Abstract
Eukaryotic RNA quality control (RQC) uses both endonucleolytic and exonucleolytic degradation to eliminate dysfunctional RNAs. In addition, endogenous and exogenous RNAs are degraded through post-transcriptional gene silencing (PTGS), which is triggered by the production of double-stranded (ds)RNAs and proceeds through short-interfering (si)RNA-directed ARGONAUTE-mediated endonucleolytic cleavage. Compromising cytoplasmic or nuclear 5′–3′ exoribonuclease function enhances sense-transgene (S)-PTGS in Arabidopsis, suggesting that these pathways compete for similar RNA substrates. Here, we show that impairing nonsense-mediated decay, deadenylation or exosome activity enhanced S-PTGS, which requires host RNA-dependent RNA polymerase 6 (RDR6/SGS2/SDE1) and SUPPRESSOR OF GENE SILENCING 3 (SGS3) for the transformation of single-stranded RNA into dsRNA to trigger PTGS. However, these RQC mutations had no effect on inverted-repeat–PTGS, which directly produces hairpin dsRNA through transcription. Moreover, we show that these RQC factors are nuclear and cytoplasmic and are found in two RNA degradation foci in the cytoplasm: siRNA-bodies and processing-bodies. We propose a model of single-stranded RNA tug-of-war between RQC and S-PTGS that ensures the correct partitioning of RNA substrates among these RNA degradation pathways.
INTRODUCTION
Eukaryotic gene expression produces large amounts of both protein-coding and non-coding RNA species. To ensure proper cellular function and viability, a high level of fidelity must be sustained. To tackle this challenge, RNA surveillance and decay serve three main purposes: first, to ensure RNA quality control (RQC) mechanisms that scrutinize RNA integrity and eliminate defective messenger RNA (mRNA), thus dampening the production of potentially toxic proteins, second, to regulate mRNA turnover to control protein abundance and third, to detect invading RNAs, to defend the cell against them (1–4) and to regulate selected endogenous mRNAs through an endonucleolytic cleavage process called post-transcriptional gene silencing (PTGS) (5–8). How RQC and PTGS pathways interact and the processes that regulate the partitioning of RNA substrates into these pathways are not well understood.
Nonsense-mediated decay (NMD) is an extensively studied RQC pathway involved in the genome-wide suppression of transcripts (9–11) in which translation is arrested either owing to the presence of a premature termination codon or owing to excessive 3′untranslated region (UTR) length (12–16). Although there are several different mechanisms by which NMD can be triggered, once instigated, NMD generally involves the recruitment and activation of conserved UPFRAMESHIFT 1 (UPF1), UPF2 and UPF3 proteins to defective transcripts that are translationally stalled. However, the presence of an exon junction complex (EJC) is not always required to evoke NMD because it can target intronless transcripts in yeast, mammals, flies and plants (17–21). This recruitment, either by invoking decapping and deadenylation pathways or via endonucleolytic cleavage, as is the case in Drosophila and humans, generates aberrant RNAs [RNAs lacking a 5′-cap structure or a 3′-poly(A) tail] that are subsequently degraded through exonucleolytic cleavage [for reviews see (2,22,23)].
Exonucleolytic RNA degradation in Arabidopsis exploits a suite of processes including, but not limited to, the shortening of the 3′-poly(A) tail (deadenylation), which is catalysed by the conserved 3′–5′ POLY(A)-SPECIFIC RIBONUCLEASE (PARN) as well as by the conserved CARBON CATABOLITE REPRESSOR 4 (CCR4) complex (24–27). It also involves the removal of the 5′-cap structure, which is accomplished by a set of conserved decapping proteins: DCP1, DCP2 (TDT), DCP5, VARICOSE (VCS) and possibly DEA(D/H)-box RNA HELICASE 1 (DHH1) (28–30). Decapping and deadenylation are a prerequisite for most RNA to be degraded by 5′–3′ XRN exoribonucleases and the multimeric 3′–5′ exoribonuclease exosome complex. Arabidopsis expresses three XRN proteins, the nuclear XRN2 and XRN3 and the cytoplasmic XRN4 (31). Biochemical and molecular characterization of the Arabidopsis exosome core complex revealed the subunits RRP4, RRP40, RRP41, RRP42, RRP43, RRP45 (CER7), RRP46, CSL4 and MTR3 (32). Additional components likely involved in exosome function include RRP44, RRP6L1, RRP6L2, RRP6L3 and MTR4 (32–35).
In addition to these RNA degradation mechanisms, plants and other eukaryotes use PTGS to defend against foreign invading RNAs, such as viruses and high levels of transgenic mRNAs (36–40). PTGS also is required to modulate the abundance or expression of cellular mRNAs important during developmental transitions, such as the mRNAs targets of the trans-acting small interfering (ta-si)RNA pathway (41,42). Double-stranded (ds)RNA is the priming trigger of PTGS and is generated though several processes such as viral replication, sense-antisense transcription or transcription of inverted-repeat (IR) sequences, whose transcripts are self-complementary and thus fold-back on themselves to form dsRNA. It can also be produced by the cellular RNA-DEPENDENT RNA POLYMERASE 6 (RDR6/SGS2/SDE1), which is coupled to the RNA stabilizing protein SUPPRESSOR OF GENE SILENCING 3 (SGS3). Once the dsRNA is produced, it is processed by DICER-LIKE (DCL) enzymes into 21–22-nt siRNAs, which serve as sequence-specific guides for ARGONAUTE 1 (AGO1)-dependent endonucleolytic cleavage of complementary transcripts (6,43,44). AGO1-mediated cleavage generates RNAs that are, in most cases, subjected to XRN- and exosome-mediated degradation (45). In the case of viruses, once PTGS is instigated, amplification of the siRNAs ensures that tissues are primed against subsequent infection by the same virus or expression of a transgene bearing virus sequences (46,47).
Previous data suggested that defects in RNA processing and degradation that lead to the accumulation of decapped and deadenylated RNA, including mutations in RNA splicing, 3′-end formation and 5′–3′ exoribonuclease XRN-mediated degradation, promote PTGS (48–50). Moreover, removing transgene 3′-terminator sequences enhanced PTGS, while having multiple terminators reduced PTGS (51). Here, we explore the ways in which an array of nuclear and cytoplasmic RQC factors and PTGS interact mechanistically and spatially in plants. Impairing either nuclear or cytoplasmic NMD UPF1 and UPF3, deadenylation PARN and CCR4a and exosome RRP4, RRP6L1, RRP41 and RRP44A components enhanced sense (S)-PTGS but had no effect on an IR-PTGS system. In the cytoplasm, RQC factors localized in siRNA-body and processing (P)-body RNA degradation foci. These findings show that nuclear and cytoplasmic aberrant RNAs are instrumental during this type of RNA silencing process, as opposed to IR-PTGS, which produces dsRNA, a direct template for the DCLs. The correct partitioning of aberrant RNA substrates among these RNA degradation mechanisms ensures the discrimination of dysfunctional self-RNA and invading non–self-RNA from functional self-RNA and acts as a barrier to prevent the undesired triggering of PTGS of self-RNA.
MATERIALS AND METHODS
Plant material
All Arabidopsis thaliana are in the Columbia accession (52). The JAP3 line was the kind gift of D. Baulcombe and the inducible RNA interference (iRNAi) lines rrp41iRNAi and rrp4iRNAi (32) were the kind gift of J. Ecker. The parn [fast neutron mutant ahg2-1; (53)] was kindly provided by T. Hirayama. The upf1-5 (SALK_112922, insertion located in the 3′UTR) was obtained from NASC. Homozygous ccr4a (SAIL_784_A07, insertion located in intron 9/10), ccr4b (SAIL_635_B07, insertion located in exon 2/11), upf1-6 (SAIL_1295_E07, insertion located 148 bp upstream of the ATG), upf3-3 (SAIL_122_G02, insertion located 183 bp upstream of the ATG), upf3-1 (SALK_025175, insertion located in exon 5/12) and rrp6L1 (rrp6A; SAIL_1306_C10 insertion located in intron 12/13) mutants were generated during this study (see Supplementary Figure S1 for molecular characterization). Seeds were obtained from NASC.
Generation of artificial miRNA lines
The artificial miRNA amiR-RRP44Aa (5′-UAUGAGUAUACAGGCGUGCUG-3′) was generated using the WMD3 microRNA designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and expressed under the ubiquitin promoter in the context of the MIR319a backbone. PTGS reporter lines were transformed using the floral dip methods (54) and transformed plants were selected on 15 μg/ml of glufosinate. PTGS was analysed in the progeny of 3 T2 lines harbouring a single UB::amiR-RRP44a insertion.
RNA extraction and RNA gel blot analysis
For RNA gel blot analyses, frozen tissue was homogenized in a buffer containing 0.1 M NaCl, 2% sodium dodecyl sulphate (SDS), 50 mM Tris–HCl (pH 9.0), 10 mM ethylenediaminetetraacetic acid (pH 8.0) and 20 mM β-mercaptoethanol, and RNAs were extracted two times with phenol and recovered by ethanol precipitation. To obtain high molecular weight (HMW) RNA, total RNA was precipitated overnight in 2 M LiCl at 4°C and recovered by centrifugation. For low molecular weight (LMW) RNA analysis, total RNA was separated on a 15% denaturing polyacrylamide gel electrophoresis gel, stained with ethidium bromide and transferred to nylon membrane (HybondNX, Amersham). LMW RNA and U6 hybridizations were at 50°C with hybridization buffer containing 5× saline-sodium citrate (SSC), 20 mM Na2HPO4, pH 7.2, 7% SDS, 2× Denhardt’s solution and denatured sheared salmon sperm DNA (Invitrogen). HMW RNA hybridization was at 37°C in SigmaPerfectHyb buffer (Sigma). Blots were hybridized with a radioactively labelled random-primed DNA probes for beta-glucuronidase (GUS) mRNA and GUS siRNAs, and end-labelled oligonucleotide probes for TAS1 ta-sRNA, TAS2 tasRNA and U6 detection.
GUS activity quantification
With the exception of amiR-RRP44A, rrp41iRNAi and rrp4iRNAi lines, plants were grown on Bouturage 2 medium (Duchefa Biochemie) in standard long-day conditions (16 h light, 8 h dark at 20–22°C), transferred to soil after 2 weeks and grown in controlled growth chambers in standard long-day conditions. To induce expression of the RNAi lines, rrp41iRNAi and rrp4iRNAi plants were grown on Bouturage media containing 8 μM estradiol for 12 days in standard long-day conditions, and then transferred to soil and grown in controlled growth chambers in standard long-day conditions. Total protein was extracted from cauline leaves of flowering plants and GUS activity was quantified as in (49) by measuring (Fluoroscan II; Thermo Scientific) the quantity of 4-methylumbelliferone produced from the substrate 4-methylumbelliferyl-b-d-glucuronide (Duchefa Biochemie).
Semi-quantitative reverse transcriptase-polymerase chain reaction
RNAs were extracted using the RNeasy plant mini kit (Qiagen), and 1 μg of RNA was reverse transcribed using oligo dT and Super ScriptII reverse transcriptase (Invitrogen). Twenty-seven cycles of polymerase chain reaction (PCR) were used to amplify RRP44A, CCR4a, CCR4b and EF1-alpha, and 28 cycles of PCR were used to amplify UPF1 and UPF3 to non-saturation. The number of cycles used to amplify RRP4 and RRP6L1 to non-saturation is indicated above each lane in Supplementary Figure S1. EF1-alpha amplification was used as a control.
Nicotiana benthamiana agro-infiltration
Agrobacterium (ASE or Agl0 strains) carrying plasmids of interest were grown overnight at 30°C in 3 ml Lysogeny Broth (LB) medium containing the appropriate antibiotics to a final OD600 of between 1.0 and 2.0. The bacteria were pelleted and resuspended in 1 ml of infiltration medium (10 mM MgCl2, 10 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.2, 150 mM acetosyringone) to a final OD600 of 0.1. The bacterial solution containing the plasmid(s) of interest was coinfiltrated with a bacterial solution expressing HELPER COMPONENT-PROTEINASE (HC-Pro), a viral suppressor of silencing, into the abaxial side of leaves using a 1 ml syringe, and samples were assayed 3 days after infiltration. HC-Pro was used to better visualize the fluorescent signals and did not have an observable impact on the localization pattern of the tested RQC and PTGS components.
Confocal imaging
For confocal imaging, agro-infiltrated tobacco leaves (mounted in water) were directly imaged on a Leica Confocal TCS SP2 (Leica Microsystems). The CFP was imaged with 458 nm excitation using the dichroic mirror DD458/514 and detection window of 465–505 nm; the GFP was imaged with 488 nm excitation using the dichroic mirror DD488/543 and a detection window of 500–580 nm; the RFP was imaged with 543 nm excitation using the dichroic mirror DD488/543 and a detection window of 580–670 nm. For the co-localizations, all of the images were taken by sequential acquisition. Image analysis was performed using the National Institute of Health ImageJ (http://rsb.info.nih.gov/ij/) software.
Cloning procedures
All the clones were made using the Gateway technology (Invitrogen) and planned using Geneious (http://www.geneious.com). A list of the oligonucleotides used for cloning is provided in Supplementary Table S2. UPF3 (AT1G33980), SGS3 (AT5G23570) and RRP41 (AT3G61620) were PCR amplified from complementary DNA (cDNA) and cloned into the vector pDONR221 to generate entry clones, whereas PARN (AT1G55870), CCR4a (AT3G58560) and RRP4 (AT1G03360) were PCR amplified from genomic DNA and cloned into the vector pENTR-D to generate entry clones. To obtain the GFP fusions under the control of the 35S promoter, SGS3, CCR4a, PARN and RRP41 entry clones were recombined in the expression vector pH7WGF2, whereas the UPF3 entry clone was recombined in the expression vector pH7FWG2. To obtain the RFP fusion proteins under the control of the 35S promoter, the entry clone containing PARN was recombined in the expression vector pB7WGR2, and the one containing RRP4 in the expression vector pB7RWG2. For the GFP fusion under the control of the Ubiquitin10 promoter, the RRP4 entry clone was recombined in the expression vector pUBN-GFP (55). The 35S:RFP:DCP1 and 35S:CFP:DCP1 constructs were made by recombination of an entry clone containing DCP1 (AT1G08370, gift from C. Antonelli) in the expression vectors pB7WGR2 and pB7WGC2, respectively (56). The 35S:RDR6:GFP construct was made by PCR amplifying RDR6 (AT3G49500) from cDNA and adding the restriction sites SalI and NotI to each terminus to generate a Gateway entry clone in the plasmid pENTR1A that was then recombined in the expression vector pH7FWG2. The construct pGFP-N-Bin:UPF1 (AT5g47010) was generously provided by A. Pendle and J. Brown. The construct 35S:RFP:UPF1 was obtained by recombining the entry clone UPF1 cDNA pDONR207 (kindly provided by A. Pendle and J. Brown) into the expression vector pB7WGR2. The 35S::HC-Pro plasmid was the kind gift of J. Carrington.
RESULTS
To investigate the possible crosstalk between PTGS and other RNA degradation pathways, we isolated loss-of-function Arabidopsis mutants in many key components of RQC and RNA turnover pathways and characterized their impact on S-PTGS. In the cases where loss-of-function caused lethality, we examined the impact of partial-loss-of-function mutants when possible. The effect of RQC and RNA turnover mutants on S-PTGS was determined using the well-characterized Arabidopsis reporter lines Hc1 and 6b4. Both lines carry a 35S::GUS transgene, but whereas 6b4 stably produces GUS, silencing of the GUS transgene is spontaneously triggered in 20% of Hc1 plants at each generation add (57,58). These reporter systems allowed us to reveal both positive and negative effects of the RQC mutations on S-PTGS. To avoid the 35S interference phenomenon reported to occur when introducing the 35S::GUS transgene carried by the 6b4 and Hc1 into mutants already carrying a 35S T-DNA insertion (59), we analysed S-PTGS uniquely in mutants containing either 35S-free T-DNA insertions or fast neutron-generated mutations.
Mutations in NMD, deadenylation and exosome factors enhance S-PTGS
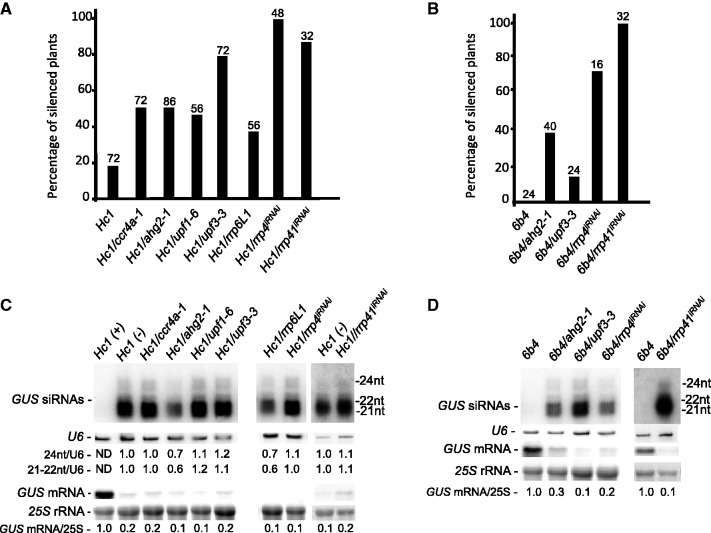
To examine the contribution of NMD to PTGS, we searched publicly available mutant collections and identified upf1 (SAIL_1295_E07, hereafter referred to as upf1-6), and upf3 (SAIL_122_G02, hereafter referred to as upf3-3) partial-loss-of-function mutants (Supplementary Figure S1A), and these mutants were crossed with Hc1 and 6b4 lines. Quantitative GUS assays performed on the progeny of plants homozygous for both the Hc1 locus and either the upf1 or upf3 mutation indicated that Hc1 silencing was enhanced from 20% in line Hc1 to 44% in Hc1/upf1-6 and to 78% in Hc1/upf3-3 (Figure 1A). To determine the strength of the silencing enhancement, we also analysed the effect of these mutations on line 6b4. The upf3-3 mutation triggered silencing in 13% of the 6b4 plants analysed (Figure 1B), whereas upf1-6 did not appear to have an effect on 6b4 silencing (0/32 plants were silenced at the 6b4 locus). Characteristic of PTGS, GUS siRNAs accumulated and GUS mRNA levels were reduced to nearly undetectable levels in the silenced Hc1/upf1-6, Hc1/upf3-3 and 6b4/upf3-3 lines (Figure 1C and D), indicating that both UPF1 and UPF3 are endogenous PTGS suppressors.
Figure 1.
NMD, deadenylation and exosome mutants enhance transgene S-PTGS. (A and B) The percentage of silenced plants determined by GUS quantitative protein assays in the indicated mutant and control lines. The number of plants analysed is indicated above each bar. (C and D) RNA gel blot analyses of the indicated mutant and control lines. High molecular weight RNA and siRNA gel blots were hybridized with a GUS DNA probe. 25S ribosomal RNA (rRNA) and U6 small nucleolar RNA (snRNA) served as loading controls, respectively. Hc1 plants that were expressing (+) and silenced (−) for GUS were analysed. The position of GUS 24, 22 and 21 nt siRNAs is noted. Normalized values of GUS mRNA to 25S rRNA (with either Hc1 (+) or 6b4 levels set at 1.0) and GUS 24 nt and GUS 21–22 nt siRNA to U6 snRNA [with Hc1 (−) levels set at 1.0] are indicated. ND = non-detectable.
Arabidopsis PARN has poly(A) RNA degradation activity and complete loss-of-function parn mutants are lethal, indicating that it is an essential ribonuclease (13). Nevertheless, a fast neutron-generated partial-loss-of-function alternative splicing parn mutant ahg2-1 has been described (60). Quantitative GUS assays on plants homozygous for both the Hc1 transgene and the ahg2-1 (parn) mutation indicated that silencing of Hc1 was increased from 20% to nearly 50% (Figure 1A). In addition, ahg2-1 triggered silencing in nearly 40% of 6b4 plants (Figure 1B), indicating that PARN is a suppressor of PTGS. Like the parn mutant that negatively impacts deadenylation, a mutation in the putative deadenylation factor CCR4a enhanced Hc1 silencing from 20% to nearly 60% (Figure 1A); however, unlike the parn mutant, the ccr4a mutation did not trigger silencing of line 6b4 (0/30 of 6b4/ccr4a plants were silenced). Silencing triggered by both parn and ccr4a deadenylation mutants led to the accumulation of GUS siRNAs and a reduction in GUS mRNA levels (Figure 1C and D). In contrast to ccr4a, a mutation in the related CCR4b gene, which is located adjacent to the CCR4a gene, did not impact Hc1 or 6b4 silencing (18%, 10/56 Hc1/ccr4b plants and 0%, 0/39 6b4/ccr4b plants were silenced), suggesting that CCR4b could be partially redundant with CCR4a. Both ccr4a and ccr4b mutants appeared to be full-loss-of-function mutants, as they did not produce detectable CCR4a and CCR4b transcripts, respectively (Supplementary Figure S1B), but additional work will be required to determine whether these proteins are partially redundant.
The multimeric exosome complex contains 3′–5′ exoribonucleases that degrade RNA with unprotected 3′-ends. To determine if perturbations in exosome function could influence PTGS, we characterized the impact on S-PTGS in the Hc1 and 6b4 reporter lines of mutants defective in the Arabidopsis core exosome subunits RRP4 and RRP41, the latter of which exhibits catalytic 3′–5′ RNAse activity, unlike the yeast and human RRP41 (61). We also examined the impact on S-PTGS of impairing RRP44A, the Arabidopsis homolog of the RRP44 (DIS3) 3′–5′ RNAse responsible for nearly all of the catalytic activity of the yeast exosome (62,63). Finally, we examined the impact on S-PTGS of a mutation in RRP6L1 [also known as RRP6A; Supplementary Figure S1C (32,64)], one of two Arabidopsis homologs of the yeast and human RRP6 exoribonuclease. Although the nuclease function of Arabidopsis RRP6L1 has not been confirmed, expression of Arabidopsis RRP6L1 complements the growth defects of a yeast rrp6 mutant strain (64). Because rrp4 and rrp41 mutants are seedling lethal, we analysed PTGS in the previously characterized rrp4 and rrp41 iRNAi lines, which silence RRP4 and RRP41 after estradiol treatment owing to the induced expression of an IR transgene targeting RRP4 and RRP41, respectively [Supplementary Figure S1D and (32)]. Furthermore, because 35S-free loss-of-function mutants in rrp44A were not available, we generated Arabidopsis lines expressing an artificial miRNA (65) (amiR-RRP44A) under the ubiquitin promoter, and analysed PTGS in line Hc1.
Quantitative GUS assays indicated that loss-of-function of rrp6L1 and down-regulation of rrp4iRNAi and rrp41iRNAi enhanced PTGS in line Hc1 from the 20% baseline to 30, 90 and 80%, respectively (Figure 1A). Furthermore, analysis of S-PTGS in Hc1/amiR-RRP44A plants revealed that line 46, which accumulated more amiR-RRP44A (Figure 2A) and less RRP44A mRNA (Figure 2B) than lines 43 and 53, triggered PTGS in 100% of Hc1 plants analysed (Figure 2C) and accumulated GUS siRNAs (Figure 2B). Moreover, the rrp4iRNAi and rrp41iRNAi lines triggered PTGS in nearly 70 and 100% of 6b4 plants, respectively (Figure 1B), whereas rrp6L1 mutants did not trigger silencing of 6b4 (GUS silencing was not observed in 64 6b4/rrp6L1 plants). The effect of the expression of the artificial amiR-RRP44A on 6b4 PTGS was not tested. Collectively these data indicate that mutations in a variety of components involved in RQC and exonucleolytic RNA turnover have the capacity to enhance S-PTGS. As all these pathways act on single-stranded (ss)RNA, these results suggest that modulation of ssRNA abundance is a key element controlling entry into PTGS.
Figure 2.
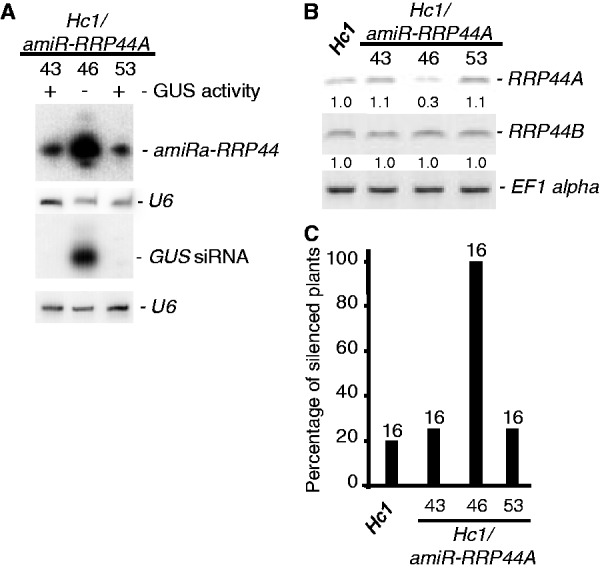
Expression of an artificial miRNA targeting RRP44A leads to enhanced S-PTGS. (A) RNA gel blot analyses of three different Hc1 plant lines expressing the artificial RRP44A miRNA amiR-RRP44A. Small RNA gel blots were hybridized with a GUS DNA probe or an oligonucleotide antisense to the amiR. U6 served as a loading control for small RNA. (B) Reverse transcriptase-PCR of RRP44A and RRP44B transcripts in the corresponding Hc1/amiR-RRP44A and control Hc1 seedlings. EF1alpha was used as an amplification control. Normalized values of RRP44A and RRP44B mRNA to EF1 alpha mRNA (with Hc1 levels set at 1.0) are indicated. (C) The percentage of silenced plants determined by GUS quantitative protein assays in the indicated mutant and control lines. The number of plants analysed is indicated above each bar.
To broaden our S-PTGS analysis to an endogenous silencing system that, like S-PTGS, requires RDR6 and SGS3 for dsRNA production, we examined the effect of RQC mutants on the ta-siRNA pathway (66–69). We did not observe any changes in tasiRNA levels arising from the TAS1 or TAS2 locus in any of our RQC mutants (Supplementary Figure S2).
Mutations in NMD, deadenylation and exosome components do not impact IR-PTGS
Next, we examined the impact of mutations in these NMD, deadenylation and exosome components on a PTGS system that produces a stem-loop dsRNA directly through transcription and, thus, does not rely on the RDR6- and SGS3-dependent conversion of ssRNA to dsRNA to become a substrate of DCL proteins. The line JAP3 expresses a PHYTOENE DESATURASE (PDS) inverted repeat under the control of the phloem-specific Suc2 promoter (70) and initiates from the veins PDS silencing, which can be easily traced owing to the photobleaching phenotype.
Mutations in NMD, deadenylation and the core exosome complex did not appear to impact the initiation or spreading of IR-PTGS in the line JAP3 (Figure 3). It was shown previously that UPF1 influenced RNAi persistence in Caenorhabditis elegans and IR-PTGS in Arabidopsis, but UPF1 did not appear to affect RNAi in Drosophila (71–73). The analysis in Arabidopsis examined the effect of the upf1-5 mutant, a SALK T-DNA insertion line containing a 35S promoter, on an IR of the endogenous APETALA 3 (AP3) gene that was expressed under the 35S promoter (71). Given the report of 35S interference on PTGS observed when combining two transgenes each containing the 35S promoter (59), we re-examined the effect of the upf1-5 mutant on IR-PTGS using the JAP3 35S-free IR-PTGS system. Similar to what we observed for the upf1-6 mutant, the upf1-5 mutant did not appear to impact the initiation or spreading of JAP3 IR-PTGS (Figure 3), indicating that UPF1 likely does not play a role in IR-PTGS in Arabidopsis and that the initial report likely was hampered by 35S interference.
Figure 3.

NMD, deadenylation and exosome mutants do not impact JAP3 IR-PTGS. Eighteen-day-old control JAP3 plants and JAP3 plants containing the indicated mutations. The photo is representative of a minimum of 20 plants screened for each genotype.
These results indicate that deadenylation, NMD and exosome components impinge on PTGS at a step unique to S-PTGS that is not required for IR-PTGS. It is interesting to speculate that this step is linked to aberrant ssRNA protection or dsRNA generation, processes accomplished by the SGS3 and RDR6 proteins, respectively (74–76).
Both nuclear and cytoplasmic RNA decay proteins regulate S-PTGS
To determine where within the cell the different exonucleolytic RNA decay and S-PTGS pathways could overlap, we first expressed a subset of the components for which mutations were shown to alter S-PTGS as translational fusions to fluorescent reporters in N. benthamiana leaves (Figure 4). The S-PTGS components RDR6 and SGS3 were confirmed to localize in cytoplasmic foci. We also confirmed the previously reported subcellular localization of UPF3 and UPF1 in the nucleus and cytoplasmic foci, respectively (77,78). RRP44A was previously reported to be predominantly nuclear (35), and we observed that the core subunits of the exosome, RRP4 and RRP41, also were detected primarily in the nucleus, with only a weak diffuse signal present in the cytoplasm (Figure 4). Finally, we showed that the deadenylation factors CCR4a and PARN both accumulated in cytoplasmic foci (Figure 4). The subcellular localizations of UPF3, RRP41, CCR4a, PARN, RDR6 and SGS3 observed in transient expression were confirmed in stable Arabidopsis lines expressing the fusion proteins at low levels (Supplementary Figure S3), indicating that the subcellular localizations observed in N. benthamiana leaves are not artifacts caused by over-expression.
Figure 4.
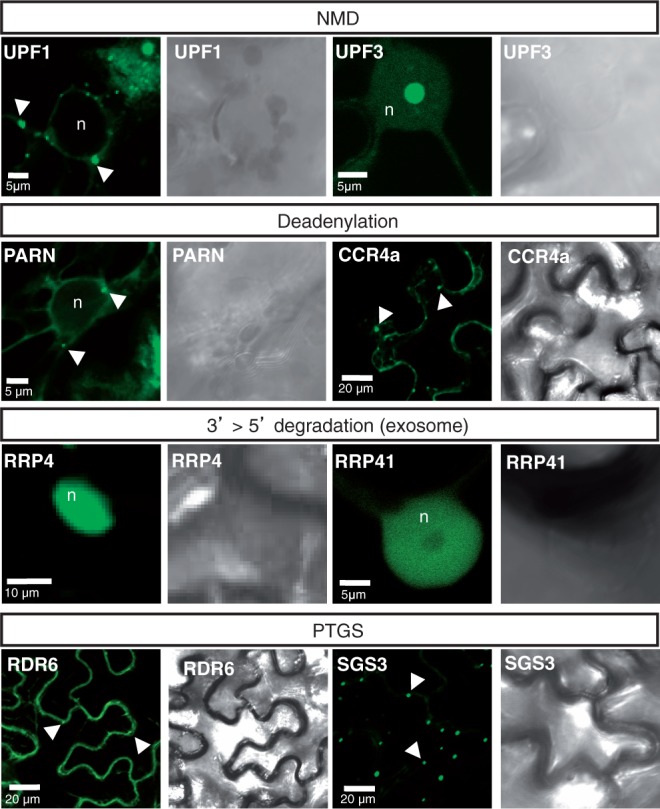
Subcellular localization of NMD, deadenylation, exosome and PTGS components. Confocal sections and their corresponding bright-field images of N. benthamiana leaves expressing the indicated proteins fused to GFP. The arrowheads indicate cytoplasmic foci, whereas ‘n’ labels the nucleus. Scale bars are shown on the images.
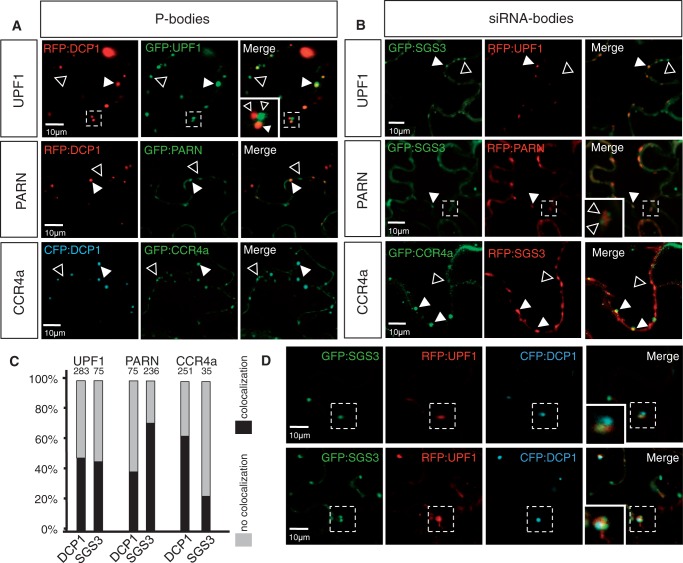
Although RDR6 has been reported in both the nucleus and the cytoplasm, it only co-localizes with SGS3 in cytoplasmic foci called siRNA-bodies (75,79–81). Another class of cytoplasmic foci involved in mRNA degradation, distinct from siRNA-bodies, is the P-bodies where the decapping complex protein DCP1 accumulates (29,80). We therefore investigated whether the cytoplasmic foci observed for UPF1, PARN and CCR4a were siRNA-bodies or P-bodies or these proteins shuttle between them. To this end, we co-expressed these tagged proteins with either fluorescently tagged DCP1 or SGS3. We observed that tagged UPF1, PARN and CCR4a co-localized with both DCP1 and SGS3 (Figure 5A and B). Quantification of the proportion of UPF1, PARN and CCR4a bodies co-localizing with DCP1 (P-bodies) or SGS3 (siRNA-bodies) indicated that while nearly 70% of PARN foci co-localized with siRNA-bodies and >60% of CCR4a foci were associated with P-bodies (Figure 5A–C), UPF1 was found nearly equally associated with both siRNA- and P-bodies. The fraction of UPF1 that co-localized with P- or siRNA-bodies nearly equaled the fraction of UPF1 that was non-co-localized to the other body (siRNA- or P-bodies, respectively, Figure 5C); thus, we more precisely examine these associations through a triple localization experiment among UPF1, DCP1 and SGS3. In the triple localization, we examined 32 adjacent P- and siRNA-body clusters and observed that for a given group of closely associated P- and siRNA-bodies, the UPF1 protein was either associated with the P-body or the siRNA-body but never with both bodies in the same cluster at the same time (Figure 5D and Supplementary Table S1).
Figure 5.
UPF1, PARN and CCR4a associate with both P- and siRNA-bodies. (A and B) Confocal sections of N. benthamiana leaves co-expressing the indicated fluorescent fusion proteins. Co-expression of UPF1, PARN and CCR4a with DCP1, a P-bodies marker (A), or with SGS3, a siRNA-bodies marker (B). White arrowheads indicate co-localization, and open arrowheads indicate foci positive for only one of the two fusion proteins. The area enclosed in the dashed box is shown in the close-up view. (C) Percentage of UPF1, PARN and CCR4a foci that co-localize with either P-bodies (as marked by DCP1) or siRNA-bodies (as marked by SGS3). Percentage of foci co-localizing (black) or not co-localizing (grey) with DCP1 and SGS3. The total number of foci counted is indicated above each bar. DCP1 and SGS3 foci were never observed to overlap. (D) Confocal sections of N. benthamiana leaves co-expressing SGS3, DCP1 and UPF1 fluorescent fusion proteins. Upper row: UPF1 is associated with a siRNA-body that is located adjacent to a P-body. Lower row: UPF1 is associated with a P-body that is located adjacent to two siRNA-bodies. The area enclosed in the dashed box is shown in the close-up view. Scale bars are shown on the images.
DISCUSSION
Our results hint to a multi-layered regulatory network governing RQC and PTGS in different subcellular compartments. It was shown previously that mutations in the cytoplasmic exoribonuclease XRN4, the cytoplasmic decapping component DCP2 and the nuclear exoribonuclease XRN2 and XRN3 enhance PTGS (49,82). Here, we show that, in addition to mutations in several cytoplasmic deadenylation and NMD components, mutations in essentially nuclear RQC components (UPF3, RRP44A and RRP6L1) enhance PTGS. These results are in agreement with the existence of both a cytoplasmic and a nuclear arm to the PTGS pathway (79,83,84) and suggest that nuclear RNAs, in addition to cytoplasmic RNAs, are instrumental during S-PTGS. However, it remains unknown if these nuclear localized proteins are spatially associated with nuclear components of PTGS. Indeed, the DCL proteins responsible for siRNA generation are nuclear localized (85). Additional work is needed to examine these putative associations.
Although it is intriguing to imagine a nuclear interface among these pathways, we cannot exclude the possibility that RNA substrates that evade elimination by these nuclear RQC components are exported from the nucleus where they trigger S-PTGS in the cytoplasm. Moreover, it is also possible that at least a fraction of these primarily nuclear RQC proteins can be shuttled to the cytoplasm at certain times. Indeed, in yeast, UPF3 is shuttle protein operating in NMD, which involves both nuclear-localized steps and a cytoplasmic-localized translation termination coupled step (86).
Our observations that UPF1, CCR4a and PARN co-localize with both P- and siRNA-body markers suggest that exchange of ribonucleoparticle substrates between the two RNA degradation bodies can occur. We propose a model of ssRNA tug-of-war between RQC and S-PTGS that ensures the correct partitioning of aberrant RNA substrates among these RNA degradation mechanisms, potentially contributing to the discrimination of dysfunctional self-RNA and invading non–self-RNA from functional self-RNA (87). We assert that this discrimination allows a cell to efficiently clear undesirable RNAs without triggering PTGS, which, owing to the amplification of siRNA production, would lead to the unregulated trans-degradation of any RNA transcripts sharing homology with the dsRNA trigger. Indeed, it is interesting to speculate that the existence of the S-PTGS pathway may serve to reinforce the efficiency of RQC pathways to eliminate defective RNAs.
We recognize that this system of checks and balances between PTGS and RQC pathways was revealed in RQC mutant plants, and, thus, contend that these pathways may normally act independently, and that RNA substrate sharing may only occur when RQC pathways are rendered inefficient or compromised. Indeed, it is highly plausible that, in normal conditions, defective endogenous transcripts are eliminated efficiently by RQC pathways so as to prevent their ‘auto-death’ by PTGS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
Agence Nationale de la Recherche [ANR-08-BLAN-0082 to A.C.M. and A.M. and ANR-10-LABX-40 to M.C. and H.V.]; Land Baden-Württemberg, Chica und Heinz Schaller Stiftung and the CellNetworks cluster of excellence (to A.M.). Funding for open access charge: Agence Nationale de la Recherche [ANR-08-BLAN-0082] the Chica und Heinz Schaller Stiftung.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bruno Letarnac, Herve Ferry, Philippe Marechal and Fabrice Petitpas for plant care. This work has benefited from the facilities and expertise of the Imagif Cell Biology Unit of the Gif campus (www.imagif.cnrs.fr), which is supported by the Conseil Général de l'Essonne. A.B.M., F.B. and A.M. carried out the cloning and the imaging experiments; A.E.Md.A., H.V. and A.C.M. carried out the genetic and molecular PTGS analyses; M.D.C., A.C.M., A.M. and H.V. designed and coordinated the study. All authors contributed to the writing and editing of the manuscript and read and approved the final manuscript.
REFERENCES
- 1.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2012;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- 5.Chen X. Small RNAs in development – insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parent JS, Martínez de Alba AE, Vaucheret H. The origin and effect of small RNA signaling in plants. Front. Plant Sci. 2012;3:179. doi: 10.3389/fpls.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 2009;12:622–627. doi: 10.1016/j.pbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Voinnet O. Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility. Curr. Opin. Plant Biol. 2008;11:464–470. doi: 10.1016/j.pbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 13.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 14.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 16.van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- 17.Buhler M, Paillusson A, Muhlemann O. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 2004;32:3304–3315. doi: 10.1093/nar/gkh651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan MT, Yu SM. The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl Acad. Sci. USA. 1998;95:6543–6547. doi: 10.1073/pnas.95.11.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 20.LeBlanc JJ, Beemon KL. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 2004;78:5139–5146. doi: 10.1128/JVI.78.10.5139-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajavel KS, Neufeld EF. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 2001;21:5512–5519. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 23.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9–25. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–1214. doi: 10.1261/rna.7540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010;152:866–875. doi: 10.1104/pp.109.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 33.Hooker TS, Lam P, Zheng H, Kunst L. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell. 2007;19:904–913. doi: 10.1105/tpc.106.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange H, Gagliardi D. The exosome and 3′-5′ RNA degradation in plants. Adv. Exp. Med. Biol. 2011;702:50–62. doi: 10.1007/978-1-4419-7841-7_5. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl Acad. Sci. USA. 2010;107:15981–15985. doi: 10.1073/pnas.1007060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallory AC, Elmayan T, Vaucheret H. MicroRNA maturation and action–the expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol. 2008;11:560–566. doi: 10.1016/j.pbi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 38.van Mierlo JT, van Cleef KW, van Rij RP. Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol. Biol. 2011;721:3–22. doi: 10.1007/978-1-61779-037-9_1. [DOI] [PubMed] [Google Scholar]
- 39.Wang MB, Masuta C, Smith NA, Shimura H. RNA silencing and plant viral diseases. Mol. Plant Microbe Interact. 2012;25:1275–1285. doi: 10.1094/MPMI-04-12-0093-CR. [DOI] [PubMed] [Google Scholar]
- 40.Waterhouse PM. Defense and counterdefense in the plant world. Nat. Genet. 2006;38:138–139. doi: 10.1038/ng0206-138. [DOI] [PubMed] [Google Scholar]
- 41.Skopelitis DS, Husbands AY, Timmermans MC. Plant small RNAs as morphogens. Curr. Opin. Cell Biol. 2012;24:217–224. doi: 10.1016/j.ceb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Willmann MR, Poethig RS. Time to grow up: the temporal role of smallRNAs in plants. Curr. Opin. Plant Biol. 2005;8:548–552. doi: 10.1016/j.pbi.2005.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 44.Chen X. Small RNAs - secrets and surprises of the genome. Plant J. 2010;61:941–958. doi: 10.1111/j.1365-313X.2009.04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brosnan CA, Voinnet O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr. Opin. Plant Biol. 2011;14:580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 49.Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herr AJ, Molnar A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl Acad. Sci. USA. 2006;103:14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Z, Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura N, Okamoto M, Narusaka M, Yasuda M, Nakashita H, Shinozaki K, Narusaka Y, Hirayama T. ABA hypersensitive germination2-1 causes the activation of both abscisic acid and salicylic acid responses in Arabidopsis. Plant Cell Physiol. 2009;50:2112–2122. doi: 10.1093/pcp/pcp146. [DOI] [PubMed] [Google Scholar]
- 54.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 55.Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64:355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 56.Karimi M, Bleys A, Vanderhaeghen R, Hilson P. Building blocks for plant gene assembly. Plant Physiol. 2007;145:1183–1191. doi: 10.1104/pp.107.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 58.Elmayan T, Balzergue S, Beon F, Bourdon V, Daubremet J, Guenet Y, Mourrain P, Palauqui JC, Vernhettes S, Vialle T, et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I. Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 2008;13:4–6. doi: 10.1016/j.tplants.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005;44:972–984. doi: 10.1111/j.1365-313X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- 61.Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem. 2000;275:33158–33166. doi: 10.1074/jbc.M005493200. [DOI] [PubMed] [Google Scholar]
- 62.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 63.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 64.Lange H, Holec S, Cognat V, Pieuchot L, Le Ret M, Canaday J, Gagliardi D. Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:3038–3044. doi: 10.1128/MCB.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felippes FF, Wang JW, Weigel D. MIGS: miRNA-induced gene silencing. Plant J. 2012;70:541–547. doi: 10.1111/j.1365-313X.2011.04896.x. [DOI] [PubMed] [Google Scholar]
- 66.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 68.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 72.Domeier ME, Morse DP, Knight SW, Portereiko M, Bass BL, Mango SE. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- 73.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 75.Elmayan T, Adenot X, Gissot L, Lauressergues D, Gy I, Vaucheret H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276:835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- 76.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 77.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merai Z, Benkovics AH, Nyiko T, Debreczeny M, Hiripi L, Kerenyi Z, Kondorosi E, Silhavy D. The late steps of plant nonsense-mediated mRNA decay. Plant J. 2013;73:50–62. doi: 10.1111/tpj.12015. [DOI] [PubMed] [Google Scholar]
- 79.Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. Posttranscriptional gene silencing in nuclei. Proc. Natl Acad. Sci. USA. 2011;108:409–414. doi: 10.1073/pnas.1009805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012;31:1704–1713. doi: 10.1038/emboj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 82.Thran M, Link K, Sonnewald U. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012;72:368–377. doi: 10.1111/j.1365-313X.2012.05066.x. [DOI] [PubMed] [Google Scholar]
- 83.Le Masson I, Jauvion V, Bouteiller N, Rivard M, Elmayan T, Vaucheret H. Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell. 2012;24:3603–3612. doi: 10.1105/tpc.112.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 85.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shirley RL, Ford AS, Richards MR, Albertini M, Culbertson MR. Nuclear import of Upf3p is mediated by importin-alpha/-beta and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics. 2002;161:1465–1482. doi: 10.1093/genetics/161.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christie M, Brosnan CA, Rothnagel JA, Carroll BJ. RNA decay and RNA silencing in plants: competition or collaboration? Front Plant Sci. 2011;2:99–106. doi: 10.3389/fpls.2011.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.