding genes present in each serovar, only ∼350 were sufficiently depleted in transposon insertions to be classified as required for growth in rich media. This means that >92% of the coding genome has sufficient insertion density to be queried in future assays. Dense transposon mutagenesis libraries have been used to assay gene requirements under conditions relevant for infection, including S. Typhi survival in bile (35), Mycobacterium tuberculosis catabolism of cholesterol (108), drug resistance in Pseudomonas aeruginosa (109) and Haemophilus influenzae survival in the lung (110). We expect that parallel experiments querying gene requirements under the same conditions in both serovars examined in this study will yield further insights in to the differences in the infective process between Typhi and Typhimurium and ultimately the pathways that underlie host-adaptation.
Both serovars possess substantial complements of horizontally acquired DNA. We have been able to use TraDIS to assay these recently acquired sequences. In particular, we have been able to identify, on a chromosome wide scale, active prophage through the requirement for their repressors. The P4 phage uses an RNA-based system to make decisions regarding cell fate, and structurally similar systems are used by P1, P7 and N15 phage (111,112). C4-like transcripts have been regarded as the primary repressor of lytic functions, though the IsrK-like sequence is known to be essential to the establishment of lysogeny in P4 and is transcribed in at least two phage types (60,112). Our observations in S. Typhi suggest an important role for the IsrK-like sequence in maintenance of the lysogenic state in P4-like phage, though the mechanism remains unclear.
Recent advances in high-throughput sequencing have greatly enhanced our ability to detect novel transcripts, such as ncRNAs and short open reading frames (sORFs). Our ability to identify these transcripts now far out-strips our ability to experimentally characterize these sequences. There have been previous efforts at high-throughput characterization of bacterial sRNAs and sORFs in enteric bacteria; however, these have relied on labour-intensive directed knockout libraries (47,113). Here, we have demonstrated that TraDIS has sufficient resolution to reliably query genomic regions as short as 60 bases, in agreement with a recent high-throughput transposon mutagenesis study in the alphaproteobacteria Caulobacter crescentus (38). Our method has the major advantage that library construction does not rely on genome annotation, and newly discovered elements can be surveyed with no further laboratory work.
We have been able to assign putative functions to a number of ncRNAs using TraDIS though consideration of their genomic and experimental context. In addition, ncRNA characterization generally is done in model organisms like E. coli or S. Typhimurium, and it is unclear how stable ncRNA regulatory networks are over evolutionary time. By assaying two serovars of Salmonella with the same method under the same conditions, we have seen hints that there may be differences in sRNA regulatory networks between S. Typhi and S. Typhimurium. In particular, we have found that under the same experimental conditions, S. Typhi appears to rely on the σE stress response pathway, whereas S. Typhimurium does not; it is tempting to speculate that this difference in stress response is mediated by the observed difference in requirement for two sRNAs, RybB and RseX. We believe that this combination of high-throughput transposon mutagenesis with a careful consideration of the systems context of individual genes provides a powerful tool for the generation of functional hypotheses. We anticipate that the construction of TraDIS libraries in additional organisms, as well as the passing of these libraries through relevant experimental conditions, will provide further insights into the function of bacterial ncRNAs in addition to the protein-coding gene complement.
ACCESSION NUMBERS
ERA000097, ERA000217.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–2 and Supplementary Data Set.
FUNDING
Wellcome Trust [WT076964, WT079643 and WT098051]; Medical Research Council. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Leopold Parts for assistance with the statistical analysis of required genes under laboratory conditions, Derek Pickard for discussions of phage biology and Amy Cain for comments on the manuscript. The S. Typhimurium nucleotide sequencing data have been deposited in the European Short Read Archive under accession number ERA000217. The S. Typhi data can be accessed at ERA000097.
REFERENCES
- 1.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 2.Bhutta ZA, Threlfall J. Addressing the global disease burden of typhoid fever. JAMA. 2009;302:898–899. doi: 10.1001/jama.2009.1259. [DOI] [PubMed] [Google Scholar]
- 3.Kothari A, Pruthi A, Chugh TD. The burden of enteric fever. J. Infect. Dev. Ctries. 2008;2:253–259. doi: 10.3855/jidc.218. [DOI] [PubMed] [Google Scholar]
- 4.Soper GA. The curious career of typhoid mary. Bull N.Y. Acad. Med. 1939;15:698–712. [PMC free article] [PubMed] [Google Scholar]
- 5. CDC. (2009) Salmonella surveillance: Annual summary.
- 6.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 7.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 8.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 9.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 10.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Liou SR, Plunkett G, 3rd, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulig PA, Curtiss R., 3rd Plasmid-associated virulence of Salmonella Typhimurium. Infect. Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulig PA, Doyle TJ. The Salmonella Typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan MD, Kidgell C, Nair S, Holt KE, Turner AK, Hinds J, Butcher P, Cooke FJ, Thomson NR, Titball R, et al. Variation in Salmonella enterica serovar Typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob. Agents Chemother. 2009;53:716–727. doi: 10.1128/AAC.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wain J, Diep TS, Ho VA, Walsh AM, Nguyen TT, Parry CM, White NJ. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J. Clin. Microbiol. 1998;36:1683–1687. doi: 10.1128/jcm.36.6.1683-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta N. Transmissible drug resistance in an epidemic strain of Salmonella Typhimurium. J. Hyg. (Lond.) 1962;60:301–310. doi: 10.1017/s0022172400020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt KE, Thomson NR, Wain J, Phan MD, Nair S, Hasan R, Bhutta ZA, Quail MA, Norbertczak H, Walker D, et al. Multidrug-resistant Salmonella enterica serovar Paratyphi A harbors IncHI1 plasmids similar to those found in serovar Typhi. J. Bacteriol. 2007;189:4257–4264. doi: 10.1128/JB.00232-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cain AK, Hall RM. Evolution of a multiple antibiotic resistance region in IncHI1 plasmids: reshaping resistance regions in situ. J. Antimicrob. Chemother. 2012;67:2848–2853. doi: 10.1093/jac/dks317. [DOI] [PubMed] [Google Scholar]
- 19.Darwin KH, Miller VL. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl Acad. Sci. USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella Typhimurium. Proc. Natl Acad. Sci. USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickard D, Wain J, Baker S, Line A, Chohan S, Fookes M, Barron A, Gaora PO, Chabalgoity JA, Thanky N, et al. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 2003;185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seth-Smith HM. SPI-7: Salmonella's Vi-encoding Pathogenicity Island. J. Infect. Dev. Ctries. 2008;2:267–271. doi: 10.3855/jidc.220. [DOI] [PubMed] [Google Scholar]
- 24.Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, et al. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18:1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt KE, Thomson NR, Wain J, Langridge GC, Hasan R, Bhutta ZA, Quail MA, Norbertczak H, Walker D, Simmonds M, et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics. 2009;10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 28.Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Baumler AJ. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 30.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007;65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 31.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 32.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl Acad. Sci. USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebrard M, Kroger C, Srikumar S, Colgan A, Handler K, Hinton J. sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:437–445. doi: 10.4161/rna.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter AS, Backofen R. Accessibility and conservation: General features of bacterial small RNA-mRNA interactions? RNA Biol. 2012;9:954–965. doi: 10.4161/rna.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 41.Chinni SV, Raabe CA, Zakaria R, Randau G, Hoe CH, Zemann A, Brosius J, Tang TH, Rozhdestvensky TS. Experimental identification and characterization of 97 novel npcRNA candidates in Salmonella enterica serovar Typhi. Nucleic Acids Res. 2010;38:5893–5908. doi: 10.1093/nar/gkq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 47.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009;5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong X, Campbell JW, Balazsi G, Kay KA, Wanner BL, Gerdes SY, Oltvai ZN. Genome-scale identification of conditionally essential genes in E. coli by DNA microarrays. Biochem. Biophys. Res. Commun. 2004;322:347–354. doi: 10.1016/j.bbrc.2004.07.110. [DOI] [PubMed] [Google Scholar]
- 51.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 52.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Echols H, Green L. Establishment and maintenance of repression by bacteriophage lambda: the role of the cI, cII, and c3 proteins. Proc. Natl Acad. Sci. USA. 1971;68:2190–2194. doi: 10.1073/pnas.68.9.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson N, Baker S, Pickard D, Fookes M, Anjum M, Hamlin N, Wain J, House D, Bhutta Z, Chan K, et al. The role of prophage-like elements in the diversity of Salmonella enterica serovars. J. Mol. Biol. 2004;339:279–300. doi: 10.1016/j.jmb.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 55.Kropinski AM, Sulakvelidze A, Konczy P, Poppe C. Salmonella phages and prophages–genomics and practical aspects. Methods Mol. Biol. 2007;394:133–175. doi: 10.1007/978-1-59745-512-1_9. [DOI] [PubMed] [Google Scholar]
- 56.Lemire S, Figueroa-Bossi N, Bossi L. Bacteriophage crosstalk: coordination of prophage induction by trans-acting antirepressors. PLoS Genet. 2011;7:e1002149. doi: 10.1371/journal.pgen.1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briani F, Deho G, Forti F, Ghisotti D. The plasmid status of satellite bacteriophage P4. Plasmid. 2001;45:1–17. doi: 10.1006/plas.2000.1497. [DOI] [PubMed] [Google Scholar]
- 58.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella Typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forti F, Dragoni I, Briani F, Deho G, Ghisotti D. Characterization of the small antisense CI RNA that regulates bacteriophage P4 immunity. J. Mol. Biol. 2002;315:541–549. doi: 10.1006/jmbi.2001.5274. [DOI] [PubMed] [Google Scholar]
- 60.Sabbattini P, Forti F, Ghisotti D, Deho G. Control of transcription termination by an RNA factor in bacteriophage P4 immunity: identification of the target sites. J. Bacteriol. 1995;177:1425–1434. doi: 10.1128/jb.177.6.1425-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 62.Iyer LM, Babu MM, Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- 63.Kuhle V, Jackel D, Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–370. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 64.Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella Typhimurium. Proc. Natl Acad. Sci. USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freeman JA, Ohl ME, Miller SI. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 2003;71:418–427. doi: 10.1128/IAI.71.1.418-427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manna D, Porwollik S, McClelland M, Tan R, Higgins NP. Microarray analysis of Mu transposition in Salmonella enterica, serovar Typhimurium: transposon exclusion by high-density DNA binding proteins. Mol. Microbiol. 2007;66:315–328. doi: 10.1111/j.1365-2958.2007.05915.x. [DOI] [PubMed] [Google Scholar]
- 68.Hensel M, Shea JE, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden DW. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella Pathogenicity Island 2. Mol. Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 69.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 70.Ohlson MB, Fluhr K, Birmingham CL, Brumell JH, Miller SI. SseJ deacylase activity by Salmonella enterica serovar Typhimurium promotes virulence in mice. Infect. Immun. 2005;73:6249–6259. doi: 10.1128/IAI.73.10.6249-6259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 72.Dillon SC, Cameron AD, Hokamp K, Lucchini S, Hinton JC, Dorman CJ. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 2010;76:1250–1265. doi: 10.1111/j.1365-2958.2010.07173.x. [DOI] [PubMed] [Google Scholar]
- 73.Rabsch W, Voigt W, Reissbrodt R, Tsolis RM, Baumler AJ. Salmonella Typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 1999;181:3610–3612. doi: 10.1128/jb.181.11.3610-3612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edwards MF, Stocker BA. Construction of delta aroA his delta pur strains of Salmonella Typhi. J. Bacteriol. 1988;170:3991–3995. doi: 10.1128/jb.170.9.3991-3995.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella Typhimurium in the mouse. Infect. Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 77.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic. Acids. Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nasvall SJ, Chen P, Bjork GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- 82.Frank DN, Pace NR. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 83.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 84.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aseev LV, Levandovskaya AA, Tchufistova LS, Scaptsova NV, Boni IV. A new regulatory circuit in ribosomal protein operons: S2-mediated control of the rpsB-tsf expression in vivo. RNA. 2008;14:1882–1894. doi: 10.1261/rna.1099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer MM, Ames TD, Smith DP, Weinberg Z, Schwalbach MS, Giovannoni SJ, Breaker RR. Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique'. BMC Genomics. 2009;10:268. doi: 10.1186/1471-2164-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benard L, Philippe C, Ehresmann B, Ehresmann C, Portier C. Pseudoknot and translational control in the expression of the S15 ribosomal protein. Biochimie. 1996;78:568–576. doi: 10.1016/S0300-9084(96)80003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 92.Diwa A, Bricker AL, Jain C, Belasco JG. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 93.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 94.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 96.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 98.Zurawski G, Brown K, Killingly D, Yanofsky C. Nucleotide sequence of the leader region of the phenylalanine operon of Escherichia coli. Proc. Natl Acad. Sci. USA. 1978;75:4271–4275. doi: 10.1073/pnas.75.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naville M, Gautheret D. Premature terminator analysis sheds light on a hidden world of bacterial transcriptional attenuation. Genome Biol. 2010;11:R97. doi: 10.1186/gb-2010-11-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 102.De Las Penas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J. Bacteriol. 1997;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santiviago CA, Toro CS, Hidalgo AA, Youderian P, Mora GC. Global regulation of the Salmonella enterica serovar Typhimurium major porin, OmpD. J. Bacteriol. 2003;185:5901–5905. doi: 10.1128/JB.185.19.5901-5905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bossi L, Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 106.Jones AM, Goodwill A, Elliott T. Limited role for the DsrA and RprA regulatory RNAs in rpoS regulation in Salmonella enterica. J. Bacteriol. 2006;188:5077–5088. doi: 10.1128/JB.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010;76:467–479. doi: 10.1111/j.1365-2958.2010.07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2:e00315–e00310. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl Acad. Sci. USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Citron M, Schuster H. The c4 repressors of bacteriophages P1 and P7 are antisense RNAs. Cell. 1990;62:591–598. doi: 10.1016/0092-8674(90)90023-8. [DOI] [PubMed] [Google Scholar]
- 112.Ravin NV, Svarchevsky AN, Deho G. The anti-immunity system of phage-plasmid N15: identification of the antirepressor gene and its control by a small processed RNA. Mol. Microbiol. 1999;34:980–994. doi: 10.1046/j.1365-2958.1999.01658.x. [DOI] [PubMed] [Google Scholar]
- 113.Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J. Bacteriol. 2010;192:59–67. doi: 10.1128/JB.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.