Abstract
Stress response mechanisms that modulate the dynamics of tRNA degradation and accumulation from the cytoplasm to the nucleus have been studied in yeast, the rat hepatoma and human cells. In the current study, we investigated tRNA degradation and accumulation in HeLa cells under various forms of stress. We found that initiator tRNAMet (tRNA(iMet)) was specifically degraded under heat stress. Two exonucleases, Xrn1 and Xrn2, are involved in the degradation of tRNA(iMet) in the cytoplasm and the nucleus, respectively. In addition to degradation, we observed accumulation of tRNA(iMet) in the nucleus. We also found that the mammalian target of rapamycin (mTOR), which regulates tRNA trafficking in yeast, is partially phosphorylated at Ser2448 in the presence of rapamycin and/or during heat stress. Our results suggest phosphorylation of mTOR may correlate with accumulation of tRNA(iMet) in heat-treated HeLa cells.
INTRODUCTION
Cells are often exposed to various kinds of stress such as heat, oxidation, nutrient starvation or UV irradiation. To combat stress, cells have developed tolerance mechanisms such as repair of DNA damage, cell cycle arrest and alterations in translation (1). Important components of the stress response mechanism include up-regulation of heat shock proteins, modification of specific proteins, as well as modulation of RNA metabolism and/or localization (1,2).
Transfer RNAs (tRNAs) are a fundamental component of the translation machinery and play a key role in modulating translation during cellular stress. For instance, non-methionine-tRNAs [e.g. tRNA(Leu)] are aminoacylated with methionine under conditions of oxidative stress (3). Thus, the rate of methionyl-misacylation increases, and the build-up of mischarged tRNAs leads to misincorporation of methionine during protein synthesis in HeLa cells (3). Furthermore, tRNA transcription has been shown to be suppressed by various forms of stress (4). Moreover, fragments produced from multiple tRNAs, so called tiRNAs, are generated in mammalian cells under arsenite-induced oxidative stress, and contribute to translational arrest (5,6). The tRNA fragments are also produced in response to nutrient starvation and hydrogen peroxide-induced oxidative stress (7). In mammalian cells, the cleavage of multiple tRNAs is catalysed by an endonuclease, angiogenin, which is a member of the RNase A family (7). However, in yeast, the tRNA fragment is detected in oxidative stress and produced by an endonuclease Rny1, which is a member of the RNase T2 family (4,8,9). Furthermore, it has been reported that in heat-stressed yeast cells, hypomodified tRNA and unstable tRNA, induced by mutation at the acceptor and/or T-stem, promotes degradation via nuclear exosome pathway-mediated TRAMP (Trf4/5-Air1/2-Mtr4) complex and the rapid tRNA decay (RTD) pathway (10–12). In the nuclear exosome pathway, hypomodified precursor-tRNA(iMet) is recognized by the TRAMP complex, polyadenylated and then degraded by the nuclear exosome (9). In the RTD pathway, specific hypomodified species of tRNA and destabilized tRNAs are degraded by exonucleases, Rat1 in the nucleus and Xrn1 in the cytoplasm (11,12).
In addition to the degradation of the tRNAs, nuclear accumulation of tRNAs is known to occur under the environmental stress (i.e. nutrient starvation, oxidative stress, heat stress). For instance, nuclear accumulation of tRNAs occurs in Saccharomyces cerevisiae during nutrient starvation and oxidative stress (13,14), and in rat hepatoma cells during nutrient starvation (15). However, a more recent study showed that during amino acid starvation, nuclear accumulation of tRNAs does not occur in Kluyveromyces lactis, Schizosaccharomyces pombe, rat hepatoma cells and HeLa, HEK293 cells (14). Thus, the relationship between stress response and the dynamics of tRNA in mammalian cells remain unclear under conditions of environmental stress. We have attempted to reveal the mechanism of tRNA degradation and the dynamics of nuclear accumulation in various types of mammalian cells during environmental stress.
In this study, we initially examined tRNA degradation in HeLa cells under different forms of stress such as heat, oxidation, low pH and cycloheximide-induced translational inhibition. As a result, we found that tRNA(iMet) is specifically degraded by the exonuclease Xrn1 in the cytoplasm and Xrn2 in the nucleus under heat stress. Additionally, heat stress also induced nuclear accumulation of tRNA(iMet). Here, we report a new degradation pathway of tRNA(iMet) and its nuclear accumulation in heat-stressed HeLa cells.
MATERIALS AND METHODS
Cell culture, cell treatment and RNA isolation
HeLa cells were maintained in RPMI1640 medium with 10% fetal bovine serum (FBS) (BioWest) and antibiotic-antimycotic (GIBCO) at 37°C under an atmosphere of 5% CO2, and were grown until 70–80% confluent. The cells were given fresh medium in the presence of RNA polymerase III inhibitor (Merck), which was used at a final concentration of 50 µM, and pre-treated at 37°C for 30 min. Heat stress was achieved by incubating cells in a 43°C water bath for 0–6 h. Other forms of stress were induced by adding ethanol to the growth medium to a final concentration at 2%, 100 µM hydrogen peroxide, low pH (pH 5.0), 35 µM cycloheximide (Sigma-Aldrich) or addition of 150 mM NaCl. Cells were heat treated in the absence or presence of rapamycin (Sigma-Aldrich) (final concentration of 5 nM) and incubated at 37°C for 30 min. Rapamycin was added to the medium at the beginning of the heat stress. The cells were then washed with 1× PBS and suspended in RNA iso (TaKaRa) for total RNA extraction according to the manufacturer’s instructions.
RNA analysis by Northern blot
Total RNAs were separated by 10% PAGE in 7 M urea and TBE buffer, and then blotted onto a nylon membrane. RNA was detected using 5′-end [γ-32P]-ATP-labelled oligonucleotide probe, and the blot was scanned using an Imaging analyzer (Fujifilm). The tRNA probe sequences used in this study were tRNA(iMet): 5′-GCA GAG GAT GGT TTC GAT CCA TC-3′, tRNA(eMet): 5′-CCC CGT GTG AGG ATC GAA CTC AC-3′, tRNA(Leu): 5′-CGA ACC CAC GCC TCC ATC CGG AGA C-3′, Lys: 5′-GAA CCC TGG ACC CTC AGA-3′, tRNA(Pro): 5′-CTC GTC CGG GAT TTG AAC CCG GGA CC-3′, 5S: 5′-CAG GGT GGT ATG GCC GTA GAC-3′. Quantification of band intensity was calculated by Multi Gauge (Fujifilm).
Purification of tRNA(iMet) and tRNA(eMet) using a solid-phase DNA hybridization probe
The 3′-biotinylated DNA oligomer (5′-GCA GAG GAT GGT TTC GAT CCA TCG ACC TC-biotin-3′) was used to purify tRNA(iMet). The 3′-biotinylated DNA oligomer (5′-CCC CGT GTG AGG ATC GAA CTC ACG ACC TT-biotin-3′) was used to purify tRNA(eMet). Both oligomers were used as hybridization probes. HeLa cells were either incubated at 37°C or 43°C for 6 h. The tRNAs were prepared using Solution D, which included guanidinium isothiocyanate. Samples were Phenol/Chloroform extracted and partially purified by ion-exchange column chromatography. Purification of matured tRNA(iMet) and tRNA(eMet) was performed using a solid-phase DNA probe as described in Ishida’s report (16).
Melting profile analyses of tRNA(iMet) and tRNA(eMet)
tRNA(iMet) and tRNA(eMet) were annealed in buffer A (50 mM Tri-HCl pH 7.6, 5 mM MgCl2, 100 mM NaCl) and then the melting curves were recorded on a spectrophotometer, UV-1650PC (Shimadzu). The temperature was increased from 25°C to 95°C over a period of 70 min.
HPLC analysis
tRNA(iMet) prepared from non-treated or heat-treated (0.05 A260 unit) HeLa cells were digested with 2 µg snake venom phosphodiesterase (Sigma-Aldrich), 20 µg RNaseA (Qiagen) and 0.125 U bacterial alkaline phosphatase (TaKaRa) in 20 µl of 50 mM Tris-HCl (pH 8.0) at 37°C overnight. Nucleosides were analysed on an HPLC (Hitachi L-2000 system) equipped with a reverse phase C18 column (NUCLEOSIL 100 C18). The solvent consisted of buffer, as detailed in our recent report (17).
Western blot analysis
HeLa cells were treated at 43°C for 0–6 h, or treated with specific siRNAs. The cells were washed with 1× PBS, suspended by buffer A [50 mM Tris-HCl pH 7.6, 6 mM β-mercaptoethanol, 150 mM KCl and protease inhibitor cocktail (Merck)] and then sonicated. Concentration of total protein was measured by Protein assay Reagent (Invitrogen). Total protein of 15 µg was separated by 8% SDS-PAGE and blotted onto a PVDF membrane (Millipore). After blotting, the membrane was blocked in Tris-buffered saline with 0.2% Tween 20 (TBST) containing 3% BSA for 1 h. The membrane was incubated with primary antibody (α-tubulin, mTOR, phopho-mTOR at Ser2448, eIF2 α and phosho-eIF2 α were purchased from Cell Signaling, and Xrn1 and Xrn2 from Abcam) in TBST containing 3% BSA. The membrane was washed using TBST, and incubated with second antibody (Millipore). Target protein was detected by imaging analyzer (Fujifilm). The band intensity was quantified using the Multi Gauge program (Fujifilm).
RNA fluorescence in situ hybridization (FISH), immunofluorescence, imaging analysis and statistical analysis of fluorescent intensity
FISH analyses were performed according to our recent report, using a DIG-labelled RNA probe (Roche) (18). Localization of Xrn1 and Xrn2 were determined by immunofluorescence analyses. After the cells were heat treated, they were fixed with 4% paraformaldehyde phosphate buffer solution (Wako). Fixed cells were rinsed, using 1× PBS and permeabilized in 1× PBS containing 0.5% Triton X-100 on ice for 10 min. The cells were washed with 1× PBS three times. Then, the cells were blocked with blocking buffer [10% FBS (BioWest), 1× PBS, 0.01% tween20] at room temperature for 1 h and incubated with 2 µg/ml anti-Xrn1 antibody (Abcam) or anti-Xrn2 antibody (Abcam) with blocking buffer for 1.5 h. Unbound antibodies were removed with PBST (1× PBS, 0.2% tween20) for 15 min. The cells were incubated with 2 µg/ml alexa647-conjugated secondary antibodies (Invitrogen) for 1 h. After washing, the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent intensity was quantified using LAS AF Lite software (Leica), and the numerical data were analysed by student’s t-test (for localization quantification: n = 40).
Treatment with siRNA
For Xrn1 and Xrn2 knock-down, Xrn1 and Xrn2 siGENOME SMART pools for each molecule were purchased from Thermo Fisher Scientific. HeLa cells were transfected with 10 nM of siRNA using Lipofectamin RNAi max (Invitrogen) for 24 h at 37°C under an atmosphere of 5% CO2. On the next day, the cells were replated and then cultured for 48 h.
Quantitative PCR
To monitor the knock-down levels by siRNAs, total RNA (500 ng) from siRNA-treated cells was used for Real-time PCR. The mRNAs were reverse-transcribed into cDNAs using RT reagent (TaKaRa). qPCR (SYBR Green 1 dye detection, TaKaRa) was performed on a detection system (TaKaRa) using specific oligo primers (Xrn1 sense 5′-GGA TTT TGC ACT ATT ACT ATC ATG GA-3′ and antisense 5′-GGA AAG GTG CAT AAT GAT AAG GA-3′, Xrn2 sense 5′-GAA GCC ATT CTT CCA GAT CAA-3′ and antisense 5′-CTG TGT CAG ATC CCT TAA CAT AGG-3′, GAPDH sense 5′-AGC CAC ATC GCT CAG ACA C-3′ and antisense 5′-GCC CAA TAC GAC CAA ATC C-3′). The cDNA of GAPDH was used as an internal control. The levels of expression of Xrn1 and Xrn2 were normalized against the GAPDH expression level. We independently repeated the experiments two or more times.
RESULTS
tRNA(iMet)-specific degradation during heat stress
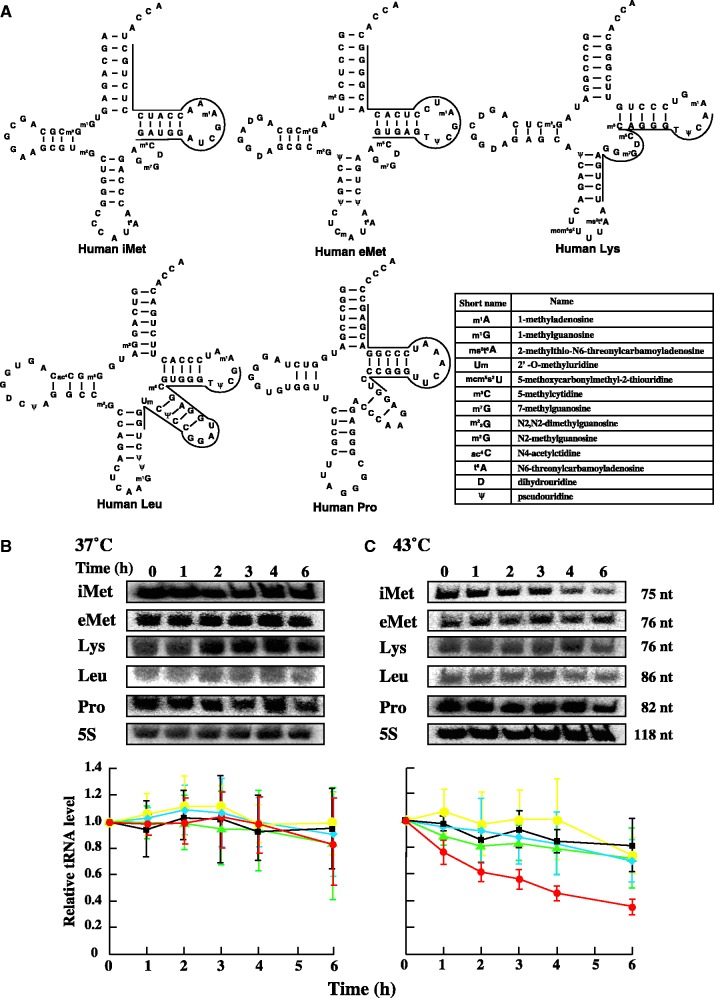
Initially, we used northern blot analysis to analyse the degradation of tRNAs in HeLa cells under heat stress (43°C), low pH stress, ethanol- and hydrogen peroxide-induced oxidative stress and translation inhibition by cycloheximide or high concentrations of sodium chloride (Figure 1 and Supplementary Figure S1). To specifically detect the degradation of tRNA, HeLa cells were pretreated with RNA polymerase III inhibitor. RNA polymerase III inhibitor was reported to have an IC50 of 27 µM in HEK293 (19). The IC50 is the concentration of compound that inhibits growth by 50%. Furthermore, RNA polymerase III activity is 90% inhibited in the presence of 200 µM RNA polymerase III inhibitor by in vitro analysis (19). Thus, RNA polymerase III inhibitor was used at a concentration of 50 µM. Several candidates were selected for the analysis of tRNAs—tRNA(iMet), elongator tRNAMet (tRNA(eMet)), tRNALys (tRNA(Lys)), tRNALeu (tRNA(Leu)) and tRNAPro (tRNA(Pro)) (Figure 1A). These tRNA species were not degraded under ethanol- and hydrogen peroxide-induced oxidative stress, low pH stress and cycloheximide- and high concentration of NaCl-induced translational inhibition (Supplementary Figure S1). By contrast, tRNA(iMet) was specifically degraded when the cells were subjected to heat stress. As shown in Figure 1B, none of the tRNAs were degraded when the cells were cultured at 37°C without heat shock. In contrast, when the cells were subject to heat stress by incubation at 43°C for up to 6 h, tRNA(iMet) was extensively degraded while the other tRNAs, including tRNA(eMet), displayed only slight degradation (Figure 1C). We independently repeated the hybridization experiments three times in order to confirm the reproducibility of the heat-induced tRNA(iMet)-specific degradation (Figure 1B and C, lower panels). Furthermore, the expression level of tRNA(iMet) was evaluated in the absence or presence of RNA polymerase III inhibitor during heat stress (Supplementary Figure S2). As shown in Supplementary Figure S2, the expression levels of tRNA(iMet) in the presence of RNA polymerase III inhibitor remained less than the expression levels of tRNA(iMet) in the absence of inhibitor. This result indicates that supply of tRNA(iMet) by RNA polymerase III was suppressed by RNA polymerase III inhibitor. In summary, transcription of tRNAs by RNA polymerase III in HeLa cells was suppressed at a concentration of 50 µM RNA polymerase III inhibitor.
Figure 1.
Sequence and degradation of tRNAs in HeLa cells with or without heat stress. (A) Cloverleaf structures of our selected tRNAs are shown. The complimentary region to the DNA probe is illustrated, and the sequence of the DNA probe is given in Supplementary information. Northern blot analysis of tRNAs degradation at 37°C (B) and 43°C (C). 7.5 µg total RNA was loaded and blotted. Selected tRNAs were iMet (red), eMet (black), leucine (yellow), lysine (blue), proline (green). 5S rRNA transcribed by RNA polymerase III was used as a control. tRNAs and 5S rRNA were detected by a specific probe. Data represent the mean±SD for three independent experiments.
Although tRNA(iMet) and tRNA(eMet) both transport methionine to the ribosome, the two tRNA species have different characteristics. tRNA(iMet) functions in translational initiation and forms a specific complex with eukaryotic initiation factor 2α (eIF2α). In contrast, tRNA(eMet) functions in the translational elongation step and forms a ternary complex with eukaryotic elongation factor 1α (eEF1α). Therefore, tRNA(iMet) degradation induced by heat stress directly leads to a defect in the translational initiation step of protein biosynthesis.
Suppression mechanism of tRNA(iMet) degradation
Although tRNA(iMet) was subject to significant degradation after heat treatment, there was, nonetheless, a residual amount of tRNA(iMet) detectable in heat-stressed cells (Figure 1C). We reasoned that HeLa cells may possess a suppression mechanism associated with the degradation of tRNA(iMet). However, such a suppression mechanism has not been reported in the literature. To clarify the mechanism, we envisaged four working hypotheses as follows. First, the structural stability of tRNA(iMet) is altered by heat stress, which affects its degradation pattern. Second, the amount(s) and/or composition of modified nucleoside(s) within tRNA(iMet) are changed by heat stress, thereby adjusting its pattern of degradation. Third, the heat stress induces a decrease in the expression level of eIF2α, resulting in an accumulation of free tRNA(iMet). This accumulation of free tRNA(iMet) causes the specific degradation. Fourth, localization of tRNA(iMet) is altered by heat stress in mammalian cells, as reported for S. cerevisiae, which suppresses its degradation. We reasoned that the degradation of tRNA(iMet) and the subsequent suppression of its degradation could be a result of a combination of these proposed mechanisms.
The first hypothesis can be addressed by measuring the melting temperature of tRNA(iMet) prepared from normal and heat-stressed cells. If the L-shaped structure of tRNA(iMet) is partially disordered by heat stress, tRNA(iMet) may be recognized by nuclease(s). To confirm this idea, tRNA(iMet) was purified from HeLa cells with or without heat stress using the solid-phase DNA probe method (16). The purity of tRNA(iMet) was confirmed by gel electrophoresis and HPLC nucleoside analyses. The HPLC analysis relies on the fact that tRNA(iMet) is an exceptional tRNA, which does not contain pseudouridine modification at any position (Figure 1A). The purified tRNA(iMet) was digested with RNase A, phosphodiesterase and alkaline phosphatase and then the modified nucleosides were analysed. As shown in Supplementary Figure S3, a peak corresponding to pseudouridine was not observed in the nucleosides, suggesting that we successfully purified tRNA(iMet). The nucleosides in tRNA(iMet) from the heat-stressed cells were then compared with those from the non-heated cells: the calculated ratios are given in Supplementary Figure S3, lower panel. The amount of modified nucleosides in both samples of tRNA(iMet) was almost identical, suggesting that heat stress does not influence its pattern of modification. Next, we measured the melting temperature of tRNA(iMet) isolated from normal and heat-stressed cells, which were determined to be 76.2°C and 77.3°C, respectively (Figure 2A, B and D). Thus, we confirmed that there is no significant difference in the melting temperatures of tRNA(iMet) derived from heat-treated and non–heat-treated cells. Furthermore, our experimental results showed that the melting temperature of tRNA(iMet) is no different from that of general tRNAs (e.g. melting temperature of yeast tRNAPhe is 76.0°C), although human tRNA(iMet) has an exceptional sequence (A54U55) in the T-loop. Moreover, we purified tRNA(eMet) from non–heat-stressed HeLa cells, and the melting temperature was calculated to be 76.6°C (Figure 2C and D). Hence, the melting temperature of tRNA(iMet) was similar to that of tRNA(eMet). We conclude that there is no significant change in the stability of tRNA(iMet) after heat stress. Moreover, because the melting temperature of tRNA(iMet) is similar to that of tRNA(eMet), nuclease(s) probably recognize the sequence of tRNA(iMet).
Figure 2.
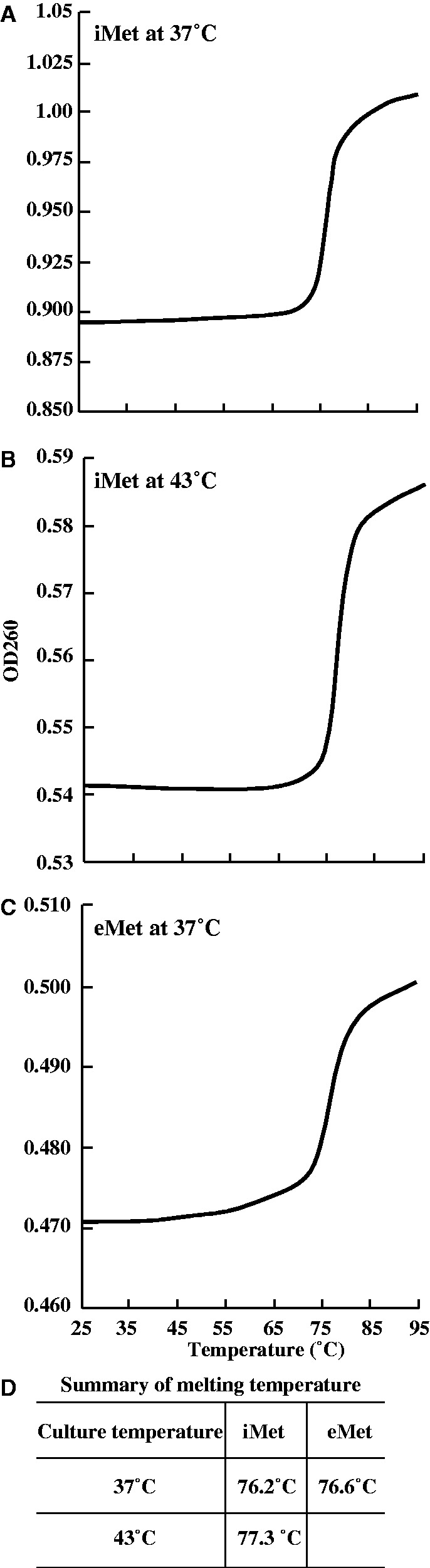
Melting profile and melting temperature of tRNA(iMet) and tRNA(eMet). Melting profile and melting temperature of tRNA(iMet) under normal conditions (A) and heat stress conditions (B) were calculated. Additionally, melting profile and melting temperature of tRNA(eMet) from normal culture conditions (C) was calculated. The calculated melting temperatures are summarized in (D).
Our second hypothesis suggests that the amount of modified nucleoside in tRNA(iMet) may change upon heat stress. Recent reports have shown that hypomodified tRNAs are subject to degradation in heat-stressed S. cerevisiae (10,11). Thus, if the degree of modification in tRNA(iMet) changes when HeLa cells are heat stressed, nuclease(s) may not recognize the modified nucleosides in tRNA(iMet), and this could account for the observed suppression in its degradation. Therefore, the amounts of modified nucleoside of tRNA(iMet) purified from the HeLa cells with or without heat stress were analysed by HPLC (Supplementary Figure S3). However, the amount of modified nucleosides was unchanged, showing that the heat stress did not influence the modification pattern of tRNA(iMet).
Our third hypothesis suggests that eIF2α is denatured (or degraded) by heat stress, leading to degradation of free tRNA(iMet). We examined the level of eIF2α in heat-stressed and non–heat-stressed HeLa cells by western blot analysis (Supplementary Figure S4A and B). Contrary to our expectation, the level of eIF2α slightly increased as a result of heat stress. Furthermore, we analysed the phosphorylation level of eIF2α (Supplementary Figure S4A and C). When eIF2α is phosphorylated under various forms of stress, translation is arrested (1). As shown in Supplementary Figure S4A and C, the level of phosphorylation of eIF2α rapidly increased shortly after the onset of heat stress, reached a maximum some 2–3 h later, and then gradually decreased. Phosphorylation of eIF2α is linked to a variety of stress signalling pathways, which leads to translation arrest. The decrease in the concentration of cytoplasmic tRNA(iMet) could be associated with dephosphorylation of eIF2α. These results suggest that the mechanism of suppression of tRNA(iMet) degradation does not involve thermal denaturation or degradation of eIF2α under heat stress, and that tRNA(iMet) degradation may be linked to the phosphorylation status of eIF2α. Based on these experimental results, we confirmed that the degradation of tRNA(iMet) cannot be explained by the first, second or third of our proposed mechanisms.
The fourth hypothesis suggests that the localization of tRNA(iMet) is altered by heat stress. In a recent report by Murthi, shuttling of tRNAs was shown to constitutively occur in S. cerevisiae (20). Furthermore, nuclear accumulation of tRNAs is known to result from nutrient starvation and methylmethane sulfonate (MMS)-induced DNA damage in S. cerevisiae, and puromycin-induced translation arrest in Chinese hamster ovary cells (CHO) (13,14,21,22).
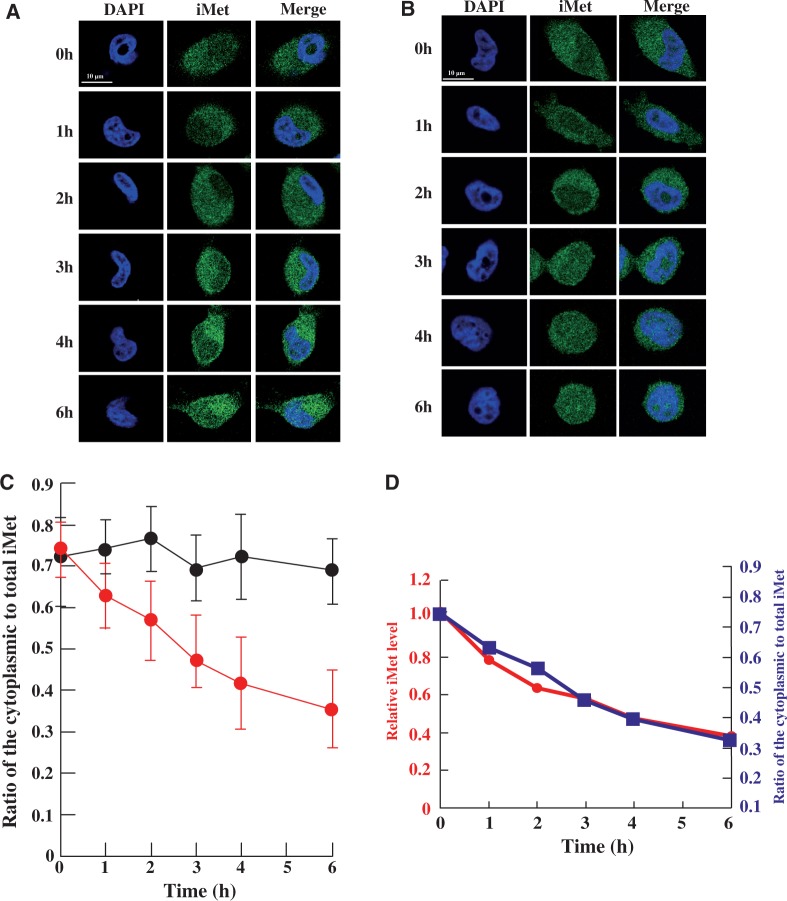
Thus, we reasoned that heat stress-induced localization of tRNA(iMet) was a likely mechanism to explain the suppression in its degradation. To address this issue, we analysed the accumulation of tRNA(iMet) using FISH under heat stress (Figure 3). To inhibit transcription in the nucleus, HeLa cells were pretreated with RNA polymerase III inhibitor. The HeLa cells were then incubated in a water bath at 37°C or 43°C for 0–6 h. When the cells were cultured at 37°C, the ratio of cytoplasmic to total tRNA(iMet) was calculated to be 72% at 0 h, and no accumulation of tRNA(iMet) in the nucleus was observed during the time course of the experiment (Figure 3A and C). In contrast, heat stress induced nuclear accumulation of tRNA(iMet) (Figure 3B and C). At the start point (0 h), 73% of tRNA(iMet) was present in the cytoplasm. However, after the temperature shift, accumulation of tRNA(iMet) in the nucleus was detected in a time-dependent manner. The ratio of tRNA(iMet) in the cytoplasm to total tRNA(iMet) decreased during the first 3 h after the initial heat stress, although the rate of accumulation then slowed considerably (Figure 3B and C).
Figure 3.
Heat stress caused nuclear accumulation of tRNA(iMet). Nuclear accumulation of tRNA(iMet) was monitored by FISH in HeLa cells at 37°C (A) and 43°C (B) for the time indicated. (C) Ratio of cytoplasmic to nuclear tRNA(iMet) at 37°C (black) and at 43°C (red) is shown. Quantification of fluorescence was performed in 40 cells per hour by using the LAS program (Leica) and the results were averaged. Data represent the mean±SD (n = 40). (D) Superposition of degradation and accumulation of tRNA(iMet). The circles show the degradation of tRNA(iMet). The squares show accumulation of tRNA(iMet) from the cytoplasm to the nucleus.
In the recent report, when tRNA(iMet) with three extra nucleotide at its 5′ end and the mature 3′ end was injected into the nuclei of Xenopus oocyte, matured tRNA(iMet) appeared rapidly in the cytoplasm after processing of the 5′ end (23). Thus, mature tRNA(iMet) cannot accumulate in the nucleus under normal conditions. In our experiment where new transcription was suppressed by addition of an RNA polymerase III inhibitor, mature tRNA(iMet) was mostly observed by northern blot analysis (Figure 6A). Additionally, FISH analysis confirmed that tRNA(iMet) did not accumulate in the nucleus under non-stress conditions (Figure 3A). These observations indicate that the mature form of tRNA(iMet) accumulated in the cytoplasm under normal conditions, but was probably transported into the nucleus when HeLa cells were exposed to heat stress.
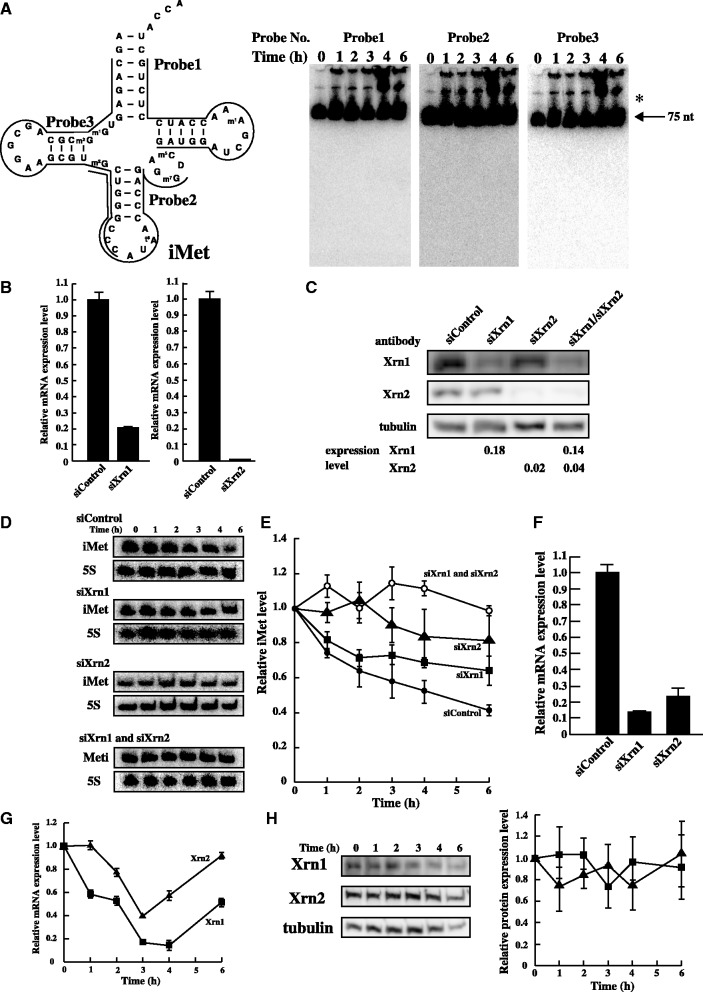
Figure 6.
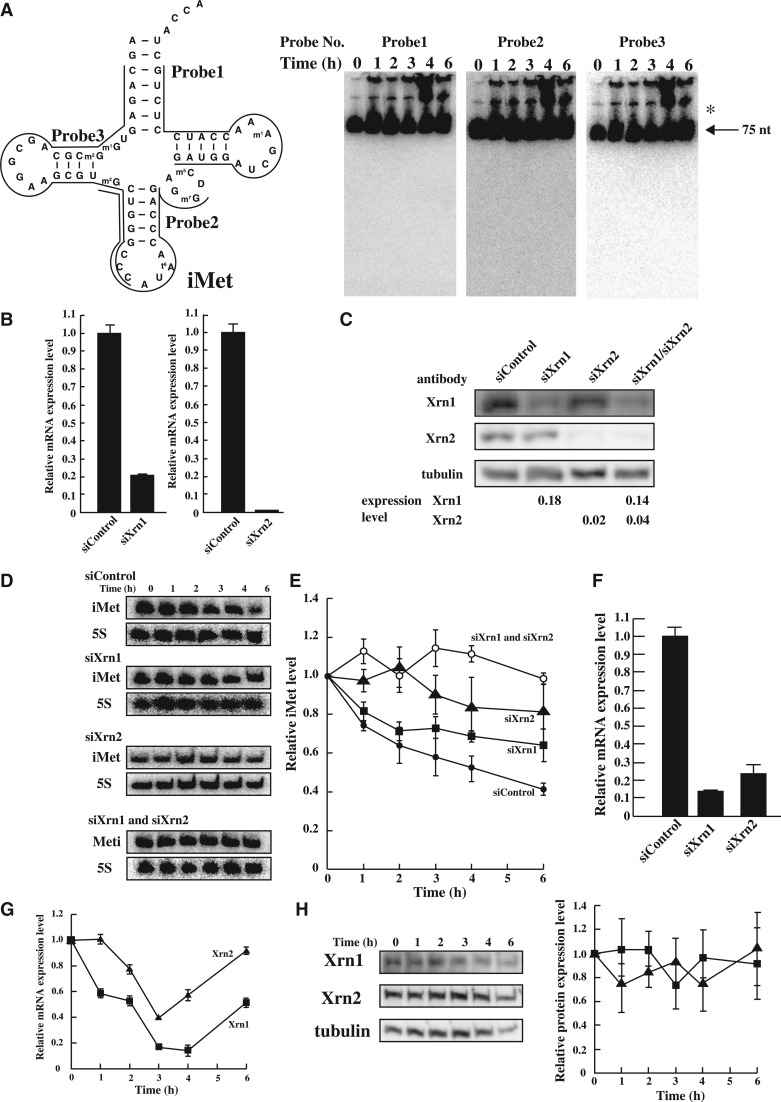
Degradation of tRNA(iMet) by Xrn1 and Xrn2 under heat stress. (A) To identify nuclease(s), a large amount of total RNA (50 µg) was loaded. tRNA(iMet) sequence and the three probe positions that were targeted are shown (left). tRNA(iMet) was detected by northern blot analysis using the three probes (right). Arrow shows mature tRNA(iMet) and asterisk shows the precursor-tRNA(iMet). (B) mRNA level of Xrn1 or Xrn2 by knock-down using specific siRNA was analysed by Real-time PCR. HeLa cells were transfected with non-targeting control siRNA, siXrn1 (left) and siXrn2 (right). (C) Efficiency of knock-down using specific siRNA was determined by western blot analysis. Tubulin was used as a control. (D) Quantification of tRNA(iMet) and 5S rRNA in heat-treated HeLa cells. Knock-down of Xrn1, Xrn2 or Xrn1/2 was detected by northern blot analysis. (E) Degradation profile of tRNA(iMet) was calculated in heated HeLa cells treated with specific siRNA. Filled circles, non-targeting control; squares, siXrn1 treated; triangles, siXrm2 treated; opened circles, double knock-down after treatment with siXrn1 and siXrn2. Degradation of tRNA(iMet) was normalized by 5S rRNA. Data represent the mean±SD for three independent experiments. (F) Double knock-down level of Xrn1 and Xrn2 was analysed by Real-time PCR. (G) mRNA expression levels of Xrn1 (square) and Xrn2 (triangle) were analysed by Real-time PCR under heat stress. (H) Protein expression level of Xrn1 (square) and Xrn2 (triangle) was analysed by western blotting. Tubulin was used as a control.
In our previous report, we showed that the ratio of cytoplasmic to total tRNA(eMet) was slightly decreased after heat stress (18). Taken together, these results suggest that tRNA(eMet) may be degraded in the cytoplasm at 43°C. Indeed, a slight decrease of tRNA(eMet) was observed by northern blot analysis (Figure 1C).
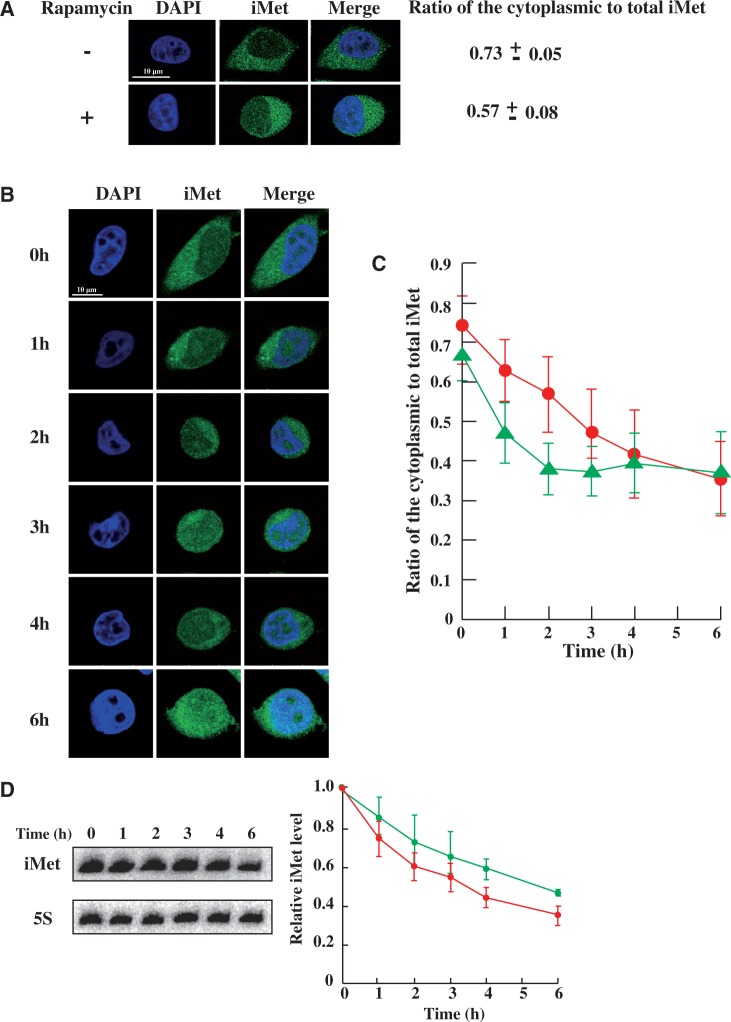
The accumulation pattern of tRNA(iMet) in the nucleus was similar to the degradation pattern of tRNA(iMet) (Figure 3D). This observation suggests that degradation of tRNA(iMet) is prevented by accumulation in the nucleus. If this hypothesis is correct, we might anticipate that the degradation of tRNA(iMet) would be suppressed more rapidly by promotion of nuclear accumulation of tRNA(iMet) using a suitable chemical reagent. It has been reported that nuclear accumulation of tRNAs is inhibited in the presence of rapamycin under amino acid starvation in S. cerevisiae (13). Furthermore, the nuclear accumulation of tRNA(Tyr) is promoted by rapamycin in normal cultured S. cerevisiae (24). To examine whether rapamycin affects the nuclear accumulation of tRNA(iMet) in the HeLa cells, we treated HeLa cells with rapamycin for 30 min and then measured the ratio of cytoplasmic to total tRNA(iMet). The ratio of cytoplasmic to total tRNA(iMet) decreased from 0.73 to 0.57 in HeLa cells after exposure to rapamycin (Figure 4A), similar to the results reported for cytoplasmic tRNA(Tyr) in S. cerevisiae (24). Thus, a rapamycin-dependent tRNA accumulation system is present in mammalian cells.
Figure 4.
Nuclear accumulation of tRNA(iMet) was accelerated by rapamycin, which suppressed its degradation. Nuclear accumulation of tRNA(iMet) in the presence or absence of rapamycin was monitored by FISH. (A) HeLa cells were incubated in the absence (upper) or presence of 5 nM rapamycin (lower) for 30 min. Data represent the mean±SD (n = 40). (B) HeLa cells were heat-shocked in the presence of rapamycin for the time indicated. (C) Ratio of cytoplasmic to nuclear tRNA(iMet) under heat-shock in the absence (red) or presence of rapamycin (green) is shown. Quantification of fluorescence was performed in 40 cells per hour by using the LAS program and the results were averaged. Data represent the mean±SD (n = 40). (D) Degradation profile of tRNA(iMet) in heat-treated cells in the absence (red) or presence (green) of rapamycin. 5S rRNA was used as a control. Data represent the mean±SD of three independent experiments.
Next, we investigated the effect of heat stress in the presence of rapamycin and an RNA polymerase III inhibitor. As shown in Figure 4B and C, rapamycin accelerated the nuclear accumulation of tRNA(iMet) more rapidly under heat stress. To our surprise, the ratio of cytoplasmic to total tRNA(iMet) did not fall below ∼35% even in the presence of rapamycin (Figure 4C). Thus, although we confirmed a similar effect of rapamycin in mammalian cells as reported for yeast cells, there is an additional unknown mechanism(s) that controls the localization or degradation of tRNA(iMet). If all the tRNA(iMet) is accumulated in the nucleus during heat stress, protein expression should be completely arrested. However, a complete cessation of protein biosynthesis would block the expression of heat shock proteins, which are essential factors in the tolerance mechanism of cells. Therefore, it is essential that a constant amount of tRNA(iMet) is retained in the cytoplasm. We reasoned that an unknown mechanism must regulate the accumulation of tRNA(iMet) to maintain a ratio of cytoplasmic to total tRNA(iMet) of 35%.
To test whether the degradation was suppressed by the rapamycin-induced nuclear accumulation, HeLa cells were treated with rapamycin at 43°C and then the degradation of tRNA(iMet) was detected by northern blot analysis (Figure 4D). As expected, the degradation of tRNA(iMet) was suppressed by treatment of the heat-stressed cells with rapamycin. This result indicates that tRNA(iMet) degradation probably occurs in the cytoplasm. In summary, heat stress induces nuclear accumulation of tRNA(iMet), thereby blocking its degradation in the cytoplasm. Therefore, HeLa cells may display a suppression mechanism for tRNA(iMet) degradation.
Nuclear accumulation of tRNA(iMet) is correlated with the mTOR signalling pathway
The mammalian target of rapamycin (mTOR) was the first identified target protein of rapamycin (25). The mTOR signalling pathway, which includes mTOR, p70S6K kinase, eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and others, plays an important role in the regulation of multiple intracellular processes such as cell growth, cell proliferation, autophagy and translational arrest (26–29). Furthermore, it is suggested that tRNA trafficking is controlled by the mTOR signalling pathway in S. cerevisiae (30).
In the current study, we showed that rapamycin accelerated the nuclear accumulation of tRNA(iMet) (Figure 4A–C). This finding indicates that the nuclear accumulation signal of tRNA(iMet) may be correlated with the mTOR signalling pathway under heat stress. The mTOR signalling pathway has been shown to be regulated by phosphorylation of mTOR at Ser2448, mediated by p70S6K kinase (31). Furthermore, phosphorylation of mTOR at a different Ser residue, Ser2481, is known to be the result of self-phosphorylation (32). Therefore, we initially tested whether Ser2448 of mTOR is phosphorylated in rapamycin-treated HeLa cells using western blot analysis (Figure 5). Chiang and Abraham (31) have reported that the phosphorylation of mTOR at Ser2448 is reduced in the presence of rapamycin after 30 min. However, under our experimental conditions, the phosphorylation of mTOR at Ser2448 was slightly increased in the presence of rapamycin after 30 min (Figure 5A). To confirm this result, we independently carried out the experiment twice and the results were averaged. The apparent discrepancy between our results and those of the Abraham group may be due to a difference in the concentration of rapamycin (5 nM under our condition, 100 nM under Abraham group’s condition) and the type of mammalian cells that were used (HeLa cells in our study, MCF-7 in Abraham group’s study). In summary, these results suggest that the phosphorylation of mTOR at Ser2448 may depend on the concentration of rapamycin and on the mammalian cell line.
Figure 5.
Ser2448 phosphorylation level of mTOR. (A) HeLa cells were treated in the absence or presence of rapamycin for 30 min. Phosphorylation level of mTOR at Ser2448 was evaluated in the absence (right) or presence (left) of rapamycin by western blot analysis. Phospho-mTOR was normalized according to the expression level of mTOR. (B) Ser2448 phosphorylation of mTOR was detected under heat stress by western blotting analysis. mTOR was used as a control. (C) Circles, cells heat treated in the absence of rapamycin. Squares, cells heat treated in the presence of rapamycin. The ratio of phospho-mTOR was calculated as 1.0 for none treated zero time point. The expression of mTOR was used as a control. (D) Phosphorylation of mTOR under heat shock in the presence of rapamycin was detected.
Nuclear accumulation of tRNA(iMet) is accelerated in the presence or absence of rapamycin when the cells are heat stressed (Figures 3B and 4B). Next, we tested whether phosphorylation of mTOR at Ser2448 occurs under heat stress. HeLa cells were heat treated at 43°C in the absence of rapamycin (Figure 5B and C). To confirm the phosphorylation level of mTOR at Ser2448, we repeated the western blot analysis twice and the results were averaged. As shown in Figure 5B and C, the level of phosphorylation of mTOR at Ser2448 increased slightly after the onset of heat stress, reaching a maximum at 3 h, and decreased thereafter. The level of phosphorylation at Ser2448 of mTOR increased slightly upon heat stress irrespective of the presence/absence of rapamycin (Figure 5B and C). Moreover, as shown in Figure 4B and C, nuclear accumulation of tRNA(iMet) was accelerated by rapamycin under heat stress. Is the nuclear accumulation of tRNA(iMet) correlated to the phosphorylation of Ser2448? To address this question, we analysed whether the phosphorylation level of Ser2448 was synergistically increased when the cells were heat treated in the presence of rapamycin. Thus, we treated HeLa cells with rapamcyin under heat stress, and assessed the Ser2448 phosphorylation level by western blot analysis (Figure 5C and D). As anticipated, the level of Ser2448 phosphorylation under heat stress in the presence of rapamycin increased more rapidly than when the experiment was conducted in the absence of rapamycin (Figure 5C and D). These results suggest that the nuclear accumulation of tRNA(iMet) may be correlated to Ser2448 phosphorylation of mTOR.
tRNA(iMet) degradation by Xrn1 and Xrn2
tRNA(iMet) was specifically degraded in heat stress-induced HeLa cells (Figure 1C). However, the nuclease(s) responsible for the degradation of tRNA(iMet) during heat stress was/were not identified. To clarify whether tRNA(iMet) was degraded by an endonuclease(s) and/or exonuclease(s), northern blot analysis was performed using total RNAs and three separate probes, designed to hybridize to different regions of tRNA(iMet) (Figure 6A). To detect the fragment(s) of tRNA(iMet), we used a large amount of total RNAs (50 µg) in each lane (Figure 6A). The northern blot confirmed degradation of tRNA(iMet) (data not shown), but no breakdown products were identified. Hence, we tried to detect the fragments of tRNA(iMet) by deliberately overexposing the imaging plate (Figure 6A). However, despite repeated attempts, we could not detect any fragment or ladder derived from tRNA(iMet) (Figure 6A). The fact that we were unable to detect tRNA(iMet) fragments indicates that it was degraded to the nucleotide level within a short period of time. This result strongly suggests that HeLa cells possess a specific and positive degradation mechanism for tRNA(iMet). Two independent studies have detected fragments of tRNAs from HeLa cells under nutrient starvation and oxidative stress (5,7). However, because the heat stress-induced degradation was tRNA(iMet) specific and extremely rapid, our results indicated the presence of a novel degradation pathway.
From the results shown in Figure 6A, we were unable to judge whether the nuclease(s) responsible for the degradation of tRNA(iMet) was an endonuclease and/or exonuclease. We therefore assumed that the candidate exonuclease was a member of the Xrns family (Xrn1, Xrn2 and others) (33,34). The Xrns members of the family are 5′–3′ exonucleases and are highly conserved in eukaryotes. It is well-known that members of the Xrns family mediate various kinds of RNA degradation and have important functions in transcription, RNA metabolism and RNA interference (34,35). Xrn1 is localized to the cytoplasm and is responsible for degrading decapped mRNAs, mRNA lacking a termination codon, mRNAs harbouring a premature termination codon and microRNA species (35–39). Rat1 in S. cerevisiae, the counterpart of Xrn2 in mammals, is localized to the nucleus, and is thought to have a role in mRNA trafficking from the nucleus to the cytoplasm (40,41). In more recent reports, Xrn2 was identified as an exonulcease that degrades mRNA, rRNA and microRNA in S. cerevisiae and mammalian cells (35,39,42,43). Furthermore, Rat1 and Xrn1 promote degradation of hypomodified tRNAs and destabilized tRNAs in S. cerevisiae under heat stress (10,11). Xrn1 and Xrn2 degrade small RNAs (e.g., miRNA) and large RNAs (e.g., rRNA).
Thus, we considered whether the degradation of tRNA(iMet) could be suppressed by knock-down of Xrn1 or Xrn2. To knock-down Xrn1 or Xrn2, HeLa cells were treated with Xrn1- and Xrn2-specific siRNAs. The mRNA expression levels of Xrn1 and Xrn2 were then measured by quantitative PCR (Figure 6B). We independently repeated the experiments twice. Furthermore, the protein expression level of Xrn1 and Xrn2 using specific siRNAs, siXrn1/siXrn2, was determined by western blot analysis (Figure 6C). The experiments were independently repeated twice. We confirmed that specific siRNA species reduced the mRNA and protein expression levels of Xrn1 and Xrn2 to <20% those of the control cells, which were treated with a non-targeting siRNA (Figure 6B and C).
Next, we tested whether degradation of tRNA(iMet) under heat stress was suppressed by treatment with siRNA, siXrn1 or siXrn2, using northern blot analysis (Figure 6D). The degradation pattern of tRNA(iMet) treated with the non-targeting siRNA was similar to that observed in the absence of siRNA treatment (Figure 6D and E). In contrast, the degradation of tRNA(iMet) was significantly suppressed by the knock-down of Xrn1 (Figure 6D and E) showing that one of the responsible nucleases was Xrn1. The degradation curve of tRNA(iMet) in Xrn1 knock-down cells was similar to that in rapamycin-treated cells (Supplementary Figure S5A). Thus, the degradation of tRNA(iMet) was not completely suppressed in HeLa cells treated with siXrn1, suggesting that another nuclease(s) in addition to Xrn1 is probably involved in its breakdown. By contrast, the degradation of tRNA(iMet) was drastically reduced in HeLa cells treated with siXrn2, showing that tRNA(iMet) is principally degraded by Xrn2 in the nucleus (Figure 6D and E). However, the ratio of total tRNA(iMet) was increased in the nucleus (Figure 3B and C). This indicates that tRNA(iMet) may be translocated into the nucleus where the degradation took place. The combined degradation profiles of tRNA(iMet) by Xrn1 and Xrn2 resembled the degradation profile in control cells treated with non-targeting siRNA (Supplementary Figure S5B). This observation suggests that tRNA(iMet) is specifically degraded by both Xrn1 and Xrn2. To confirm this idea, the double knock-down of Xrn1 and Xrn2 was performed (Figure 6B and F). As shown in Figure 6E (siXrn1 and siXrn2), tRNA(iMet) was not degraded in the double knock-down cells under heat stress. Thus, Xrn1 and Xrn2 are the main nucleases responsible for the degradation of tRNA(iMet) during heat stress. Until now, tRNAs have been reported to undergo cleavage by angiogenin in the cytoplasm during oxidative/starvation stress in mammalian cells (5,7). In the current study, it was found that mammalian cells possess different stress-response mechanisms according to the type of stress the cells are subjected. Moreover, we identified a novel degradation pathway that operates in mammalian cells under heat stress.
We investigated whether heat stress has an effect on the transcription of mRNA of Xrn1 or Xrn2. As shown in Figure 6F, the mRNA expression levels of Xrn1 and Xrn2 were decreased by heat stress. Furthermore, the effect of heat stress on the protein expression levels of Xrn1 and Xrn2 was examined by western blot analysis (Figure 6H). We independently repeated the experiments four times. Our results show that the expression levels of Xrn1 and Xrn2 remained unchanged upon heat stress. These results suggest that Xrn1 and Xrn2 proteins may be more stable than Xrn1 and Xrn2 mRNAs during heat shock. Additionally, Xrn1/2 mRNA may be decreased with translational arrest during heat stress.
Finally, we tested whether the localization of Xrn1 and Xrn2 were altered during heat stress by immunofluorescence measurements (Figure 7). As shown in Figure 7A, there was slight translocation of Xrn1 from the cytoplasm to the nucleus when the cells were subjected to heat stress. However, no such translocation of Xrn2 was observed (Figure 7B). Previous reports suggest Xrn1 localizes in processing bodies (so called P-bodies) during normal culture (44,45). However, cytoplasmic foci of Xrn1 were not observed under our experimental set-up (Figure 7A). P-bodies are undetectable in yeast during the exponential phase of growth when cultured under normal conditions (46,47). Because the culture condition of HeLa cells in our study was exponential phase, cytoplasmic foci of Xrn1 may be undetectable. Furthermore, the number of P-bodies in yeast increased over 10–20 min during heat stress (47). However, no P-bodies could be observed when the cells were held under conditions of heat stress for 60 min (47). Because HeLa cells were treated for over 1–6 h in our experimental conditions, cytoplasmic foci of Xrn1 may be undetectable. These results indicate that the activation mechanism of Xrn1 and Xrn2 is not straightforward. Specifically, this activation is not controlled by transcription of Xrn1 and Xrn2 mRNAs, protein expression or major localization changes of Xrn1 and Xrn2.
Figure 7.
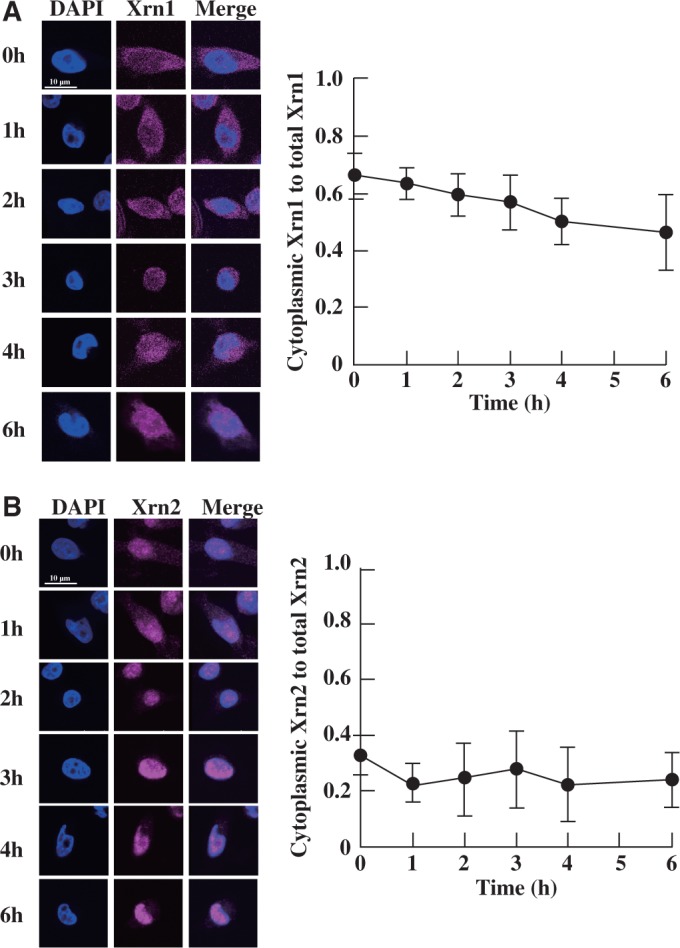
Immunofluorescence of Xrn1 and Xrn2 during heat stress. HeLa cells were heat shocked for different lengths of time, and the localization of Xrn1 and Xrn2 was determined at each time point. Altered localization of Xrn1 (A, left) or Xrn2 (B, left) was analysed by immunofluorescence. Ratio of cytoplasmic Xrn1 to total Xrn1 (A, right) or Xrn2 to total Xrn2 (B, right) is shown. Quantification of fluorescence was performed in 40 cells per hour, and the results were averaged. Data represent the mean±SD (n = 40).
DISCUSSION
Herein, we found that when HeLa cells are exposed to heat, a stress response mechanism is activated that causes tRNA(iMet) to undergo specific nuclease-mediated degradation with concomitant nuclear accumulation (Figure 8). Furthermore, we found that rapamycin accelerates nuclear accumulation of tRNA(iMet). This phenomenon in HeLa cells is similar to that reported in yeast (24). Moreover, Ser2448 phosphorylation of mTOR by heat stress may correlate with the accumulation of tRNA(iMet) in the nucleus. Additionally, using a knock-down procedure mediated by specific siRNA species, we identified the nucleases responsible for the degradation of tRNA(iMet) to be Xrn1 in the cytoplasm and Xrn2 in the nucleus.
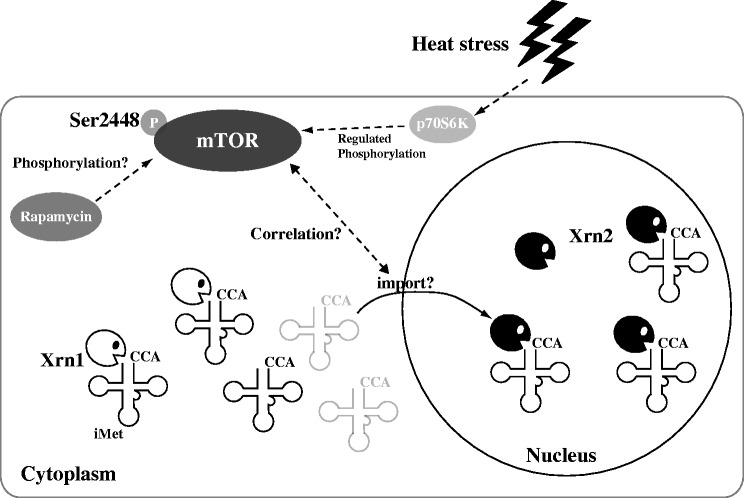
Figure 8.
Summary of the heat stress response in HeLa cells. When HeLa cells are exposed to heat stress, tRNA(iMet) is specifically degraded by Xrn1 in the cytoplasm and Xrn2 in the nucleus, and the rate of nuclear accumulation of tRNA(iMet) is accelerated. Furthermore, nuclear accumulation of tRNA(iMet) is accelerated by rapamycin. Ser2448 of mTOR is slightly phosphorylated by heat stress and/or treatment with rapamycin. The phosphorylation of Ser2448 may correlate with the concentration of tRNA(iMet).
What regulates activation of Xrn1 and Xrn2? Recent reports indicate that Rny1 and angiogenin are key proteins involved in the cleavage of tRNA (4,48). Rny1 is segregated from cytoplasmic tRNAs in the absence of stress, because Rny1 is localized to the vacuole (9). However, Rny1 and other vacuolar proteins appear to exit the vacuole and move into the cytoplasm under oxidative stress, allowing access to cytoplasmic RNAs (9). Furthermore, Xrn1 and Rat1 are regulated in heat stress-induced S. cerevisiae by Met22, which is involved in salt tolerance, methionine biogenesis and the dephosphorylation of 3′-phosphoadenosine-5′-phosphate (11). As shown in Figure 7B, to our surprise, a signal of Xrn2 was slightly detected in the nucleolus at 0 h. However, time-dependent changes were not observed with this signal in the nucleolus, and Xrn2 was uniformly distributed throughout the nucleus. This result suggests that altered localization of Xrn2 may be a heat stress-induced activation mechanism of Xrn2, akin to the activation mechanism of Rny1. Additionally, as demonstrated in our previous report, after tRNA(iMet) and tRNA(eMet) has accumulated in the nucleus during heat stress, these tRNAs form nuclear tRNA granules around the nucleolus (18). This phenomenon suggests that localization of tRNA(iMet) in nuclear tRNA granules may protect it from Xrn2-mediated breakdown, thereby repressing its degradation.
The melting temperature of tRNA(iMet) from normally cultured HeLa cells was similar to that of tRNA(eMet) (Figure 2D). This result suggests that Xrn1 and Xrn2 may also recognize the sequence of tRNA(iMet). Measurement of melting temperature did not reveal the partial stability of tRNA. It was recently reported from a nuclease sensitivity assay with RNase T2 that the RTD pathway that includes Rat1 and Xrn2, monitors the structural integrity of the acceptor and T-stem of tRNA and degrades unstable tRNA species (12). The structure of the combined acceptor and T-stem of tRNA(iMet) destabilized the structure of tRNA(eMet) by 0.6 kcal/mol, as calculated using the RNA structure program (49). Because the energy difference of 0.6 kcal/mol between tRNA(iMet) and tRNA(eMet) by calculation could be related to recognition of Xrn1 and Xrn2, it is necessary to clarify the problem of the structural stabilization of the acceptor and T-stem in tRNA(iMet).
Our results indicate that accumulation of tRNA(iMet) may be correlated with the mTOR signalling pathway (Figures 4A, 5A–D and 8). In a recent report, the phosphorylation level at Ser2448 of mTOR was shown to be regulated by the expression level of exportin-t (Xpo-t) (50). Xpo-t binds newly synthesized tRNAs in the nucleus and mediates their transport to the cytoplasm. In yeast, knock-out of LOS1, which is the yeast counterpart of mammalian Xpo-t, leads to impaired tRNAs splicing and accumulation in the nucleus (50). In nutrient starvation-induced fibroblasts, the degree of phosphorylation of mTOR at Ser2448 is lowered by knock-down of Xpo-t. Thus, phosphorylation of mTOR is regulated by the expression level of Xpo-t. Our current results and the report by Huynh suggest that phosphorylation of mTOR at Ser2448 may be controlled by Xpo-t in mammalian cells.
Recently, accumulation of tRNA in the nucleus was shown to occur under starvation of amino acids or glucose in S. cerevisiae (13). However, tRNA accumulation is not accelerated in the presence of rapamycin under glucose starvation (13). This result indicates that stress responses triggered by amino acid starvation and glucose starvation operate via distinct pathways in S. cerevisiae (13). In this study, nuclear accumulation of tRNA(iMet) is accelerated in the presence of rapamycin under heat stress (Figure 3B and C). Our results, together with those reported by Whitney et al. (13), suggest that the nuclear accumulation pathway during heat stress in mammalian cells may be similar to the stress response initiated by amino acid starvation in S. cerevisiae.
Why is tRNA(iMet) degraded and accumulated in the nucleus during heat stress? Under heat stress, translational arrest is brought about by phosphorylation of eIF2α (1). In a report by Barhoom et al., yeast tRNAs transfected into CHO were found to accumulate in the nucleus in the presence of puromycin, which induces translational arrest (22). In this report, tRNA(iMet) was accumulated in the nucleus (Figure 3B and C). Nuclear accumulation of tRNA(iMet) may reflect translational arrest induced by heat stress. Is translational arrest simply the result of a decrease in the level of cytoplasmic tRNA(iMet)? In a recent report, DNA damage was detected in mammalian cells after heat stress (51). Furthermore, tRNA accumulates in the nucleus as a consequence of MMS-induced DNA damage in S. cerevisiae (21). Moreover, cancer cell survival following DNA damage is regulated by mTOR in mammalian cells (52). Hence, nuclear accumulation of tRNA(iMet) not only activates translational arrest, but may also act as a sensor of DNA damage, a trigger of cell cycle arrest and autophagy as well as other cellular events.
Until now, angiogenin has been reported to be the only key endonuclease responsible for RNA degradation in mammalian cells under various forms of stress. Angiogenin cleaves the anticodon loop of mature tRNA to produce 5′- and 3′-fragments during nutrient starvation and oxidative stress (7,8). The 5′-fragments trigger the assembly of stress granules in U2OS cells (53). If angiogenin cleaves the anticodon loop of tRNA(iMet) during heat stress, the 5′-fragments may induce the assembly of stress granules, while the 3′-fragment is degraded by Xrn1/2. The full-length tRNA(iMet) was detected using this probe, but the fragment and half molecules of tRNA were not. In this study, when the expression levels of Xrn1/2 were reduced using specific siRNA, the degradation of tRNA(iMet) was almost completely abolished. Thus, the major degradation factors are Xrn1/2. Additionally, cytoplasmic foci were not observed by FISH analysis. Hence, the major factors in our identified tRNA degradation pathway were Xrn1/2. Nonetheless, based on the findings in this article, we cannot completely rule out the possibility that angiogenin might also participate in tRNA degradation. In conclusion, Xrn1/2 are the major factors in tRNA degradation although angiogenin may also be involved.
Our results suggest that tRNA(iMet) is recognized and degraded under heat stress conditions by two exonucleases, Xrn1 and Xrn2, in the cytoplasm and the nucleus, respectively. It is reported that unstable tRNAs are recognized by Xrn1 and Rat1 during heat stress in yeast (12). Moreover, Phizicky’s group used in vivo analysis to show that Xrn1 primarily degrades unstable tRNA(Ser) species (12). The identified degradation pathway in heat stress-induced human cells includes Xrn1 and Xrn2, indicating the degradation pathway in human may resemble the RTD pathway in yeast.
Our findings, together with recent reports by others, indicate that stress responses differ in terms of heat stress and nutrient starvation/oxidative stress. The current study contributes to our knowledge concerning stress-induced RNA degradation and accumulation in mammalian cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Japan Society for the Promotion of Science (JSPS) Research Fellowships for Young Scientist [11J04597 to R.M.] (in part); The Naito Foundation (to K.W.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Komiyama (Tokyo University) and Dr Aiba (Tokyo University) for their help in the measurement of melting points and Dr Hoshino (Nagoya City University) for helpful comments on the article.
REFERENCES
- 1.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol. Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell. Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 8.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell. Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes. Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol. Biol. Cell. 2007;18:2678–2686. doi: 10.1091/mbc.E07-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chafe SC, Pierce JB, Eswara MB, McGuire AT, Mangroo D. Nutrient stress does not cause retrograde transport of cytoplasmic tRNA to the nucleus in evolutionarily diverse organisms. Mol. Biol. Cell. 2011;22:1091–1103. doi: 10.1091/mbc.E09-07-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc. Natl Acad. Sci. USA. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2010;39:2304–2318. doi: 10.1093/nar/gkq1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomikawa C, Yokogawa T, Kanai T, Hori H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010;38:942–957. doi: 10.1093/nar/gkp1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagawa R, Mizuno R, Watanabe K, Ijiri K. Formation of tRNA granules in the nucleus of heat-induced human cells. Biochem. Biophys. Res. Commun. 2012;418:149–155. doi: 10.1016/j.bbrc.2011.12.150. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, et al. Novel small-molerule inhibitors of RNA polymerase III. Eukaryot Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in S. cerevisiae. Mol. Biol. Cell. 2010;21:639–649. doi: 10.1091/mbc.E09-07-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 22.Barhoom S, Kaur J, Cooperman BS, Smorodinsky NI, Smilansky Z, Ehrlich M, Elroy-Stein O. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res. 2011;39:e129. doi: 10.1093/nar/gkr601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;208:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 24.Pierce JB, Eswara MB, Mangroo D. The ins and outs of nuclear re-export of retrogradely transported tRNAs in Saccharomyces cerevisiae. Nucleus. 2010;1:224–230. doi: 10.4161/nucl.1.3.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartford CM, Ratain MJ. Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin. Pharmacol. Ther. 2007;82:381–388. doi: 10.1038/sj.clpt.6100317. [DOI] [PubMed] [Google Scholar]
- 26.Xiang X, Zhao J, Xu G, Li Y, Zhang W. mTOR and the differentiation of mesenchymal stem cells. Acta Biochim. Biophys. Sin. 2011;43:501–510. doi: 10.1093/abbs/gmr041. [DOI] [PubMed] [Google Scholar]
- 27.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrate p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 28.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1466. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuefeld TP, Arsham A. tRNA trafficking along the TOR pathway. Cell Cycle. 2010;9:3146–3150. doi: 10.4161/cc.9.16.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 32.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J. Biol. Chem. 2009;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that atXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newbury SF. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 2006;34:30–34. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 35.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5'-3' exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 2011;18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 37.Hsu CL, Stevens A. Yeast cells lacking 5′ to 3′ exoribonuclease I contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 39.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 41.Kenna M, Stevens A, McCammon M, Douglas MG. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol. Cell. Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010;38:4487–4502. doi: 10.1093/nar/gkq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. RNA. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 44.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 45.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 46.Teixeria D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowart LA, Gandy JL, Tholanikunmel B, Hannun YA. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 2010;431:31–38. doi: 10.1042/BJ20100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nawrot B, Sochacka E, Duchler M. tRNA structural and functional changes induced by oxidative stress. Cell. Mol. Life Sci. 2011;68:4023–4032. doi: 10.1007/s00018-011-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-129. 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huynh LN, Thangavel M, Chen T, Cottrell R, Mitchell JM, Praetorius-Ibba M. Linking tRNA localization with activation of nutritional stress responses. Cell Cycle. 2010;9:3112–3118. doi: 10.4161/cc.9.15.12525. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi A, Matsumoto H, Nagayama K, Kitano M, Hirose S, Tanaka H, Mori E, Yamakawa N, Yasumoto J, Yuki K, et al. Evidence for the involvement of double-strand breaks in heat-induced cell killing. Cancer Res. 2004;64:8839–8845. doi: 10.1158/0008-5472.CAN-04-1876. [DOI] [PubMed] [Google Scholar]
- 52.Back JH, Rezvani HR, Zhu Y, Guyonnet-Duperat V, Athar M, Ratner D, Kim AL. Cancer cell survival following DNA damage-mediated premature senescence is regulated by mammalian target of rapamycin (mTOR)-dependent Inhibition of sirtuin 1. J. Biol. Chem. 2011;286:19100–19108. doi: 10.1074/jbc.M111.240598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu G, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.