Abstract
Fidelity and efficiency of pre-mRNA splicing are critical for generating functional mRNAs, but how such accuracy in 5′ splice site (SS) selection is attained is not fully clear. Through a series of yeast genetic screens, we isolated alleles of prp28 that improve splicing of suboptimal 5′SS substrates, demonstrating that WT-Prp28p proofreads, and consequently rejects, poor 5′SS. Prp28p is thought to facilitate the disruption of 5′SS–U1 snRNA pairing to allow for 5′SS–U6 snRNA pairing in the catalytic spliceosome; unexpectedly, 5′SS proofreading by Prp28p is dependent on competition with the stability of the 5′SS:U6 duplex, but not the 5′SS:U1 duplex. E404K, the strongest prp28 allele containing a mutation located in the linker region between adenosine triphosphatase (ATPase) subdomains, exhibited lower RNA-binding activity and enhanced splicing of suboptimal substrates before first-step catalysis, suggesting that decreased Prp28p activity allows longer time for suboptimal 5′SS substrates to pair with U6 snRNA and thereby reduces splicing fidelity. Residue E404 is critical for providing high splicing activity, demonstrated here in both yeast and Drosophila cells. Thus, the subdomain linker in Prp28p plays important roles both in splicing efficiency across species and in proofreading of 5′SS.
INTRODUCTION
Pre-mRNA splicing, catalyzed by the spliceosome, a large dynamic RNA–protein complex composed of five small nuclear ribonucleoproteins (snRNPs) and many protein factors, excises introns and ligates exons to generate mature mRNAs (1). Three conserved cis-elements in the intron, the 5′ splice site (5′SS), branch site (BS) region and 3′ splice site (3′SS), are critical for intron recognition, spliceosome assembly and splicing catalysis. The 5′SS is recognized by U1 snRNP, the 3′SS is bound by U2AFs and U2 snRNP binds to the BS region to form a stable pre-spliceosome. Subsequently, U4/U6–U5 tri-snRNP joins, and the active spliceosome forms after release of U1 and U4 snRNPs (2,3). Many RNA–RNA and RNA–protein interactions occur transiently and are disrupted during progression of the splicing pathway, most of which are facilitated by eight adenosine triphosphatases (ATPases) (4–6). For example, the early 5′SS–U1 snRNA interaction is exchanged for a 5′SS–U6 snRNA interaction before formation of spliceosomal complex B, facilitated by Prp28p (7,8). Likewise, U4–U6 snRNA interaction in the U4/U6–U5 tri-snRNP is replaced by U2–U6 snRNA interaction during formation of the ‘activated spliceosome’, facilitated by Brr2p (9–11).
Splicing fidelity is important for generating accurate mRNAs, and thereby functional proteins. The dynamic nature of spliceosome assembly and catalysis provides many opportunities for proofreading. Two non-mutually exclusive mechanisms have been proposed for altering fidelity, a two-state model and a kinetic proofreading model (12–14). The former was based on evidence of competition between spliceosomal conformations; such as, alleles of PRP8, U6 or CEF1 that inhibited first-step catalysis also improved second-step catalysis, and vice versa (15–18). The latter was based on the idea that ATPases function as timers to reject slowly progressing suboptimal substrates or as sensors of substrate identity to reject suboptimal substrates (3,14). For example, prp16 alleles with reduced ATPase activity improve splicing of poor first-step substrates, and prp22 alleles improve second-step catalysis on suboptimal substrates, presumably by allowing longer dwell time in the first and second-step conformations, respectively (15,19–24). Prp5p modulates BS fidelity by competing with the stability of the BS:U2 snRNA duplex (25,26); Prp43p promotes discard of intermediates and establishes a pathway for turnover of stalled intermediates at multiple stages (27). In addition, the specificity of the U2AF/3′SS interaction during early assembly can be increased by competition with DEK and hnRNP A1 binding in mammals (28,29).
Prp28p is a DEAD-box protein with nine conserved motifs (30,31). Genetic studies have demonstrated that Prp28p facilitates a destabilization of the 5′SS–U1 snRNP interaction, resulting in a switch of 5′SS:U1 pairing to 5′SS:U6 pairing (8,32). Mutations in U1 snRNP components, including U1C, Prp42p, Snu71p and U1 snRNA, can bypass the requirement of Prp28p (33–35). Mammalian Prp28 is a component of U5 snRNP (36), and its phosphorylation by SRPK2 is necessary for U4/U6–U5 tri-snRNP integration into the spliceosome (37). hPrp28 interacts directly with the 5′SS, detected by cross-links between motif III and positions +7 and +8 in the 5′SS (38). However, whether Prp28p is involved in substrate proofreading at the 5′SS is not yet known.
Here, we isolated prp28 alleles that improve splicing of suboptimal 5′SS substrates using yeast genetic screens. Alteration of 5′SS–U1 and 5′SS–U6 snRNA interactions demonstrated that 5′SS proofreading was sensitive to the stability of the 5′SS:U6 duplex, but not the 5′SS:U1 duplex. The strongest allele prp28-E404K has a reduced RNA-binding activity and improves splicing of 5′SS mutants at a stage before the first step of splicing. Furthermore, the ATPase subdomain linker sequence in Prp28p is phylogenetically variable; its replacements in both yeast and fly systems with heterologous linker sequences indicate that the yeast linker sequence and E404 are critical for cell growth and splicing efficiency. Thus, the Prp28p subdomain linker plays an important role in maintaining splicing activity across species, and mutations in this region have revealed a role in proofreading of 5′SS by Prp28p.
MATERIALS AND METHODS
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1. Plasmid-borne alleles of prp28, snr19 (U1 snRNA), snr6 (U6 snRNA), U1C and ACT1-CUP1 reporters were prepared by either in vivo gap repair cloning or traditional cloning using Escherichia coli.
Screening of prp28 alleles in S. cerevisiae
WT-PRP28 on pRS313(HIS) was digested by MfeI and transformed with a pool of error-prone polymerase chain reaction (PCR) products, covering nucleotides +525 to +1680 of PRP28, for gap repair in yYZX19 strain that contained various ACT1-CUP1 reporters. Transformants were replicated to 5-Fluoroorotic Acid (FOA) plates to lose the URA marked WT-PRP28 plasmid; then replicated to select for higher resistance to copper. Subsequent steps for identification and confirmation of prp28 alleles were carried out as described previously (26). Error-prone PCR was performed using buffer containing 0.5 mM MnCl2, 0.05 U/µl of Taq DNA polymerase and 400 pg/µl templates for eight cycles to introduce 7–10 mutants per 1000 bp (39). Products from error-prone PCR were gel purified and amplified further under standard conditions.
Copper assay and primer extension
Copper assays were carried out as described previously (26,40). Plates were scored by the maximum copper concentrations of strain growth and photographed after 4 days at 30°C or 10 days at 20°C. For simplicity, growth of each strain with a reporter on two or three representative copper concentrations was chosen to illustrate altered splicing activities by prp28 alleles. Primer extensions were performed as described previously (41), using primer YAC6 5′-GGCACTCATGACCTTC, complementary to exon 2 of the ACT-CUP1 reporter for detection of splicing products, and primer y5S 5′-ACAGTTGATCGGACGGG, complementary to 5S rRNA, for a loading control. Extension products were quantified by phosphorimaging and Multi Gauge V3.0.
Purification of recombinant ScPrp28 proteins and UV cross-linking
His6-tagged Prp28p and mutant proteins were expressed in E. coli, then subsequently purified by Ni-NTA agarose (QIAGEN), CHT-I column (Bio-rad) and Superdex D75 10/60 (GE). Proteins were stored in buffer D [20 mM HEPES–KOH (pH 7.9), 0.2 mM ethylenediaminetetraacetic acid, 100 mM KCl, 0.5 mM dithiothreitol, 1 mM Phenylmethylsulfonyl fluoride and 20% glycerol]. In vitro ATP-binding assays were performed as described previously (42). Gel-purified 32P-labeled [U]10 (RiboBio Co., Ltd) was used for RNA-binding assays. Purified Prp28p was incubated with 0.1 pmole of 32P-[U]10 (5 × 104 cpm) in 20 µl with 0.05 mM of ATP. Then UV cross-linking was performed as described previously (43) using 254-nm light for 20 min on ice, separated on 10% sodium dodecyl sulfate, visualized by autoradiography and quantified by Multi Gauge V3.0.
Construction of splicing reporter system in Drosophila S2 cell line
To generate expression plasmids of DmPrp28, fragments of hygromycin resistance gene with P copia promoter and simian vacuolating virus 40 poly(A) were cloned into pMT/V5-His B vector (Invitrogen), followed by insertion of DmPrp28 open reading frame with N-terminal 3× FLAG tag. Then, the plasmids were transfected into S2 cells and selected by 100 μg/ml of hygromycin to obtain stable cell lines that express similar levels of DmPrp28 proteins after induction with 0.5 mM CuSO4. Sk2-EGFP reporters were constructed by insertion of DNA fragments with Kozak sequence (CGAAATGGGC), truncated Drosophila Sk2 gene (21 nt of exon 1; 479 nt of intron 1 and 54 nt of exon 2) into pEGFP-N1 vector (CLONTECH), which is driven by the immediate early promoter of cytomegalovirus. Detection of EGFP fluorescence by fluorescence-activated cell sorting (FACS) (BD LSR II Flow Cytometer) was performed after transfection of Sk2-EGFP reporter and 2.5 days CuSO4 induction. To visualize the expressed proteins, western blots were probed using monoclonal M2–Peroxidase (Sigma) against FLAG, monoclonal antibody against GAPDH (ImB) and mAb 7G9 (Abmart) against GFP.
Prp28 linker region
The linker region of DEAD-box proteins was defined as the loop between the subdomain 1 and subdomain 2 of ATPase/helicase domains. Linkers of Dhh1 and eIF4A were extracted from published crystal structures, and the linker of Prp5 from our unpublished structure data. The linker sequences of Prp28 and Ded1 were acquired via comparison of their protein sequences with Prp5, Dhh1 and eIF4A using Basic Local Alignment Search Tool and confirmed by secondary structure predictions using software NetSurfP (ver. 1.1).
RESULTS
Genetic screens for prp28 alleles that improve splicing at suboptimal 5′SS
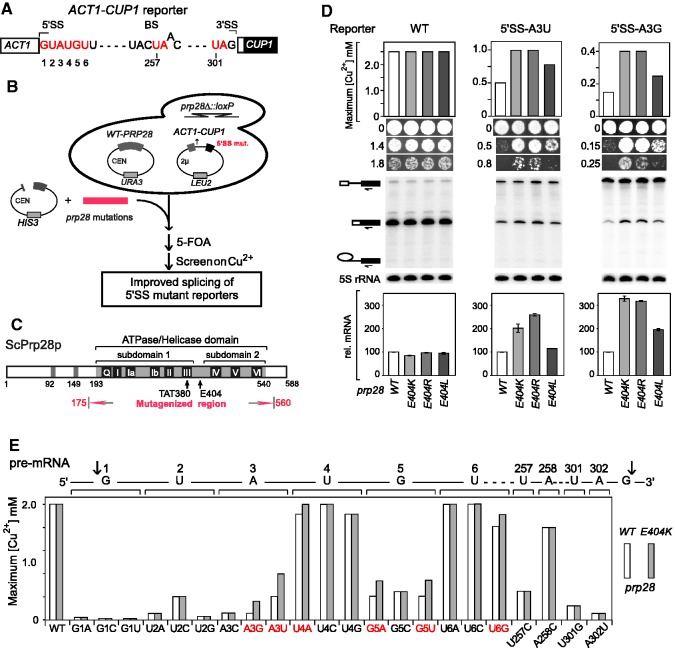
Based on the idea that most of the spliceosomal ATPases contribute to kinetic proofreading (44), we used a series genetic screens to identify alleles of prp28 that could improve splicing of suboptimal substrates. To identify substrates sensitive to altered Prp28p activity, we tested two mutants in motif III (TAT), which is thought to be required for coupling of ATP hydrolysis to conformational change: AAT and TAA. ACT1-CUP1 reporters, which confer copper tolerance to yeast lacking the chromosomal CUP1 genes, were used to monitor splicing efficiency in vivo (40). Sixteen reporters (Figure 1A), representing mutations at the 5′SS, BS and 3′SS, were tested for growth on copper in the presence of these two prp28 alleles. Subtle improvement by the two prp28 alleles was observed; however, subsequent screens using randomized motif III residues did not reveal stronger mutants. To identify alleles exhibiting more potent effects, we next mutated the entire ATPase domain (175–560 amino acids) using error-prone PCR; selection for improved growth on copper for 5′SS-A3G or A3U reporters identified several prp28 alleles (Figure 1B and C). The strongest allele, which improved A3U and A3G reporters >2-fold, contained four mutations: N178Y, M346V, L359F and E404K. To isolate the responsible residue(s) and obtain multiple prp28 mutants in that position, each of the four positions was separately randomized and reselected for improved growth for the 5′SS-A3U reporter. Only mutants at residue E404 yielded improved growth on copper, and these were comparable in potency with the originally isolated allele.
Figure 1.
Genetic screens for prp28 alleles that improve splicing of suboptimal 5′SS substrates. (A) Schematic of ACT1-CUP1 reporters used for monitoring splicing effects in vivo. Mutant sites used in this study are indicated in red. (B) Strategy of genetic screens for prp28 alleles that improve splicing of suboptimal 5′SS substrates. (C) Conserved motifs within ATPase domain of Prp28p and region of mutagenesis used. (D) Analysis of three prp28 alleles that enhanced splicing of 5′SS mutant reporters A3U and A3G. (Top) Improved splicing activities by prp28 alleles were determined by maximum copper concentration that cells could grow. Growth of each strain on three representative copper concentrations was also selected to illustrate altered splicing activities. (Middle) Analysis of in vivo RNA levels of ACT1-CUP1 by primer extension. Pre-mRNA, mRNA and lariat intermediate are indicated by icons on the left. 5S rRNA was analyzed in parallel as a loading control. (Lower) Quantitated mRNA levels from primer extension were normalized by the level of 5S rRNA and the mRNA of each reporter in the presence of WT-PRP28 allele. Error bars were calculated from two independent assays. (E) Copper assays indicated that the prp28-E404K allele specifically improved splicing of 5′SS mutants, but not BS or 3′SS mutant reporters. Improved reporters are shown in red.
Several E404 mutants were isolated and compared. prp28-E404K and -E404R alleles behaved similarly: both improved A3G and A3U growth on copper by 2- to 3-fold relative to WT-PRP28 and comparably increased A3G and A3U mRNA levels (Figure 1D). prp28-E404L, -E404V and -E404A alleles mildly improved A3G and A3U, whereas prp28-E404D mutant behaved exactly as WT (selected alleles are shown in Figure 1D). This pattern of effects suggests that disruption of the negative charge at position 404 is the key feature of this class of prp28 mutants. We chose the prp28-E404K allele for further analysis.
To investigate specificity of altered intron selection exhibited by prp28 alleles, we tested all possible 5′SS mutations at positions 1–6. The prp28-E404K allele strongly improved splicing of A3G, A3U, G5A and G5U substrates and slightly improved splicing of U4A and U6G substrates, but not of WT or other 5′SS mutant reporters nor of tested reporters having mutations within the BS region or 3′SS (Figure 1E). We conclude that our identified prp28 alleles alter substrate selectivity most strongly at +3 and +5 positions, although other 5′SS positions are also affected. As screens were performed on 5′SS mutants, the existence of prp28 alleles that improve splicing of BS or 3′SS mutants cannot be ruled out here.
Alteration of 5′SS–U1 snRNA interaction does not change the effect of prp28-E404K
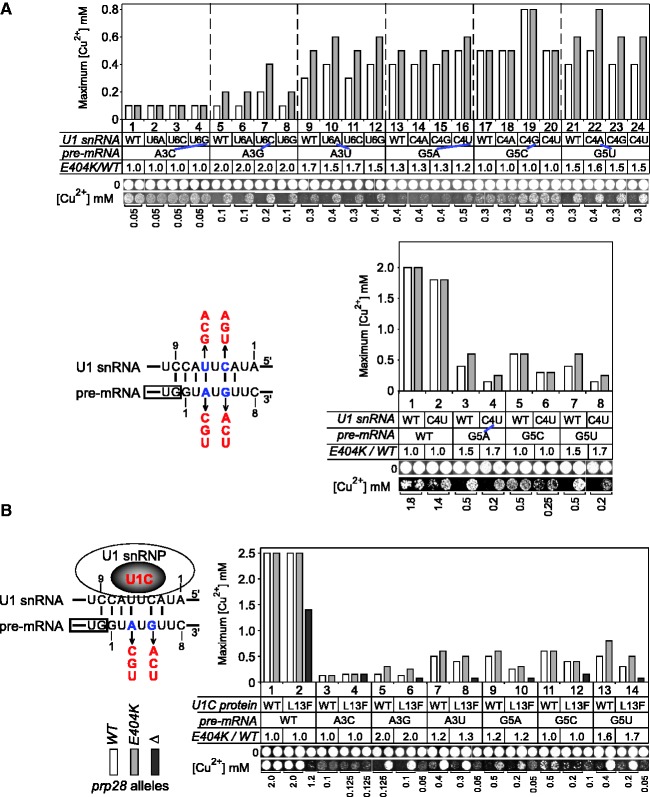
Prp28p has been proposed to dissociate the duplex formed between U1 snRNA and the 5′SS (Figure 2A), resulting in release of U1 snRNP and formation of 5′SS:U6 snRNA pairing (8,33). If the 5′SS:U1 duplex was important for altered substrate use because of prp28 alleles, then changing the stability of the duplex would be expected to alter the potency of prp28 effects. To test this, we increased duplex stability using U1 snRNA mutants and decreased stability using a U1C mutant.
Figure 2.
Alteration of 5′SS–U1 snRNA interaction does not change the effect of prp28 allele on splicing. (A) Compensatory changes in U1 snRNA do not alter the improved splicing of suboptimal 5′SS substrates by prp28 allele. Base pairs between 5′SS and U1 snRNA are indicated by lines. (Upper) Restored base pairing of 5′SS and U1 snRNA generally improved splicing efficiency, but it did not cancel or decrease the improved level of splicing by prp28-E404K. (Bottom) As the sole copy of U1 snRNA, the presence of the viable U1–C4U allele, which restored base pairing with 5′SS-G5A, did not cancel the improved splicing effects by prp28-E404K. (B) Destabilizing 5′SS–U1 snRNP interaction by mutation of U1C protein does not alter the improved splicing of suboptimal 5′SS substrates by prp28 allele. (Left) Schematic of U1C protein stabilizing 5′SS:U1 snRNA duplex. (Right) Mutant U1C–L13F slightly decreased the splicing efficiency of most substrates, but did not affect the enhanced splicing of suboptimal 5′SS substrates by prp28-E404K allele. The null prp28 allele significantly decreased splicing efficiency for most reporters in the presence of U1C–L13F. RNA mutations are indicated in red; base pairs at mutation sites are shown by blue lines. The level of improved splicing by prp28-E404K allele is calculated as E404K/WT.
We first generated strains carrying U1 snRNA mutants along with a second plasmid carrying WT U1 snRNA because of the lethality of most U1 snRNA mutants as the sole copy (33,45,46). Compensatory mutations in U1 snRNA that restore pairing with 5′SS mutants improved their splicing (Figure 2A, col. 7, 10, 16, 19 and 22), whereas non-cognate U1s did not. For example, splicing of the A3G reporter was improved in the presence of U1–U6C, but not in the presence of U1–U6A or –U6G alleles. However, compensatory changes in U1 snRNA did not alter the level of improved splicing of suboptimal 5′SS substrates by prp28-E404K allele (Figure 2A). Same assay was also performed in the presence of a single copy of U1–C4U, the only viable U1 snRNA allele with mutation at the U1–C4 or –U6 position (33,46). Consistent with the aforementioned results, improved splicing of the 5′SS–G5A reporter by prp28-E404K was not changed when U1 snRNA was mutated to U1–C4U, which restored the pairing between 5′SS-pos.5 and U1 snRNA-pos.4 (Figure 2A, bottom). Second, we replaced the yeast chromosomal U1C gene with plasmid-borne WT-U1C or U1C-L13F allele that reduces the stability of the 5′SS–U1 snRNA interaction, resulting in a bypass of the requirement for Prp28p (33). In comparison with WT–U1C, splicing of most suboptimal 5′SS mutants was decreased in the presence of the U1C–L13F allele. However, the splicing enhancement because of prp28-E404K did not change when WT–U1C was replaced with U1C–L13F (Figure 2B). For example, improvement of the A3G reporter was 2-fold in the presence of the prp28-E404K allele, which remained 2-fold with either WT–U1C or the U1C–L13F allele (Figure 2B, col. 6 to 5).
Thus, either restoration of 5′SS:U1 snRNA pairing or destabilization of 5′SS–U1 snRNP interaction does not alter the improvement of splicing of suboptimal 5′SS substrates by the prp28-E404K allele, suggesting that modulation of 5′SS selectivity by Prp28p is not dependent on 5′SS:U1 snRNA duplex stability.
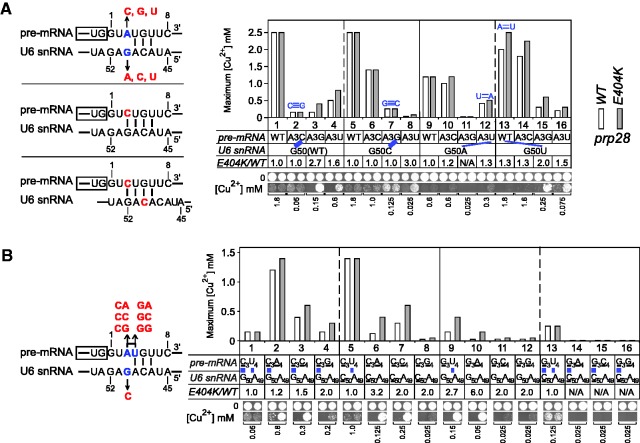
Stabilization of the 5′SS:U6 snRNA duplex results in loss of prp28-E404K effects
We next asked whether the stability of the 5′SS:U6 snRNA duplex is important for altered substrate use because of prp28 alleles. As shown earlier in the text, effects of prp28-E404K are most strongly at 5′SS-pos.A3 and G5, which are matched with U6-pos.50 and 48, respectively. Because of the inviability of U6-pos.48 mutant alleles (47), we focused on U6-pos.50 and tested all possible combinations of 5′SS-pos.3 and U6-pos.50. Previous observed insensitivity of the A3C reporter to prp28-E404K might be due to its additional CG pair with WT U6–G50 (Figure 3A). Consistent with this hypothesis, improved splicing of 5′SS-A3G and A3U by prp28-E404K was either cancelled or significantly decreased when U6-G50 was mutated to C or A, which also form an additional base pair (Figure 3A). In contrast, prp28-E404K improved the splicing of A3C reporter when U6–G50 was mutated to either A or U, disrupting the CG base pair (Figure 3A, col. 10 and 14). In all the 16 cases, improved splicing by prp28-E404K occurred when the nucleotide at 5′SS-pos.3 was mismatched with the nucleotide at U6-pos.50, except for the WT reporter that always exhibited high copper tolerance and the 5′SS-A3C reporter in the presence of U6–G50C. The inconsistency for the cases of WT reporter (Figure 3A, col. 1, 5, 9 and 13) would be due to an unknown reason. However, an early cross-linking assay demonstrated that U6-pos.51 interacts with various nucleotides in the 5′SS region, resulting in a shifted pairing between the 5′SS and U6 snRNA (48). Therefore, the inability of prp28-E404K to improve splicing of the A3C reporter in the presence of U6–G50C might be due to a shifted and stable four base pairs interaction between the 5′SS and U6 snRNA (Figure 3A left), or another unknown mechanism that we cannot address here.
Figure 3.
A hyper-stabilized 5′SS:U6 snRNA duplex results in loss of enhanced splicing by prp28 allele. (A) (Left) Base pairing between 5′SS and U6 snRNA. The 5′SS-A3C mutant reporter would form hyper-stabilized duplexes with WT U6 snRNA and U6–G50C mutant. (Right) prp28-E404K allele improves splicing of 5′SS-A3G and A3U, but not A3C, mutant reporters in the presence of WT U6 snRNA. Mutations at U6–G50 position, which disrupt its base pair with 5′SS-pos.3, altered the original effects by prp28-E404K allele, shown a pattern that hyper-stabilized 5′SS:U6 duplex cancels or significantly decreases the enhanced splicing by prp28 allele. The CG base pairs have stronger cancellation effects than AU base pairs. (B) (Left) Schematic of double mutations of 5′SS-pos.3 and 4, and in U6–G50, indicating various stabilities of 5′SS:U6 duplex. (Right) Enhanced splicing activity of suboptimal 5′SS substrates by prp28-E404K allele is cancelled by hyper-stable CG base pairs between 5′SS-pos.3 and U6-pos.50. However, improved splicing because of prp28-E404K can be restored by simultaneous disruption of the adjacent base pair between 5′SS-pos.4 and U6-pos.49. Mutations are indicated in red; base-pairs at mutation sites are shown by blue lines.
The extent of cancellation of the prp28-E404K effects by additional pairing is variable. GC base pairs fully cancelled the improvement; UG wobble or AU pairs only partly or did not cancel the improvement (Figure 3A). This might be due to either the identity or stability of CG pairs. To distinguish, we used 5′SS-A3C or A3G mutant reporters with additional mutations at the 5′SS-pos.4 and tested their splicing activities in the presence of U6 alleles (Figure 3B). First, in the presence of WT U6 snRNA, prp28-E404K allele did not improve splicing of the single mutant, A3C, as shown earlier in the text (for clarity, referred to here as C3U4), in which the duplex has four base pairs. However, splicing of reporters having additional mutations at 5′SS-pos.4 to disrupt the second base pair with U6–A49 was improved by prp28-E404K allele even if the first CG base pair was present (Figure 3B, col. 2–4 to 1). Likewise, improved splicing of A3G reporter was cancelled in the presence of U6–C50A49 that forms four base pairs; analysis was not available for reporters of A3G with additional pos.4 mutations because of their low splicing activity. Second, splicing of mutant reporters that disrupted two base pairs was even more strongly improved by prp28-E404K. For example, the improved splicing level of C3A4 reporter in the presence of U6–C50A49 is higher than the presence of WT U6 (Figure 3B, col. 6 to 2). These results strongly suggest that the base identity of CG pairs between 5′SS-pos.3 and U6-pos.50 is not important for the 5′SS selection by Prp28p, but instead that the 5′SS:U6 duplex stability is critical for the altered substrate use.
We then analyzed in vivo mRNA and lariat-intermediate products by primer extension. DBR1, the gene for debranching enzyme, was deleted to prevent degradation of lariat intermediates for accurate visualization (16,49). mRNA levels of all tested alleles by primer extension are consistent with the copper reporter assays described earlier in the text (Figure 4A, top graph). Further analysis revealed that the improved levels of mRNA by prp28-E404K allele were due to enhanced efficiency of the first step of splicing, with a level of 30–50% improvement when the mutant substrates formed only three base pairs with U6 snRNA (Figure 4A, middle graph), consistent with the observation from the gel (Figure 4A, upper). The small differences that we detected in the second step are insignificant, or alternatively might reflect an effect of prp28 on the second step that was not selected for in our screen (Figure 4A, bottom graph), suggesting that the prp28-E404K effects are before the first step of splicing.
Figure 4.
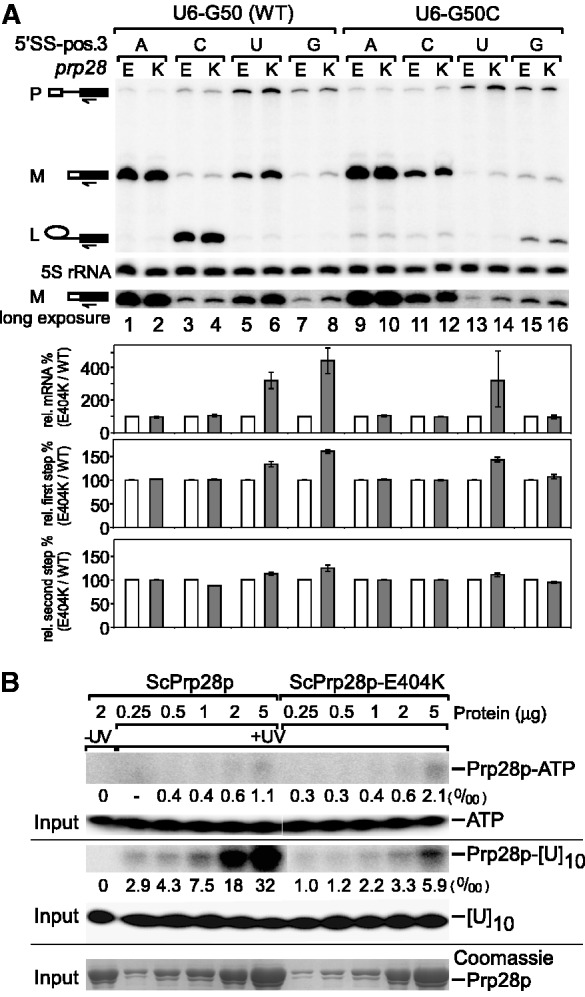
prp28-E404K allele decreases RNA-binding ability and improves the first step of splicing of suboptimal 5′SS substrates. (A) prp28-E404K allele enhances splicing for suboptimal 5′SS substrates at the stage of 5′SS–U6 snRNA association, before the first step of splicing. In vivo mRNA levels of reporters analyzed by primer extension are consistent with previous copper assays. Primer extensions of 5S rRNA were performed as a loading control. Mutant prp28-E404K improves the first step efficiency of suboptimal 5′SS substrates when it forms a less stable RNA duplex with U6 snRNA. Normalized by 5S rRNA, efficiency of the first and second step were calculated as (M+L)/(P + M + L) and M/(M + L), respectively. Relative efficiency of each step was normalized to the WT-PRP28 allele in each case. Error bars represent two independent experiments. P: pre-mRNA, M: mRNA, L: lariat intermediate. (B) Prp28p-E404K has lower RNA-binding ability than WT Prp28p. (Upper) In vitro UV cross-linking assay showed low ATP-binding activity of Prp28 proteins. (Middle) Prp28p-E404K exhibits decreased binding to 32P-[U]10. Signals were determined by phosphorimaging and normalized by the input. (Lower) Input proteins were stained using Coomassie blue.
prp28-E404K alters RNA-binding activity
Prp28p is an RNA-dependent ATPase; therefore, the E404K mutation may alter ATPase activity of Prp28p. However, ATP hydrolysis in vitro was undetectable when purified recombinant Prp28p was incubated with ATP, consistent with a previous report (7). Similarly, ATP binding by Prp28p was low; only faint bands of cross-linked 32P-ATP with Prp28p were observed by UV cross-linking (Figure 4B). To date, the direct RNA target of Prp28p is not known, as with most other spliceosomal ATPases, except Brr2p and Prp22, whose targets were revealed by cross-linking assays (9–11,50). However, we could detect RNA-binding activity of Prp28p using UV cross-linking with homopolymeric [U]10, an RNA substrate mimic. In comparison with WT Prp28p, mutant E404K protein had ∼80% reduction of binding to 32P-labeled [U]10 (Figure 4B), suggesting that reduced RNA-binding activity results in less effective conformational change facilitated by the prp28 mutant.
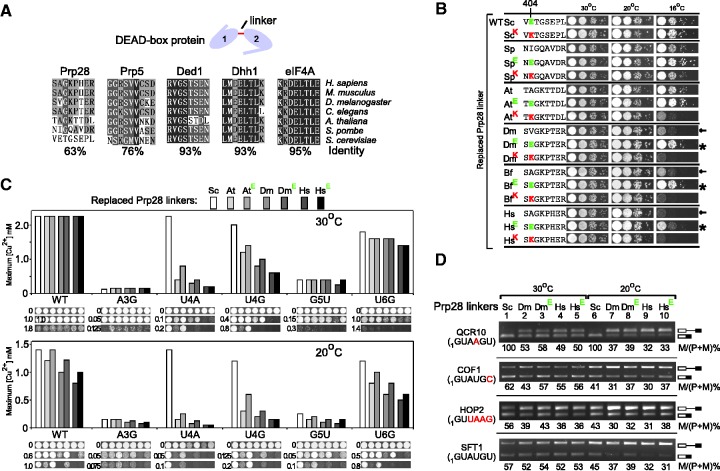
E404 of Prp28p is critical for efficient splicing in yeast
DEAD-box proteins contain an ATPase/helicase domain and various flanking domains (30), in which the core ATPase/helicase domain comprises two RecA subdomains separated by a short peptide linker (Figure 5A). Glutamic acid 404 is the second residue in a linker region between the two Prp28p ATPase subdomains. Phylogenetic conservation of this region in Prp28p is much lower than in other DEAD-box proteins, such as eIF4Ap, Ded1p and Dhh1p; in particular, E404 is unique to budding yeast (Figure 5A). The variation in Prp28p linkers might contribute to the maintenance of splicing activity or fidelity in yeast. To test this possibility, we constructed mosaic prp28 mutants in S. cerevisiae, in which the linker region was replaced by linkers from other species. Swapping the linker with metazoan Prp28ps, including linkers from Drosophila melanogaster; Branchiostoma floridae and Homo sapiens, resulted in growth defects of budding yeast, exhibited by a cs phenotype at 16°C, whereas linkers from fission yeast and plant showed no detectable growth defects. However, the cs phenotypes were rescued by a single mutation at position 404 back to Glu (Figure 5B).
Figure 5.
Residue E404 and the linker region of Prp28 are critical to maintain high splicing efficiency in budding yeast. (A) The linker region between ATPase subdomains of Prp28 is more variable across species in comparison with linkers of other DEAD-box proteins. Linker regions of each listed DEAD-box protein were either extracted from known crystal structures or predicted using software NetSurfP (ver. 1.1). (B) Swapping the ScPrp28 linker with metazoan Prp28 linkers causes cs phenotypes (arrows). However, these defects are rescued by single mutation of residue 404 to Glu (asterisk), but not rescued by mutation to Lys. (C) Replacement of Prp28p linkers with sequences derived from D. melanogaster; B. floridae or H. sapiens, decreases the splicing efficiency, as detected by copper assay. At 30°C, splicing efficiency of reporters is reduced primarily at 5′SS-pos.4 in the presence of metazoan linkers; however, at 20°C, the decreased splicing is broader to all tested pre-mRNA reporters, including WT. Single mutation of residue 404 back to Glu partially rescued these splicing defects. (D) Splicing of endogenous intron-containing genes is inhibited when the yeast Prp28p linker is replaced with metazoan linker sequences. In vivo mRNA levels of genes with non-consensus 5′SS were analyzed by reverse transcriptase–PCR, showing that replaced Prp28p linker sequences decrease splicing activity of all intron-containing genes, especially for the genes having non-consensus nucleotide at the fourth position in 5′SS. Sc: S. cerevisiae, Sp: S. pombe, At: A. thaliana, Dm: D. melanogaster, Bf: B. floridae, Hs: H. sapiens.
To address whether these cs growth defects were due to decreased splicing activity, we measured copper resistance of ACT1-CUP1 reporters and mRNA levels of endogenous genes in yeast strains. First, prp28 alleles with replaced linkers significantly inhibited splicing of 5′SS-pos.4 mutant reporters, but not WT reporter, or slightly inhibited other mutant reporters at 30°C (Figure 5C). These mosaic prp28 alleles inhibited splicing of all tested reporters at lower temperature (20°C), including the WT reporter and 5′SS-pos.4 mutants that are the most inhibited substrates. Importantly, the splicing defects were partly rescued when residue 404 was mutated back to Glu (Figure 5C). Second, we tested mRNA levels by reverse transcriptase–PCR of several endogenous intron-containing genes. The spliced mRNA levels of all tested genes, whether they contain a consensus 5′SS sequence, were decreased in the presence of mosaic prp28 alleles (exacerbated at lower temperatures) and were partly restored when residue 404 was mutated back to Glu, although the restoration was not as strong as for the reporters (Figure 5D). These results suggest that the cs phenotypes caused by replaced Prp28 linker sequences are due to a generally decreased splicing efficiency; the E404 residue is especially critical for maintenance of splicing efficiency in budding yeast.
The variation of Prp28p linkers and the sensitivity of 5′SS-pos.4 mutants to the replaced metazoan linker alleles in yeast suggested a correlation between them. Therefore, we compared the nucleotide frequency at each 5′SS position from the species we used for swapping (Figure 6A and Supplementary Table S2). Consistent with previous analysis (51), the 5′SS is highly conserved at each position with a consensus sequence 1GUAUGU6 in S. cerevisiae; nucleotides from position 3–6 are more flexible in higher eukaryotes. One notable difference is that the most common nucleotide at 5′SS-pos.4 is uridine in S. cerevisiae but adenosine in other species.
Figure 6.
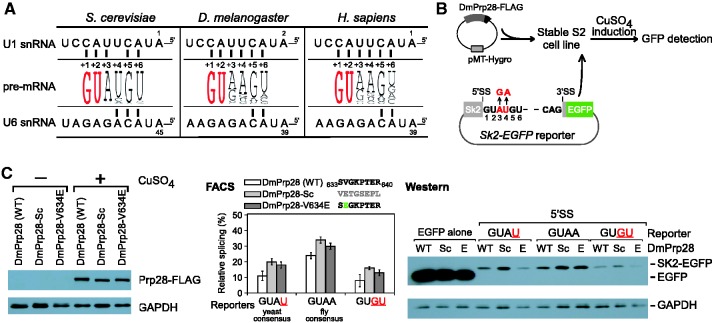
Linker switches of DmPrp28 in Drosophila S2 cell line. (A) The 5′SS region is more diverse in higher eukaryotes; see Supplementary Table S2 for details and additional species. The base pairing regions of U1 and U6 snRNAs to the 5′SS are invariable across species. (B) Strategy of constructing splicing reporter system in Drosophila S2 cell line. Stable cell lines were established by transfection and drug selection. (C) (Left) Western blot analysis indicated that FLAG-tagged DmPrp28s were expressed at similar levels in the three stable cell lines after induction with CuSO4. Switching yeast Prp28 linker to DmPrp28 or single mutation of DmPrp28-V634E increases splicing efficiency of the three tested splicing reporters, indicated both by FACS detection of EGFP fluorescence (middle) and by western analysis of fusion Sk2-EGFP protein (right). Linker region sequences of WT and mutant DmPrp28s are indicated.
Yeast Prp28p linker enhances splicing efficiency in Drosophila cells
To address whether the Prp28p linker is important for maintaining splicing efficiency in higher eukaryotes, we chose the Drosophila S2 cell line. Stable S2 cell lines carrying FLAG-tagged DmPrp28p or mutants, driven by an MT promoter, were transfected with Sk2-EGFP splicing reporters to monitor splicing activity. This reporter contains a truncated Drosophila Sk2 exon 2 and fused with EGFP (Figure 6B). Analyzed by FACS, the DmPrp28-Sc allele that contained the budding yeast linker sequence showed improved EGFP fluorescence for all three tested reporters, including 5′SS-GUAA4 (fly consensus), -GUAU4 (yeast consensus) and mutant -GUGU4 reporters, suggesting that the yeast Prp28p linker sequence provides higher splicing activity (Figure 6C). A single mutation at residue V634 in DmPrp28p to Glu also improved expression of EGFP, consistent with the previous findings in yeast. The pattern of EGFP expression analyzed by western blotting was consistent with the analysis by FACS, except that less improvement was seen (Figure 6C). In addition, splicing of the 5′SS-GUAA4 reporter was more efficient than the -GUAU4 reporter, consistent with adenosine being the favored nucleotide at 5′SS-pos.4 in higher eukaryotes. Taken together, these data demonstrate that the budding yeast linker sequence of Prp28p provides a higher splicing efficiency than the corresponding linker sequence from Drosophila Prp28p.
DISCUSSION
We have obtained prp28 alleles that specifically alter splicing of suboptimal 5′SS substrates, indicating proofreading of 5′SS by Prp28p. The mutation in the strongest prp28 allele is located in the linker region between Prp28p ATPase subdomains, close to the site of a previously observed cross-link between Prp28p and the 5′SS (38), and this mutation impaired RNA-binding ability. We propose that movement of the 5′SS across U6 snRNA facilitated by Prp28p competes with the stability of the 5′SS:U6 duplex, resulting in rejection of weak 5′SS:U6 pairs.
Supported both by genetic studies and in vitro assays, the common feature of proofreading by ATPases is that WT ATPases discriminate suboptimal substrates, whereas slower ATPase mutants allow progression of splicing by providing a longer temporal opportunity for formation of productive complexes on suboptimal substrates (20,22–27). We provide several lines of evidence that Prp28p also modulates splicing fidelity, proofreading 5′SS at the stage of 5′SS–U6 snRNA association as follows: (i) isolated prp28 alleles improved splicing of suboptimal 5′SS substrates, but not BS or 3′SS mutants; (ii) either stabilization of the 5′SS:U1 snRNA duplex, attained by complementary changes of U1 snRNA, or destabilization of the 5′SS–U1 interaction, using a mutated U1C protein, resulted in neither cancellation nor exacerbation of the improved splicing activity by prp28-E404K allele; (iii) hyper-stabilization of the 5′SS:U6 snRNA duplex, attained by either mutations in the 5′SS or in U6 snRNA, cancelled the enhanced splicing activity conferred by the prp28 allele, and this cancellation was not because of nucleobase identity in the 5′SS. Furthermore, the prp28-E404K allele that improved splicing of 5′SS mutants exhibited reduced RNA-binding activity, which presumably results in a reduced ability to promote conformational change. The simplest explanation is a model in which these alleles allow longer time for an inefficient 5′SS:U6 snRNA duplex formation. In such a scenario, WT introns and U6 snRNA would pair efficiently, and subsequent activity by either WT-Prp28p or mutant Prp28p would facilitate a conformational change that stabilizes the association of U6 snRNA to introns (Figure 7A). By contrast, a suboptimal 5′SS would pair inefficiently and WT-Prp28p would function before duplex formation, resulting in a similar, but abortive, conformational change in the 5′SS–U6 snRNA complex. The slowed activity of mutant prp28 alleles would allow more time for the association of a weakened 5′SS:U6 snRNA duplex, thereafter progressing to allow an improved first step of splicing (Figure 7B).
Figure 7.
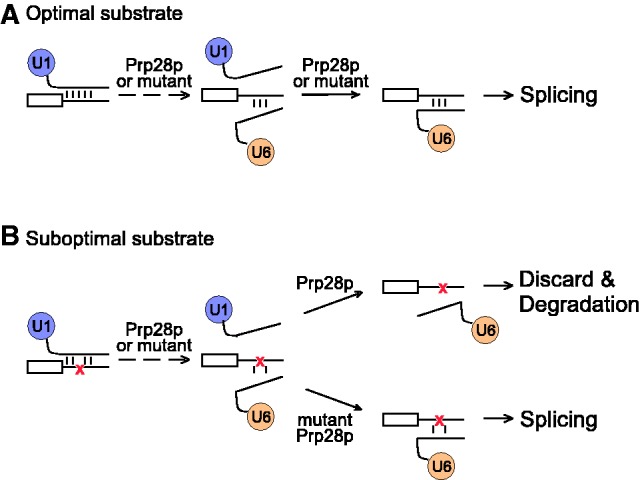
Prp28p proofreads splicing at 5′SS during the association of 5′SS with U6 snRNA. By either an unwinding activity or an RNPase activity, Prp28p disrupts the 5′SS–U1 interaction, then facilitates the 5′SS–U6 interaction and proofreads 5′SS before the first step of splicing. It is unclear whether this is a concerted one-step U1 release/U6 association or two distinct steps; therefore, disruption of 5′SS–U1 snRNA interaction by Prp28p is shown in dash. (A) Productive splicing pathway facilitated by Prp28p. Duplex formation between optimal 5′SS substrates and U6 snRNA is faster than the rate of conformational change by either WT-Prp28p or selected mutant Prp28p. (B) Conformational change by WT-Prp28p is faster than the rate of duplex formation between suboptimal 5′SS substrates and U6 snRNA, resulting in Prp28p-mediated transition prior to 5′SS:U6 pairing and thus rejecting suboptimal substrates. However, mutant Prp28p with a slower activity reduces the rate of conformational change, allowing longer temporal opportunity to achieve stable 5′SS:U6 pairing and thus progression of splicing.
We previously reported that the stability of the BS–U2 snRNA structure is in competition with the activity of Prp5p, linking an RNA duplex with the modulation of splicing fidelity by an ATPase (26). In this study, the stability of the 5′SS:U6 RNA duplex is proofread by Prp28p, providing another example of ATPase-mediated modulation of splicing fidelity being coupled to the stability of an RNA duplex. Prp28p has been proposed to mediate a switch of 5′SS:U1 pairing to 5′SS:U6 pairing, disrupting the 5′SS–U1 interaction by either its unwinding activity to separate the 5′SS:U1 duplex (8) or its RNPase activity to remove U1 proteins (33,35); Prp28p was also proposed to translocate the 5′SS into a complex to interact with U6 snRNA by base pairing (38). Our results demonstrated that the level of prp28 allele’s effect was highly dependent on the stability of 5′SS:U6 duplex, but not 5′SS:U1 duplex, suggesting that ATP hydrolysis by Prp28p destabilizes the 5′SS–U1 interaction, resulting in release of U1 snRNP; then, the 5′SS is translocated to U6 snRNA for formation of a second duplex, providing an opportunity for splicing proofreading. However, it is uncertain whether this is one coordinated step of U1 release/U6 association or two distinct steps that are facilitated by Prp28p and ATP hydrolysis, although we favor a coordinated one-step process (Figure 7).
The 5′SS sequence is more diverse in higher eukaryotes than in budding yeast, as are the BS and 3′SS (51). The base pairing partners for the 5′SS in U1 snRNA (3ACUUAC8) and U6 snRNA (47ACAGAG52) are invariant in all eukaryotes (52,53). Therefore, the 5′SS:U1 and 5′SS:U6 duplexes are generally uniform in budding yeast, but more variable and less stable in higher eukaryotes. In this study, we found that the linker region between Prp28p ATPase subdomains is variable between species and is particularly divergent in budding yeast ScPrp28p. A glutamic acid residue in the linker region is critical for maintaining high splicing efficiency, and swapping the entire linker sequence across species resulted in cell growth defects because of a general reduction of splicing activity. The 5′SS-pos.4 in budding yeast is the most sensitive position for mutant prp28s that contain replaced linker sequences. Mutant assays in Drosophila cells provided similar results. It has been demonstrated that linker regions are critical for maintenance of functional protein conformation from many studies of ATPases. For example, mutations in the linker region between the protease and helicase domains of dengue virus NS3 protein resulted in significantly lower ATPase and helicase activities and a reduction of viral RNA synthesis (54). Therefore, we propose that the linker region between Prp28p subdomains is a key element for maintenance of the active conformation of Prp28p, and its second residue Glu—the important feature being negative charge—is critical for high splicing efficiency in budding yeast. Our results also showed a strong correlation between the variation of Prp28p linkers and the fourth nucleotide in the 5′SS consensus sequence.
In addition, the more variable 5′SS sequences in higher eukaryotes, especially the fourth nucleotide, would form much different stabilities of RNA duplexes with the invariant U6 snRNA, thus, providing more opportunities for discrimination and alternative splicing by Prp28p in higher eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2.
FUNDING
National Natural Science Foundation of China [30970622, 31270842] and National Basic Research Program of China [2012CB114101 to Y.-Z.X.]; National Institutes of Health [GM57829 to C.C.Q]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Moldón, W. Shao and other members in the three groups for discussions and critical reading of the manuscript. They also thank Ms X.H. Wu and Ms R.H. Zhu for FACS detection, and Mr. Y.L. Ma for statistics of intron consensus sequences and figure drawing.
REFERENCES
- 1.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 5.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 6.Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem. Soc. Trans. 2010;38:1105–1109. doi: 10.1042/BST0381105. [DOI] [PubMed] [Google Scholar]
- 7.Strauss EJ, Guthrie C. PRP28, a ‘DEAD-box' protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994;22:3187–3193. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 9.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat. Struct. Mol. Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26:2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26:2422–2434. doi: 10.1101/gad.200949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egecioglu DE, Chanfreau G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA. 2011;17:383–389. doi: 10.1261/rna.2454711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability": spliceosome dynamics and the regulation of splicing. Mol. Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 2012;37:263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 16.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat. Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 18.Query CC, Konarska MM. CEF1/CDC5 alleles modulate transitions between catalytic conformations of the spliceosome. RNA. 2012;18:1001–1013. doi: 10.1261/rna.029421.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 20.Burgess S, Couto JR, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 21.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 22.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol. Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perriman R, Ares M., Jr Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell. 2010;38:416–427. doi: 10.1016/j.molcel.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl Acad. Sci. USA. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol. Cell. 2012;45:314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 29.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 30.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 31.Strauss EJ, Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991;5:629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 32.Murray HL, Jarrell KA. Flipping the switch to an active spliceosome. Cell. 1999;96:599–602. doi: 10.1016/s0092-8674(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 33.Chen JY, Stands L, Staley JP, Jackups RR, Jr, Latus LJ, Chang TH. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 34.Hage R, Tung L, Du H, Stands L, Rosbash M, Chang TH. A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction. Mol. Cell Biol. 2009;29:3941–3952. doi: 10.1128/MCB.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 36.Teigelkamp S, Mundt C, Achsel T, Will CL, Luhrmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 38.Ismaili N, Sha M, Gustafson EH, Konarska MM. The 100-kda U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA. 2001;7:182–193. doi: 10.1017/s1355838201001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson DS, Keefe AD. Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb0803s51. Chapter 8, Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 40.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004;23:376–385. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 44.Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez CJ, Romfo CM, Vanhoy RW, Porter GL, Wise JA. Mutational analysis of U1 function in Schizosaccharomyces pombe: pre-mRNAs differ in the extent and nature of their requirements for this snRNA in vivo. RNA. 1996;2:404–418. [PMC free article] [PubMed] [Google Scholar]
- 46.Seraphin B, Rosbash M. Mutational analysis of the interactions between U1 small nuclear RNA and pre-mRNA of yeast. Gene. 1989;82:145–151. doi: 10.1016/0378-1119(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 47.Madhani HD, Bordonne R, Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- 48.Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 50.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 52.Wolff T, Menssen R, Hammel J, Bindereif A. Splicing function of mammalian U6 small nuclear RNA: conserved positions in central domain and helix I are essential during the first and second step of pre-mRNA splicing. Proc. Natl Acad. Sci. USA. 1994;91:903–907. doi: 10.1073/pnas.91.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee C, Jaladat Y, Mohammadi A, Sharifi A, Geisler S, Valadkhan S. Metal binding and substrate positioning by evolutionarily invariant U6 sequences in catalytically active protein-free snRNAs. RNA. 2010;16:2226–2238. doi: 10.1261/rna.2170910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.