Abstract
Through controlling the nuclear level of active positive transcription elongation factor b (P-TEFb), the 7SK small nuclear RNA (snRNA) functions as a key regulator of RNA polymerase II transcription. Together with hexamethylene bisacetamide-inducible proteins 1/2 (HEXIM1/2), the 7SK snRNA sequesters P-TEFb into transcriptionally inactive ribonucleoprotein (RNP). In response to transcriptional stimulation, the 7SK/HEXIM/P-TEFb RNP releases P-TEFb to promote polymerase II-mediated messenger RNA synthesis. Besides transiently associating with HEXIM1/2 and P-TEFb, the 7SK snRNA stably interacts with the La-related protein 7 (Larp7) and the methylphosphate capping enzyme (MePCE). In this study, we used in vivo RNA–protein interaction assays to determine the sequence and structural elements of human 7SK snRNA directing assembly of the 7SK/MePCE/Larp7 core snRNP. MePCE interacts with the short 5′-terminal G1-U4/U106-G111 helix-tail motif and Larp7 binds to the 3′-terminal hairpin and the following U-rich tail of 7SK. The overall RNA structure and some particular nucleotides provide the information for specific binding of MePCE and Larp7. We also demonstrate that binding of Larp7 to 7SK is a prerequisite for in vivo recruitment of P-TEFb, indicating that besides providing stability for 7SK, Larp7 directly participates in P-TEFb regulation. Our results provide further explanation for the frequently observed link between Larp7 mutations and cancer development.
INTRODUCTION
Controlling the elongation capacity of RNA polymerase II (Pol II) by positive transcription elongation factor b (P-TEFb) is a fundamental step of eukaryotic transcription regulation (1–3). After transcription initiation and synthesis of the 20–30 nt long 5′-terminal RNA sequences, Pol II is arrested by negative elongation factors (4,5). To release the promoter proximally paused Pol II and to couple transcription elongation with pre-messenger RNA processing, P-TEFb phosphorylates the negative elongation factors and the heptapeptide repeats (YSPTSPS) in the carboxy-terminal domain of Pol II at serine 2. P-TEFb is a general and essential transcription factor that is composed of the cyclin-dependent kinase Cdk9 and its regulatory subunit cyclin T1 (CycT1) or less frequently CycT2 (6).
The nucleoplasmic level of active P-TEFb is controlled by the 7SK transcriptional regulatory small nuclear RNA (snRNA) (7–10). The human 7SK snRNA is an abundant, 331-nt long Pol III-synthesized nucleoplasmic RNA that is composed of a long 5′-terminal, a short 3′-terminal and two internal hairpin domains (11). Together with the homo- or heterodimer of the hexamethylene bisacetamide-inducible proteins 1 and 2 (HEXIM1 and HEXIM2), the 7SK snRNA recruits P-TEFb and inhibits its kinase activity (12–19). HEXIM1 and HEXIM2 are RNA-binding proteins, which bind to the 5′-terminal hairpin of 7SK snRNA with great specificity (13,20,21). The 5′-hairpin of 7SK carries two structurally similar HEXIM-binding motifs, which recruit HEXIM1/2 in a strictly interdependent manner in living cells (20). A conformational rearrangement induced by 7SK docking enables the C-terminal inhibitory domain of HEXIM1/2 to interact with the CycT1 regulatory subunit of P-TEFb and thereby, to inhibit the kinase activity of Cdk9 (13,15,16,22–24). Association of 7SK snRNA with HEXIM1/2 and P-TEFb is a highly dynamic process. It is controlled by a largely unknown signalling mechanism that adjusts the nuclear equilibrium of active and inactive P-TEFb to the actual transcriptional condition of the cell. On transcriptional arrest, the 7SK/HEXIM/P-TEFb transcriptional inhibitory small nuclear ribonucleoprotein (snRNP) is rapidly disassembled to release active P-TEFb and to promote Pol II transcription (7,8). At the same time, the free 7SK snRNA associates with a set of heterogeneous nuclear ribonucleoproteins (hnRNP) proteins, including hnRNP A1, A2/B1, R and Q (25–27).
In contrast to the transiently associating HEXIM1/2, CycT1, Cdk9 and hnRNP proteins, the methylphosphate capping enzyme (MePCE) (also known as BCDIN3) and La-related protein 7 (Larp7) (PIP7S) ribonucleoproteins stably interact with the 7SK snRNA to constitute the 7SK/MePCE/Larp7 core snRNP (28–33). As integral components of the 7SK snRNP, MePCE and Larp7 provide metabolic stability for the 7SK snRNA. MePCE has a methyltransferase domain, and it is responsible for monomethylation of the gamma phosphate of the 5′-terminal guanosine-triphosphate of nascent 7SK snRNA (30,33). Larp7 belongs to the family of La-related proteins, and it binds to the 3′-terminal U-rich tail of 7SK through its N-terminal La module that is composed of the conserved La motif and the following RNA recognition motif 1 (RRM1) (28,29,31,32). Diminution of 7SK snRNA increases the nuclear level of active P-TEFb that eventually leads to malignant transformation of cultured mammary epithelial cells (29). Consistent with this, Larp7 mutations or downregulation have been found to promote gastric and cervical tumorigenesis (34,35).
To understand the structural organization of the 7SK/MePCE/Larp7 transcriptional regulatory core snRNP, in this study, we have determined the structural elements of human 7SK snRNA directing the in vivo binding of MePCE and Larp7. We have also shown that binding of Larp7 to the 7SK snRNA is necessary for P-TEFb recruitment, demonstrating that in living cells, the 7SK/MePCE/Larp7 core snRNP, rather than the 7SK RNA alone, possesses the capacity to control P-TEFb activity.
MATERIALS AND METHODS
General procedures
Unless indicated otherwise, all techniques used for cloning of DNA and for manipulating RNA, oligodeoxynucleotides and proteins were performed according to standard laboratory protocols (36). Oligodeoxynucleotides were synthesized by Eurofins MWG. HeLa and G3H cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen). Expression plasmids were introduced into HeLa and G3H cells by using jetPRIME transfection reagent as recommended by the manufacturer (Ozyme).
Construction and structure of expression plasmids
Construction of the p7SK plasmid designed for transient expression of human 7SK snRNA has been described (21). Mutant 7SK expression constructs were generated by polymerase chain reaction (PCR) amplification using mutagenic oligodeoxynucleotide primers and the p7SK plasmid or its appropriate derivatives as DNA templates. The p7SKd5′h (ΔU6-A101), p7SK5′h (ΔU112-C324), p7SK5′h + 3′h (ΔU116-C295), p7SKd1 (ΔA296-C304 and ΔC320-C324), p7SKd2 (ΔG305-U319), p7SKd3 (ΔA310-G314), p7SKd4 (ΔC320-U321) and p7SKd5 (ΔC102-C114) expression plasmids carried the indicated internal deletions in the coding region of p7SK. In the p7SKm1 (A296-A301), p7SKm2 (G302-C304), p7SKm3 (G322-C324), p7SKm4 (G305-C309), p7SKm5 (G315-U319), p7SKm6 (C320-U321), p7SKm2/3 (G302-C304 and G322-C324), p7SKm4/5 (G305-C309 and G315-U319) and p7SKtag2 (C233-A245) expression plasmids, the indicated sequences of the coding region of p7SK were replaced with complementary sequences. The 7SKtag1 construct carried the C108 to G and 111-GUG-113 to ACC nucleotide alterations. The p7SK3′U construct contained the C328 to U point mutation; otherwise it was identical to p7SKtag1. The p7SKL1 (A310 to U), p7SKL2 (U311 to A), p7SKL3 (G312 to C), p7SKL4 (U313 to A) and p7SKL5 (G314 to C) plasmids was based on p7SKtag1, but they carried the indicated nucleotide transversions. The 7SK5′hm1 (G2), 7SK5′hm2 (A3), 7SK5′hm3 (U4), 7SK5′hm4 (U106-C108), 7SK5′hm5 (A109-G111) and 7SK5′hm6 (C103-C105) plasmids were generated through introduction of complementary nucleotides into the indicated positions of p7SK5′h. To obtain p7SK5′h + 3′hm7 (A310-G314) and p7SK5′hm7 + 3′hm7 (G2-U4 and A310-G314), the indicated sequences (according to the numbering of wild-type 7SK) of the p7SK5′h + 3′h expression plasmid were replaced with complementary nucleotides. To generate pU1P-7SKtag2, the 7SK coding region of the p7SKtag2 expression plasmid was PCR-amplified and used to replace the U1 snRNA coding region in the phU1 expression plasmid (37). To obtain the FL-LARP7 expression construct, the full-length complementary DNA of human Larp7 was cloned by reverse transcriptase-PCR and cloned into the p3XFLAG-CMV-10 expression vector that encodes three adjacent Flag epitopes upstream of the inserted Larp7 complementary DNA (Sigma). The pMePCE-FL expression plasmid was purchased from Origene (RC200948).
Immunoprecipitation
Extract preparation from transfected or non-transfected HeLa and G3H cells and immunoprecipitation (IP) of endogenous Larp7, CycT1 and La or ectopically expressed MePCE-FL, FL-Larp7 and HA-CycT1 proteins were performed as described before (21,26). Antibodies specific for the Flag (Sigma, M2) and HA (Roche, 12CA5) epitope tags or for the human Larp7 (Aviva Systems Biology, ARP40847-P050), La (OriGene, TA500406) and CycT1 (Abcam, ab2098) proteins have been purchased. Polyclonal anti-2,2,7-trimethyl-G (TMG) antibody was kindly provided by Dr. R. Lührmann (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany).
RNA analysis
Isolation of RNA from transfected or non-transfected HeLa and G3H cells was performed by using the guanidinium thiocyanate/phenol-chloroform extraction procedure (38). From the pellet of IP reactions, RNA was isolated by phenol–chloroform extraction. RNAs co-immunoprecipitated with Larp7 and MePCE were 3′ end-labelled with [5′-32P]pCp and T4 RNA ligase and fractionated on a 6% sequencing gel (39). For northern blot analysis, RNAs were separated on a 6% denaturing gel and electroblotted onto a Hybond-N nylon membrane (Amersham Biosciences). The filters were probed with 5′-terminally labelled oligonucleotides complementary to the human 7SK, U6 and U4 snRNAs. To generate sequence-specific antisense RNA probes for mapping of transiently expressed mutant 7SK RNAs, the corresponding 7SK expression plasmids were digested with PstI and used as templates for in vitro transcription with T7 RNA polymerase in the presence of [α-32P]CTP (3000 Ci/mmol). RNase A/T1 protection analysis has been described (38).
RESULTS
HeLa cellular RNAs associated with Larp7 and MePCE
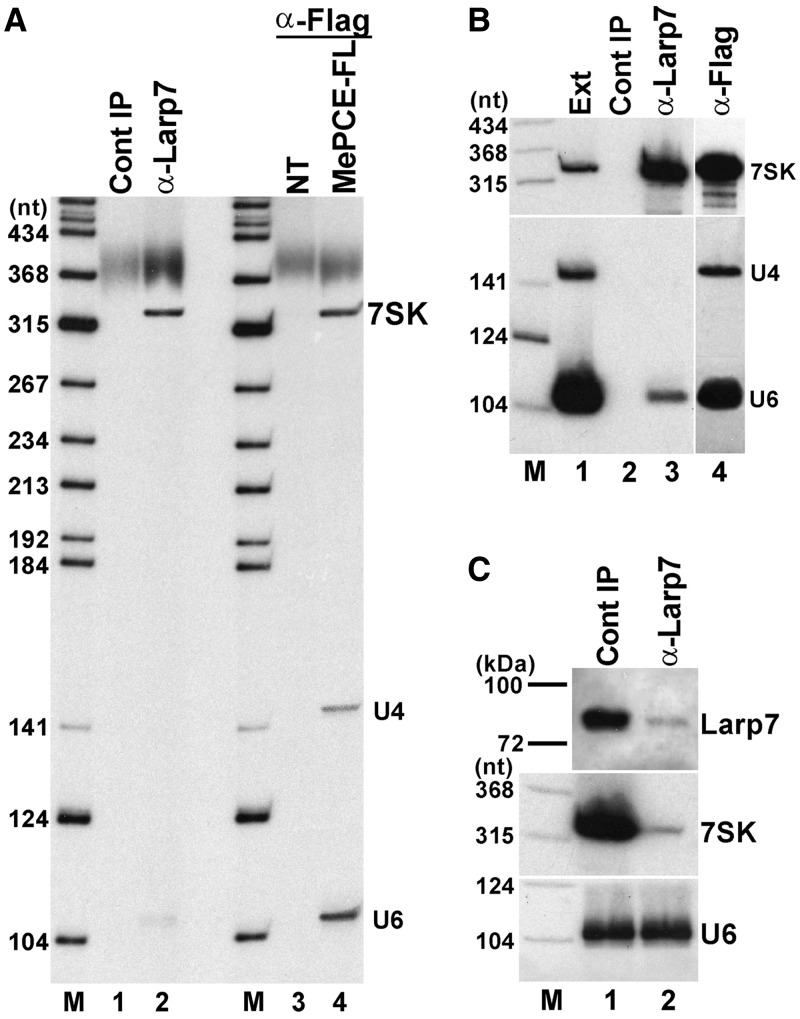
Previous works demonstrated that Larp7 and MePCE bind to the 7SK and U6 snRNAs, but it remained unclear whether they possess other target RNAs in the cell (29,31,32). To define the complete repertoire of human HeLa cellular RNAs interacting with Larp7 and MePCE, endogenous Larp7 and transiently expressed Flag-tagged MePCE were immunoprecipitated with Larp7- and Flag-specific antibodies. RNAs co-purified with Larp7 and MePCE-FL were 3′ end-labelled with [5′-32P]pCp and T4 RNA ligase and separated on a sequencing gel (Figure 1A). IP of Larp7 recovered the 7SK snRNA but, in contrast to a previous report (31), failed to pull down significant amount of U6 snRNA (lane 2). On the other hand, MePCE-FL showed a strong in vivo association with both 7SK and U6 and another HeLa RNA that, based on its gel electrophoretic mobility, was predicted to be the U4 spliceosomal snRNA that is known to form a heterodimer with U6 (lane 4). Northern blot analysis confirmed the identity of all Larp7- and MePCE-associated RNAs and further corroborated that Larp7 interacts with only a small fraction (1–2%) of HeLa U6 snRNA and that MePCE associates with not only the U6 mono-snRNP but also the U6/U4 di-snRNP (Figure 1B). To further assess the extent of Larp7 association with 7SK and U6 snRNAs, we measured the 7SK and U6 levels in HeLa cell extracts before and after immunodepletion of Larp7 (Figure 1C). Northern blot analysis demonstrated that depletion of ∼98% of Larp7 removed >96% of 7SK from the extract, but it had no detectable effect on the level of U6. Thus, we concluded that human MePCE is an integral component of both 7SK and U6 snRNPs, whereas Larp7 binds predominantly, if not exclusively (see ‘Discussion’ section), to the 7SK snRNP.
Figure 1.
In vivo association of Larp7 and MePCE with human cellular RNAs. (A) Detection of RNAs associated with Larp7 and MePCE. HeLa RNAs co-immunoprecipitated with endogenous Larp7 and transiently expressed Flag-tagged MePCE were 3′-terminally labelled and separated on a 6% sequencing gel. Control IP reactions performed from non-transfected cells (NT) or without antibody (Cont IP) are shown. Lane M, size markers in nucleotides. (B) Northern blot analysis. HeLa RNAs co-immunoprecipitated with Larp7 and MePCE-FL were separated on a sequencing gel, electroblotted onto a Nylon membrane and probed with oligodeoxynucleotide probes specific for the 7SK, U4 and U6 snRNAs. RNAs isolated from cell extracts (Ext) and pellets of control IPs (Cont IP) were also analysed. (C) The majority of HeLa 7SK snRNA is associated with Larp7. The levels of Larp7, 7SK and U6 were determined by western and northern blot analyses in Larp7-depleted (α-Larp7) and mock-depleted (Cont IP) extracts.
The 3′-hairpin and the following oligouridylate tail of 7SK work together in Larp7 binding
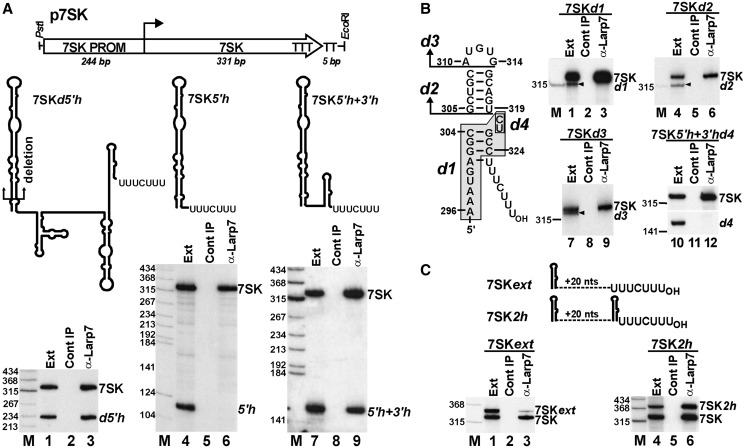
It has been demonstrated that under in vitro conditions, Larp7 binds to the 3′-terminal U-rich tail of 7SK through its La module (28,29,32). However, it remained unknown how Larp7 can distinguish between 7SK and other Pol III-transcripts, which also carry oligouridylate 3′-termini. To define 7SK elements providing specificity for Larp7 binding, we tested the in vivo Larp7-binding capacity of internally truncated 7SK RNAs, which were transiently expressed in HeLa cells by using the p7SK expression vector (21) (Figure 2A). After extract preparation, Larp7 was immunoprecipitated, and co-purification of the expressed mutant 7SK test RNAs and, as a positive control, the endogenous HeLa 7SK RNA was monitored by northern blot analysis. As expected, removal of the 5′-terminal hairpin of 7SK did not influence the Larp7-binding capacity of the resulting 7SKd5′h RNA (lane 3). However, the 3′-terminally truncated 7SK5′h RNA failed to interact with Larp7, although it retained the wild-type 3′-terminal U-rich sequence (UUUCUUU) of 7SK (lane 6). Insertion of the wild-type 3′-hairpin of 7SK before the U-rich tail of 7SK5′h fully restored the Larp7-binding ability of the resulting 7SK5′h + 3′h RNA (lane 9). These results demonstrated that all RNA elements required for specific and efficient in vivo binding of Larp7 are located in the 3′-hairpin and the following U-rich tail of human 7SK snRNA.
Figure 2.
7SK elements required for in vivo binding of Larp7. (A) Association of transiently expressed truncated 7SK RNAs with HeLa Larp7. Organization of the p7SK expression construct with the relevant sites is shown. Predicted schematic structures of the expressed internally truncated 7SK RNAs are shown. Co-IP of 7SK test RNAs and the endogenous HeLa 7SK snRNA with Larp7 was monitored by northern blot analysis. RNAs from cell extracts (Ext) and control IPs (no antibody) were also analysed. (B) Association of HeLa Larp7 with transiently expressed 7SK RNAs carrying truncated 3′-hairpins. Structure of the 3′-hairpin of 7SK test RNAs is shown. Deleted nucleotides are boxed (d1 and d4) or they are indicated by arrows (d2 and d3). Transiently expressed 7SKd1, 7SKd2, 7SKd3 and 7SK5′h + 3′hd4 RNAs were co-immunoprecipitated with Larp7 and analysed by northern blotting. Endogenous 7SK is indicated. (C) The 3′-terminal hairpin and the following U-rich tail of 7SK work together in Larp7 binding. Schematic structures of the 3′-terminal regions of 7SKext and 7SK2h RNAs are shown. Dashed lines indicate random sequences inserted between the 3′-hairpin and the terminal U-rich tail. Co-IP of transiently expressed 7SKext and 7SK2h RNAs with Larp7 was measured by northern blot analysis.
To confirm the importance of the 3′-hairpin of 7SK for Larp7 binding, we investigated the in vivo association of HeLa Larp7 with transiently expressed 7SK RNAs carrying partially truncated 3′-hairpins (Figure 2B). Northern blot analysis of RNAs co-immunoprecipitated with Larp7 revealed that deletion of the basal helix region (d1), the terminal stem-loop (d2) or the terminal loop (d3) of the 3′-hairpin of 7SK fully abolished the in vivo association of the mutant 7SK RNAs with Larp7 (lanes 3, 6 and 9). Likewise, deletion of the C320-U321 internal bulge nucleotides (d4) fully eliminated the in vivo Larp7-binding ability of the resulting 7SK5′h + 3′hd4 RNA (lane 12). It is noteworthy that all 7SK RNAs lacking Larp7-binding ability showed a reduced accumulation compared with RNAs carrying the wild-type 3′-terminal hairpin-tail region. This further supports the notion that Larp7 controls the cellular stability of 7SK snRNA.
To assay the functional interdependence of the 3′-terminal U-rich tail and the preceding 3′-hairpin, these two elements of 7SK were separated by a 20-nt long random sequence in the 7SKext test RNA (Figure 2C). When expressed in HeLa cells, the accumulating 7SKext RNA failed to efficiently interact with Larp7 (lane 3). However, insertion of another copy of the 7SK 3′-hairpin before the U-rich terminal sequence of 7SKext fully restored the Larp7-binding activity of the resulting 7SK2h RNA (lane 6). These results demonstrated that the 3′-terminal hairpin and the following U-rich tail of 7SK snRNA together constitute the docking site for Larp7.
7SK RNA elements supporting specific binding of Larp7
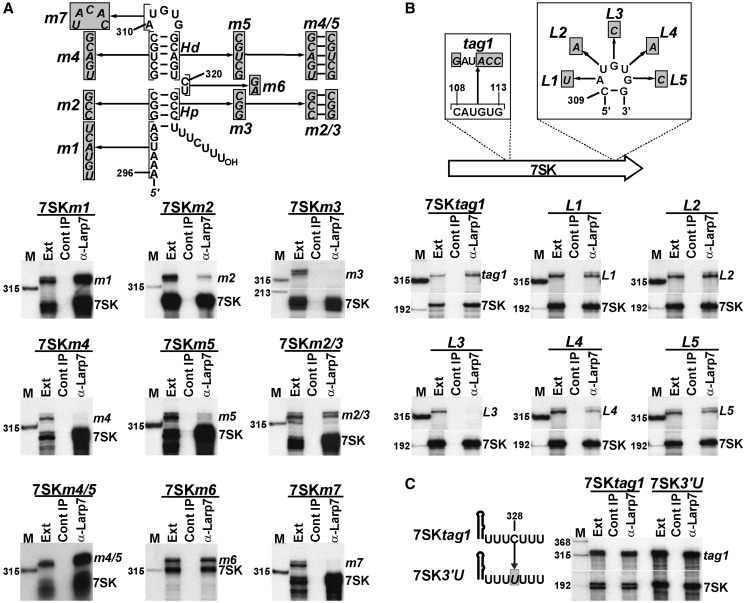
To further dissect the RNA structural determinants of Larp7 binding to 7SK, a series of sequence alterations were introduced into the 3′-terminal hairpin of 7SK snRNA (Figure 3A). The altered 7SK RNAs were expressed in HeLa cells, and their association with Larp7 was assayed. To distinguish between transiently expressed mutant and endogenous 7SK RNAs, HeLa RNAs co-immunoprecipitated with Larp7 were mapped by RNase A/T1 protection using RNA probes complementary to the test RNAs. The protected probe RNAs were separated on a sequencing gel. Alteration of the single-stranded A296-A301 sequence preceding the 3′-hairpin of 7SK failed to alter the Larp7-binding ability of the mutant 7SKm1 RNA. In contrast, disruption of the short proximal helix (Hp, 7SKm2 and 7SKm3) or the distal helix (Hd, 7SKm4 and 7SKm5) of the 3′-hairpin largely inhibited or completely abolished the Larp7-binding capacity of the Hp- and Hd-mutant RNAs, respectively. Restoration of the proximal and distal helices through combining the m2 and m3 or the m4 and m5 mutations fully re-established the in vivo interaction of 7SKm2/3 and 7SKm4/5 RNAs with Larp7, indicating that these helix structures themselves, rather than their nucleotide compositions, are important for Larp7 binding. Substitution of the C320-U321 bulge nucleotides had no significant effect on the Larp7-binding activity of 7SKm6 RNA. This indicates that the nucleotide composition of the C320-U321 internal bulge is not crucial, although its presence is essential for Larp7 binding (see Figure 2B). Replacement of the A310-G314 terminal loop sequence with its complementary sequence fully eliminated the in vivo interaction of the altered 7SKm7 RNA with Larp7, demonstrating that the nucleotide composition of the A310-G314 terminal loop is important for Larp7 binding.
Figure 3.
In vivo association of Larp7 with mutant 7SK RNAs. (A) Structural requirements of Larp7 binding to the 3′-hairpin of 7SK. Mutant 7SK RNAs carrying sequence alterations (in shaded boxes) in their 3′-hairpin were expressed in HeLa cells, and their co-IP with endogenous Larp7 was measured by RNase A/T1 mapping. For other details, see the legend to Figure 2. (B) Mutational analysis of the terminal loop of the 3′-hairpin of 7SK. Association of HeLa Larp7 with transiently expressed 7SKtag1 RNAs carrying the L1 to L5 nucleotide alterations was investigated by co-IP and RNase A/T1 mapping. (C) The C328 residue in the 3′-terminal U-rich stretch of 7SK is not required for Larp7 binding. Co-IP of transiently expressed 7SKtag1 and 7SK3′U RNAs with Larp7 was assayed by RNase A/T1 mapping.
To define the contribution of each terminal loop nucleotide to Larp7 binding, systematic base mutations (L1–L5) were introduced into terminal loop of the 7SKtag1 RNA that carried other neutral nucleotide alterations (tag1) to promote sequence-specific RNase mapping (Figure 3B). The 7SKtag1 RNA co-immunoprecipitated with Larp7 confirming that the tag1 sequence had no effect on Larp7 binding. The A310-U (L1), U311-A (L2) and G314-C (L5) nucleotide transversions failed to significantly alter the in vivo interaction of the expressed 7SKL1, 7SKL2 and 7SKL5 RNAs with HeLa Larp7. However, replacement of the G312-C (L3) and U313-A (L4) loop residues fully abolished and largely reduced the Larp7-binding capacity of the mutant 7SKL3 and 7SKL4 RNAs, respectively. We concluded that the G312 nucleotide is absolutely required, and the following U313 residue is important for efficient in vivo binding of Larp7.
The 3′-terminal oligouridylate tail of human 7SK snRNA is interrupted by a cytidine (C328) that, in principle, could contribute to the specific binding of Larp7 (Figure 3C). Conversion of the C328 residue to uridine, however, failed to alter the Larp7-binding capacity of the resulting 7SK3′U RNA, excluding a specific role for C328 in Larp7 binding. Taken together, the experiments presented thus far demonstrate that the Larp7-binding elements are confined to the 3′-terminal U-rich tail and the preceding short 3′-hairpin of 7SK. The overall structure of the 3′-hairpin and the G312 and U313 residues in its terminal loop provide the specificity for Larp7 binding.
MePCE binds to the basal part of the 5′-hairpin of 7SK snRNA
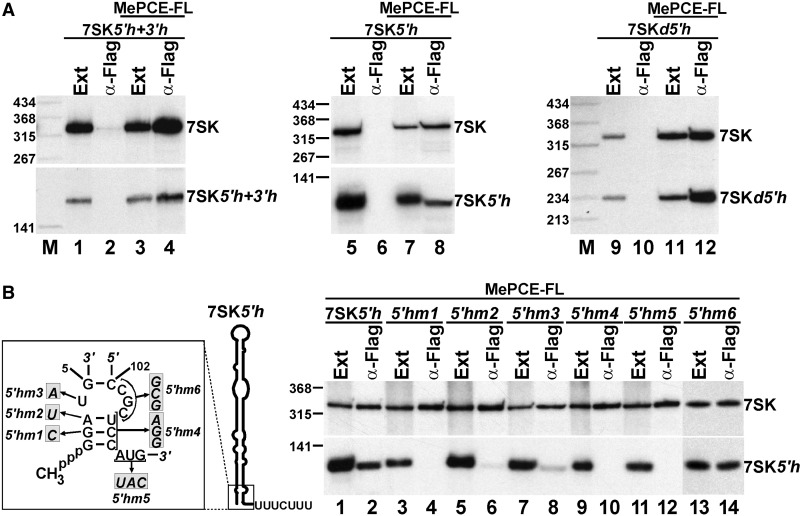
To determine the MePCE-binding region of 7SK, we first investigated the in vivo association of transiently expressed MePCE-FL with the internally truncated 7SK5′h + 3′h, 7SK5′h and 7SKd5′h RNAs (for RNA structures, see Figure 2A). On IP of MePCE-FL, northern blot analysis detected all test RNAs in the pellets of the IP reactions (Figure 4A, lanes 4, 8 and 12). This suggested that MePCE-FL binds to the basal part of the 5′-hairpin and/or to the 5′-terminal gamma-monomethyl-G cap of 7SK snRNA, as only these elements were common in three test RNAs. To confirm this assumption and to further dissect the MePCE-binding elements of 7SK, mutant versions of the 7SK5′h RNA were expressed in HeLa cells, and their association with MePCE-FL was assayed (Figure 4B). Disruption of the short G1-A3/U106–C108 terminal helix by introducing the G2-C (5′hm1) or A3-U (5′hm2) nucleotide transversions or replacing the U106-C108 sequence with its complementary sequence (5′hm4) disrupted the association of 7SK5′hm1, 7SK5′hm2 and 7SK5′hm4 RNAs with MePCE-FL (lanes 4, 6 and 10). Alteration of the U4 internal loop nucleotide (5′hm3) and the A109-G111 single-stranded sequence following the G1-A3/U106–C108 basal helix highly reduced or fully abolished MePCE-FL association with the mutant 7SK5′hm3 and 7SK5′hm5 RNAs, respectively (lanes 8 and 12). In contrast, replacement of the C103-C105 internal loop nucleotides had no detectable effect on the MePCE-binding capacity of 7SK5′hm6 (lane 14). These results demonstrated that the 5′-terminal G1-A3/U106-C108 helix and the flanking U4 and A109-G111 single-stranded nucleotides direct the in vivo binding of MePCE to the 7SK snRNA.
Figure 4.
7SK elements supporting in vivo MePCE binding. (A) MePCE binds to the basal stem region of the 5′-hairpin of 7SK. Truncated 7SK5′h + 3′h, 7SK5′h and 7SKd5′h RNAs were co-expressed with Flag-tagged MePCE (MePCE-FL) as indicated above the lanes. MePCE-FL was immunoprecipitated, and co-precipitation of mutant and wild-type 7SK RNAs was monitored by northern blotting. (B) Sequence requirements of MePCE binding to the 5′-hairpin of 7SK. Co-IP of transiently expressed MePCE-FL with mutant 7SK5′h RNAs carrying the indicated nucleotide alterations (shaded boxes) was tested by northern blotting.
MePCE can bind to 7SK in a cap-independent manner
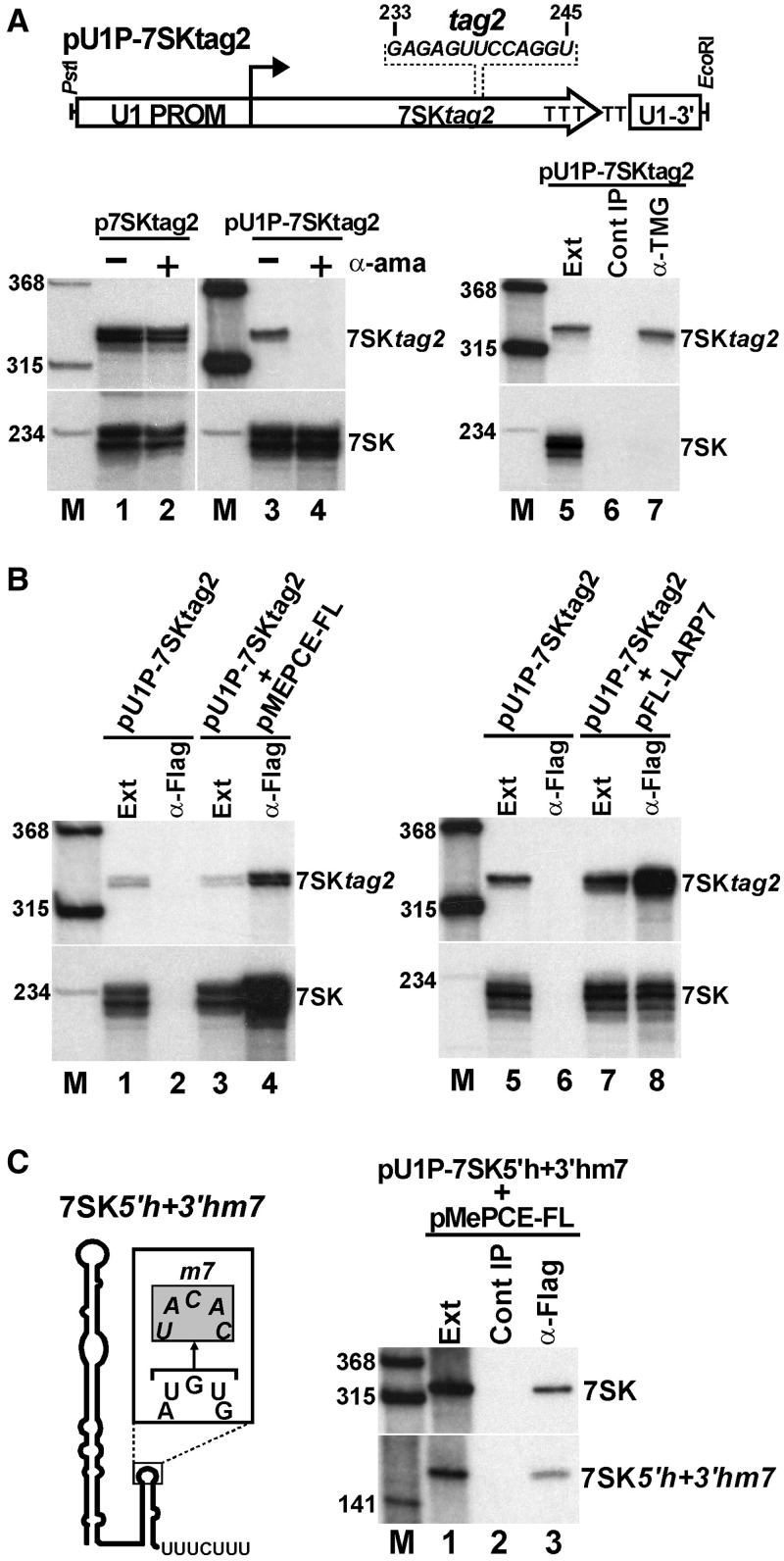
We tested whether the 5′-terminal gamma-monomethyl cap could contribute to the MePCE-binding activity of 7SK snRNA. The 7SK coding region carrying internal marker sequence (tag2) was placed under the control of the promoter and terminator of the Pol II-specific human U1 snRNA gene (Figure 5A). The 5′-termini of nascent Pol II transcripts acquire co-transcriptionally the 7-methyl-G primary cap that is further methylated to TMG in snRNAs (40,41). On transfection of the Pol II-specific pU1P-7SKtag2 and the control Pol III-specific p7SKtag2 expression plasmids into HeLa cells, the 7SKtag2 RNA accumulated in both cell lines (lanes 1 and 3). Incubation of the transfected cells with α-amanitin abolished the U1 promoter-dependent expression of 7SKtag2 (lane 4) but failed to inhibit 7SKtag2 expression from the Pol III-specific 7SK promoter (lane 2). Moreover, a TMG-specific antibody efficiently and specifically recovered the 7SKtag2 RNA synthesized from the U1 promoter but failed to recognize the endogenous HeLa 7SK snRNA (lane 7). Thus, we concluded that the pU1P-7SKtag2 expression vector was transcribed by Pol II, and the expressed 7SKtag2 RNA carried a TMG cap.
Figure 5.
Cap independent in vivo association of 7SK snRNA with MePCE. (A) Pol II-mediated expression of 7SK snRNA. Schematic structure of the pU1P-7SKtag2 expression construct with the promoter (U1 PROM) and terminator (U1-3′) of the human U1 snRNA gene is shown. Nucleotide alterations in the 7SK-coding region (tag2) are indicated. HeLa cells transfected with pU1P-7SKtag2 or p7SKtag2 were incubated with (+) or without (−) α-amanitin (α-ama). Accumulation of 7SKtag2 RNAs was measured by RNase A/T1 mapping. RNAs extracted from cells harbouring the pU1P-7SKtag2 plasmid (Ext) were reacted with an anti-trimethyl-G antibody (α-TMG). (B) Pol II-synthesized 7SK RNA associates with both MePCE and Larp7. Extracts (Ext) prepared from HeLa cells transfected with the indicated combination of the pU1P-7SKtag2, pMePCE-FL and pLARP-FL expression plasmids were subjected to IP with anti-Flag antibody. Recovery of 7SKtag2 RNA was monitored by RNase mapping. (C) In vivo association of MePCE with Pol II-synthesized 7SK RNA lacking Larp7-binding capacity. Schematic structure of the 7SK5′h + 3′hm7 RNA with the m7 nucleotide alterations is shown. Co-IP of transiently expressed 7SK5′h + 3′hm7 RNA with MePCE-FL was monitored by northern blot analysis.
Next, we assayed the association of the Pol II-synthesized TMG-capped 7SKtag2 RNA with transiently co-expressed MePCE-FL. Similar to the endogenous monomethyl-capped 7SK snRNA, the ectopically expressed TMG-capped 7SKtag2 RNA interacted with MePCE-FL (Figure 5B, lane 4), suggesting that the natural monomethyl cap of 7SK snRNA is not required for in vivo MePCE binding. We found that the Pol II-synthesized 7SKtag2 RNA also associated with transiently co-expressed FL-Larp7 (lane 8). On RNase-mediated 7SK hydrolysis, the Larp7 and MePCE snRNP proteins remain associated with each other, indicating that Larp7 and MePCE form a direct contact in the 7SK snRNP (28,33) (our unpublished results). Therefore, it is conceivable that Larp7 bound to the 3′-terminal region of the TMG-capped 7SK RNA could recruit MePCE to the 7SK snRNP without forming a direct 7SK-MePCE contact. To exclude this possibility, an altered version of the 7SK5′h + 3′h RNA carrying the m7 3′-hairpin loop mutations, which fully abolish Larp7 binding, was expressed under the control of the U1 promoter in HeLa cells also expressing MePCE-FL (Figure 5C). Similar to the endogenous 7SK snRNA, the Pol II-synthesized 7SK5′h + 3′hm7 RNA efficiently interacted with MePCE-FL (lane 3) but failed to bind Larp7 (data not shown, see Figure 3A). Thus, we concluded that the gamma-monomethyl cap of human 7SK snRNA is dispensable for in vivo MePCE binding.
Larp7 promotes MePCE recruitment to 7SK
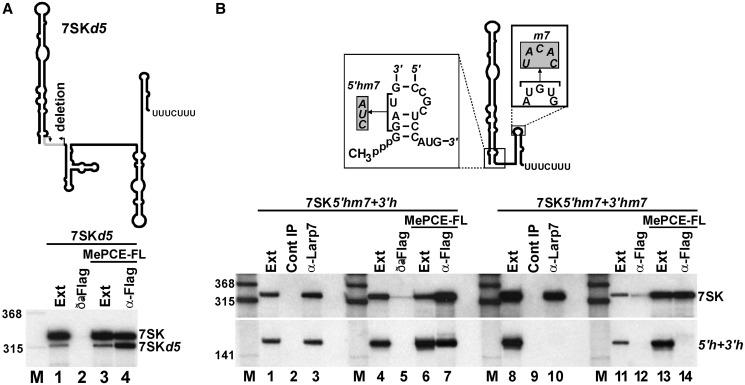
To further explore the functional importance of the observed interaction of Larp7 and MePCE (28,33) (our unpublished results), we first tested the in vivo association of MePCE with an internally truncated 7SK RNA, 7SKd5, that lacked functional MePCE docking site, but carried the wild-type Larp7-binding element (Figure 6A). The 7SKd5 RNA was expressed under the control of the cognate 7SK promoter in HeLa cells also expressing MePCE-FL. IP of MePCE-FL recovered both the transiently expressed 7SKd5 RNA and the endogenous 7SK snRNA (lane 4). This strongly suggested that MePCE can associate with 7SK RNA lacking a functional MePCE-binding site through interacting with Larp7.
Figure 6.
In vivo interaction of Larp7 and MePCE. (A) MePCE can associate with 7SK lacking functional MePCE-binding site. Extracts (Ext) prepared from HeLa cells transfected with the indicated combination of pMePCE-FL and p7SKd5 were reacted with an anti-Flag antibody. Recovery of 7SKd5 RNA was measured by northern blot analysis. (B) Larp7 can tether MePCE to the 7SK snRNP lacking functional MePCE-binding site. Nucleotide alterations (5′hm7 and 3′m7) introduced into the p7SK5′h + 3′h expression construct are indicated in shaded boxes. HeLa cells were transfected with the p7SK5′hm7 + 3′h, p7SK5′hm7 + 3′hm7 and pMePCE-FL expression constructs as indicated. Co-IP of endogenous 7SK snRNA and transiently expressed 7SK5′hm7 + 3′h and 7SK5′hm7 + 3′hm7 RNAs with MePCE-FL and Larp7 was monitored by northern blot analysis.
To confirm this notion, we investigated the in vivo association of MePCE and Larp7 with two altered versions of the 7SK5′h + 3′h RNA (Figure 6B). The 7SK5′hm7 + 3′h RNA carried the 2-GAU-4 to CUA (5′hm7) sequence alteration, which eliminates MePCE binding to its 5′-hairpin (see Figure 4B). The 7SK5′hm7 + 3′hm7 RNA, in addition to the 5′hm7 mutation, also contained the 3′hm7 loop mutation, which prevents Larp7 binding (see Figure 3A). Consistent with the observed association of 7SKd5 with both Larp7 and MePCE, the transiently expressed 7SK5′hm7 + 3′h RNA co-purified with both endogenous Larp7 and ectopically expressed MePCE-FL (Figure 6B, lanes 3 and 7). However, disruption of the Larp7-binding capacity of 7SK5′hm7 + 3′hm7 also eliminated its interaction with MePCE-FL (lanes 10 and 14), providing further support to the idea that recruitment of MePCE to 7SK RNAs lacking an active MePCE-binding motif occurred through forming an interaction with Larp7.
7SK mutations interrupting Larp7 binding also abolish P-TEFb recruitment
We have previously observed that destruction of the 3′-terminal hairpin abolishes the in vivo P-TEFb-binding activity of 7SK snRNA (21). Demonstration that the 3′-hairpin of 7SK plays a crucial role in Larp7 binding raised the possibility that the 7SK-associated Larp7 protein rather than the RNA 3′-hairpin itself supports the in vivo recruitment of P-TEFb to the 7SK snRNP. To test this idea, we investigated the in vivo association of P-TEFb with the 7SKext RNA that, although carried the wild-type 3′-hairpin, failed to efficiently bind Larp7 owing to its misplaced U-rich 3′-tail (see Figure 2C). The 7SKext RNA was transiently expressed in HeLa-based G3H cells, which stably express an HA-tagged version of CycT1 (42) (Figure 7A, lane 1). IP of HA-CycT1 failed to efficiently recover 7SKext, although it pulled down the endogenous 7SK snRNA (lane 3). This strongly supported the notion that binding of Larp7 to 7SK is a prerequisite for P-TEFb recruitment.
Figure 7.
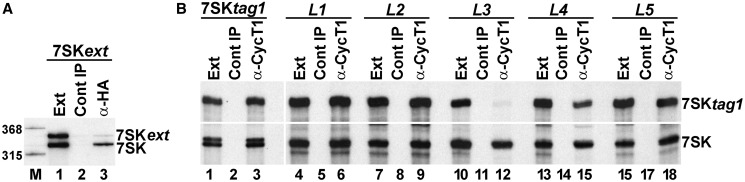
Binding of Larp7 to the 7SK snRNA is essential for P-TEFb recruitment. (A) Separation of the 3′-terminal hairpin and the U-rich tail of 7SK by sequence insertion inhibits P-TEFb binding. The 7SKext RNA (see Figure 2C) was transiently expressed in G3H cells expressing HA-CycT1. After extract preparation, HA-CycT1 was immunoprecipitated (α-HA), and co-purification of 7SKext and wild-type 7SK RNAs was tested by northern blotting. (B) The G312 and C313 loop nucleotides are important for P-TEFb binding. 7SKtag1 RNAs lacking or carrying the L1, L2, L3, L4 and L5 nucleotide transversions (see Figure 3B) were expressed in HeLa cells, and their co-IP with endogenous CycT1 was measured by RNase mapping.
Substitution of the terminal loop sequence of the 3′-hairpin of 7SK abolishes both P-TEFb and Larp7 binding (21) (Figure 3). As our current analysis identified the G312 and U313 loop nucleotides as crucial structural determinants for Larp7 binding (Figure 3B), we also tested the importance of these residues for P-TEFb recruitment (Figure 7B). HeLa endogenous CycT1 was immunoprecipitated, and co-purification of transiently expressed 7SKtag1 RNAs carrying the L1, L2, L3, L4 and L5 loop mutations was measured by RNase mapping. As it was observed for Larp7, CycT1 showed a strong association with the expressed 7SKL1, 7SKL2 and 7SKL5 RNAs (lanes 6, 9 and 18). However, the G312-C (L3) and the U313-A (L4) loop mutations, which abolished and reduced Larp7-binding, respectively, also eliminated and inhibited the CycT1-binding ability of the mutant 7SKL3 and 7SKL4 RNAs (lanes 12 and 15). These results confirmed that binding of Larp7 to the 3′-terminal hairpin-tail region of human 7SK snRNA is a prerequisite for in vivo recruitment of P-TEFb.
The La-associated fraction of 7SK does not interact with Larp7, HEXIM1 and P-TEFb
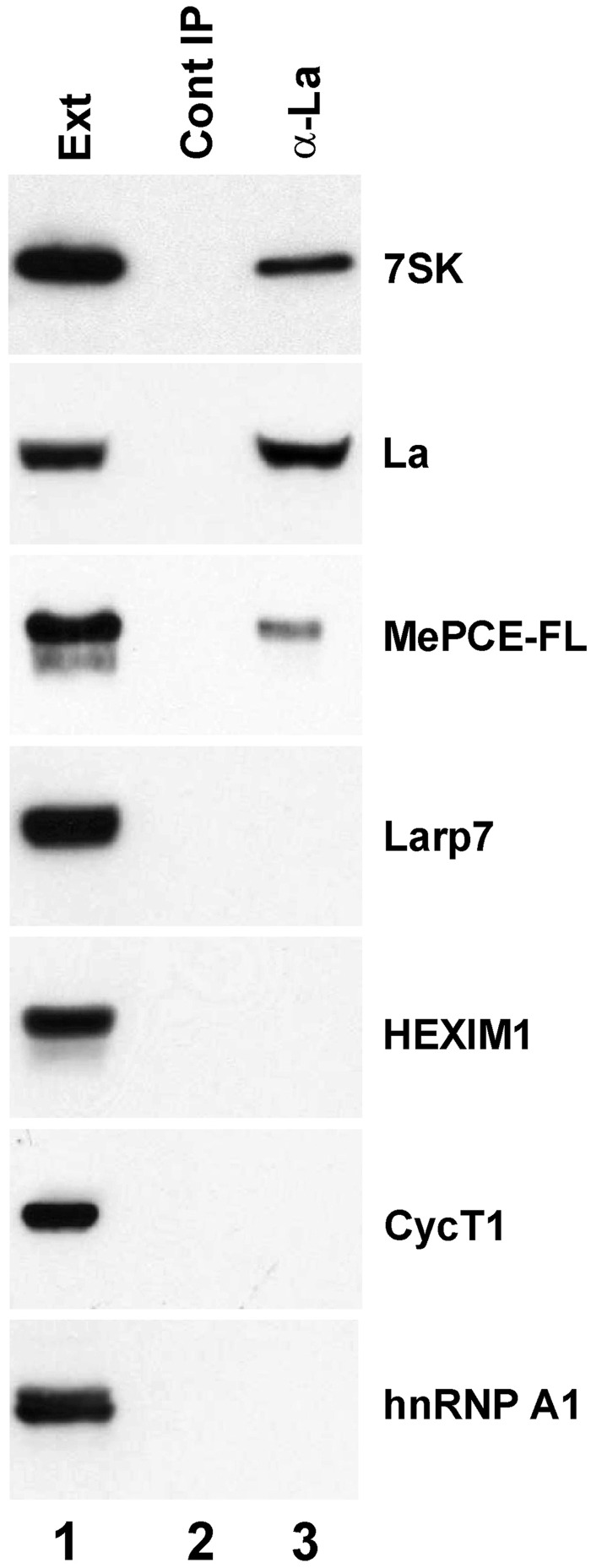
The 3′-terminal oligouridylate tail of the newly synthesized 7SK snRNA associates with the La protein that provides stability for the nascent RNA (43,44). To further confirm that docking of Larp7 is fundamental for subsequent P-TEFb recruitment, we tested whether the La-associated fraction of 7SK snRNA is able to interact with P-TEFb (Figure 8). On IP of HeLa La protein with a specific antibody, the 7SK snRNA was readily detected in the pellet (lane 3). However, apart from transiently expressed MePCE-FL, other components of the 7SK negative transcriptional regulatory snRNP, including Larp7, HEXIM1 and CyT1, were not detectable in the pellet of the IP reaction. Moreover, hnRNP A1, the most abundant component of the 7SK/hnRNP particle (26,27), also failed to co-purify with the immunoprecipitated La protein. These observations confirmed that the La-associated fraction of human 7SK RNA is inactive in transcription regulation, and they suggested that replacement of La protein with Larp7 is a prerequisite for conversion of the native 7SK RNA into functional transcriptional regulatory snRNA.
Figure 8.
Co-purification of HeLa 7SK snRNP proteins with La protein. HeLa La protein was immunoprecipitated with a La-specific antibody and co-purification of endogenous 7SK snRNA, Larp7, HEXIM1, CycT1, hnRNP A and transiently expressed MePCE-FL was monitored.
DISCUSSION
P-TEFb is a general transcription factor that regulates the processivity of Pol II and thereby controls global gene expression, cell growth and development. The nuclear activity of P-TEFb is regulated by the 7SK core snRNP that is composed of the 7SK snRNA and the MePCE and Larp7 snRNP proteins. In this study, we performed in vivo RNA–protein binding experiments to define the structural elements of human 7SK snRNA, which direct the in vivo assembly of functionally active 7SK/MePCE/Larp7 transcriptional regulatory snRNP.
Early in vitro and in vivo RNA structure probing experiments revealed that the human 7SK snRNA is composed of four major hairpin domains (11) (Figure 9). Functional analysis of compensatory base changes introduced into the predicted double-stranded regions of the 5′- and 3′-hairpins of human 7SK snRNA largely confirmed the correctness of the proposed 2D 7SK structure (20,21) (this study). The 5′- and 3′-hairpin regions of 7SK RNAs derived from evolutionarily distant species show significant structural conservation, pointing to the functional importance of these elements (20) (our unpublished data). Indeed, we demonstrated that all elements directing the in vivo assembly of the 7SK/MePCE/Larp7/HEXIM/P-TEFb transcriptional regulatory snRNP are located in the 5′- and 3′-terminal hairpin regions of human 7SK snRNA (21) (Figure 9).
Figure 9.
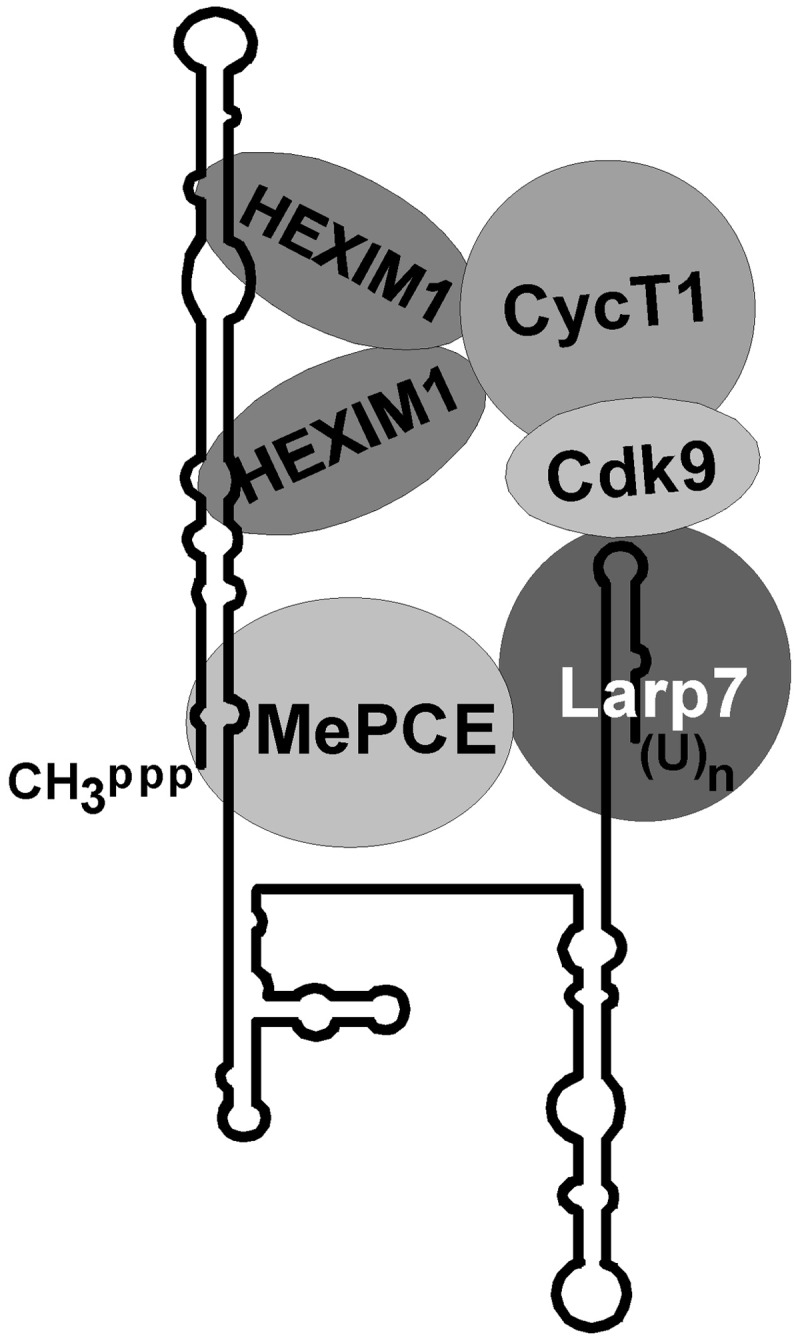
Proposed schematic structure of the 7SK transcriptional regulatory snRNP. Structure of the 7SK snRNA has been adopted from (11). The 7SK elements directing HEXIM1 binding have been reported earlier (20,21).
MePCE and 7SK co-IP experiments demonstrated that the short 5′-terminal G1-A3/U106-C108 helix and the flanking unpaired U4 and 109-AUG-11 nucleotides provide the structural information for specific binding of MePCE (Figure 4). Besides being integral component of the 7SK snRNP, MePCE methylates the gamma-phosphate of the 5′-terminal guanosine-triphosphate of the newly synthesized 7SK snRNA (30,33). Early in vitro RNA methylation studies demonstrated that monomethyl capping of the U6 and 7SK snRNAs depends on their 5′-terminal stem–loop structures and the following single-stranded sequences, which, in fact, show a similarity in U6 and 7SK snRNAs (45,46). This suggests that 7SK capping and 7SK/MePCE assembly are directed by the same or largely overlapping RNA signals and also explains the observed interaction of MePCE with both 7SK and U6 snRNAs (Figure 1). The gamma-monomethyl cap is not required for stable association of MePCE with mature 7SK snRNA; it likely protects 7SK from exonucleolytic degradation (Figure 5B).
All RNA elements directing in vivo Larp7 binding are confined to the 3′-terminal A301-U331 hairpin-tail region of 7SK snRNA (Figure 2). Although Larp7 shows a strong in vitro affinity for oligouridylate sequences (32), the U-rich tail of 7SK alone is unable to support the in vivo binding of Larp7. The U-rich terminal sequence, the stem–bulge–stem–loop structure of the preceding 3′-hairpin and the G312 and U313 terminal loop nucleotides together constitute the docking surface specific for Larp7 (Figure 3). Partial NMR and crystal structures of another member of the Larp7 family, the Tetrahymena p65 telomerase RNP protein, revealed that the C-terminal domain of p65 forms an atypical RRM, termed xRRM2, which specifically interacts with the 3′-terminal hairpin of Tetrahymena telomerase RNA (47). It has been proposed that La and Larp7 proteins follow a similar strategy for binding substrate RNAs. The N-terminal La motif and the following RRM1 of La and Larp7 proteins bind cooperatively to the 3′-terminal oligouridylate tail of the substrate RNA (44). This interaction positions their C-terminal domains, which carry the common xRRM2 cryptic RRM to specifically interact with the 3′-terminal hairpin of the target RNA (47). Consistent with this model, mutant human Larp7 proteins lacking their C-terminal regions are unable to bind 7SK in vitro (29). Our co-IP assays showed that HeLa Larp7 recognizes the 7SK snRNA with great specificity (Figure 1). The observed minor Larp7/U6 association likely represents non-specific interaction of Larp7 with U6-bound MePCE. Supporting this hypothesis, in contrast to endogenous Larp7, transiently overexpressed Larp7 shows a high level of apparently non-specific association with HeLa U6 snRNA (31) (our unpublished data).
Successful monomethyl capping of in vitro synthesized mammalian 7SK, U6, B2 and plant U3 RNAs in cell extracts and efficient incorporation of Pol II-synthesized TMG-capped 7SK RNA into 7SK/MePCE/Larp7 snRNP demonstrate that under particular conditions, MePCE function can be uncoupled from Pol III transcription (48–50) (Figure 5). However, several lines of evidence indicate that in living cells MePCE-mediated 7SK capping is functionally linked to Pol III transcription. All known RNAs carrying gamma-monomethyl-cap, such as eukaryotic U6 and 7SK snRNAs and rodent B2 RNA, are synthesized by Pol III (50–53). Even more tellingly, the U3 snoRNA, that is synthesized by Pol II and capped with a TMG cap in most eukaryotes, is a Pol III product in plants, and it carries a gamma-monomethyl cap (50,54,55). Thus, evolutionarily switching of the polymerase specificity of the U3 gene from Pol II to Pol III is accompanied with monomethyl capping of the RNA product. Finally, chromatin IP studies documented a specific enrichment of MePCE on the promoter region of human 7SK gene, providing further support to the notion that in vivo capping of 7SK snRNA is linked to Pol III transcription (33).
Similar to other nascent Pol III transcripts, the terminal oligouridylate tail of the newly synthesized 7SK RNA recruits the stabilizing La protein (43). IP of HeLa La protein recovered the 7SK snRNA and MePCE, but it failed to pull down other components of the 7SK transcriptional regulatory snRNP (Figure 8). This suggests that the La protein associated with nascent 7SK prevents premature assembly of the 7SK/HEXIM/P-TEFb negative transcriptional regulatory snRNP. The observed co-purification of MePCE with La might indicate that MePCE binds already to the La-associated fraction of 7SK and/or U6. It has been reported that methyl-phosphate capping largely reduces the affinity of U6 and B2 RNAs to the La protein (56). Hence, binding of MePCE to the nascent 7SK/La RNP and monomethyl capping of 7SK might promote the replacement of La with Larp7.
Although MePCE and Larp7 likely interact in the mature 7SK snRNP (33) (our unpublished data), they can bind to the 7SK snRNA independently from each other (Figures 4 and 6). Interestingly, Larp7 can tether MePCE to mutant 7SK RNAs lacking functional MePCE-binding sites, but on the contrary, MePCE is unable to recruit Larp7 to 7SK RNAs missing Larp7-binding ability. A possible explanation of these seemingly contradictory observations could be that binding of Larp7 to 7SK increases its affinity to MePCE. Indeed, in vitro interaction of Larp7 and MePCE is fully dependent on the presence of the 7SK snRNA (28). This could also explain the observed weak affinity of HeLa Larp7 to MePCE associated with U6. On the other hand, 7SK-independent Larp7/MePCE heterodimer formation would inhibit the methylphosphate capping activity of nuclear MePCE because binding of Larp7 to MePCE inhibits its methyltransferase activity (33).
In the test tube, in vitro synthesized 7SK RNA and recombinant HEXIM1 proved to be sufficient to recruit P-TEFb and to inhibit its carboxy-terminal domain kinase activity (13,15). A conformational rearrangement induced by 7SK binding enables the acidic C-terminal domain of HEXIM1/2 to interact with the CycT1 subunit of P-TEFb. Contrary to the fact that HEXIM1/2 binds exclusively to the 5′-hairpin of 7SK, we found earlier that the 3′-hairpin of 7SK is also required for P-TEFb binding in living cells (20,21). In accordance with this finding, here, we demonstrated that in vivo binding of Larp7 to the 3′-hairpin of 7SK is a prerequisite for in vivo P-TEFb recruitment (Figure 7). Several lines of evidence suggest that, instead of directly interacting with 7SK, P-TEFb associates with Larp7 bound to the 3′-terminal hairpin of 7SK snRNA. Point mutations abolishing the Larp7-binding capacity of 7SK snRNA also prevent P-TEFb recruitment (Figure 7B). A mutant 7SK RNA, 7SKext, although carrying the wild-type 3′-hairpin, but lacking Larp7-binding capacity owing to its misplaced 3′-terminal oligouridylate tail fails to bind P-TEFb (Figure 7A). Nascent 7SK snRNA associated with La, instead of Larp7, is unable to associate with P-TEFb (Figure 8). Finally, previous in vitro protein-binding studies revealed that the C-terminal region of GST-tagged Larp7 can specifically interact with Cdk9 (32). Thus, the available data are consistent with a model in which Larp7, besides binding to the 3′ end of 7SK snRNA, also interacts with MePCE and Cdk9, suggesting that Larp7 possesses a more complex role in 7SK-mediated P-TEFb modulation than anticipated before (Figure 9). Consistent with its central role in P-TEFb regulation, Larp7 mutations have been frequently linked to cancer development (34,35). Understanding of the molecular background of the predicted interaction of Larp7 and P-TEFb in the 7SK transcriptional regulatory snRNP will be an important task for the future.
FUNDING
La Fondation pour la Recherche Médicale (Équipes FRM to T.K.) FRM PhD fellowship (to L.M.). Funding for open access charge: FRM grant (to T.K.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Q. Zhou (University of California, Berkely, USA) and R. Lührmann (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) for providing them with human G3H cells and anti-TMG antibody, respectively.
REFERENCES
- 1.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 6.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 9.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 10.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA. 2011;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 13.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1 related protein, regulates P-TEFb through asociation with 7SK. J. Biol. Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 18.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 19.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive P-TEFb complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 20.Muniz L, Egloff S, Ughy B, Jády BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010;6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 2006;26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 2005;280:24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 24.Dames SA, Schonichen A, Schulte A, Barboric M, Peterlin BM, Grzesiek S, Geyer M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl Acad. Sci. USA. 2007;104:14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA Biol. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Herreweghe E, Egloff S, Goiffon I, Jády BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl Acad. Sci. USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biewenga P, Buist MR, Moerland PD, Ver Loren van Themaat E, van Kampen AH, ten Kate FJ, Baas F. Gene expression in early stage cervical cancer. Gynecol. Oncol. 2008;108:520–526. doi: 10.1016/j.ygyno.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y, Jin Z, Agarwal R, Ma K, Yang J, Ibrahim S, Olaru AV, David S, Ashktorab H, Smoot DT, et al. LARP7 is a potential tumor suppressor gene in gastric cancer. Lab. Invest. 2012;92:1013–1019. doi: 10.1038/labinvest.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Jobert L, Pinzon N, Van Herreweghe E, Jády BE, Guialis A, Kiss T, Tora L. Human U1 snRNA forms a novel chromatin-associated snRNP with TAF15. EMBO Rep. 2009;10:494–500. doi: 10.1038/embor.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodall GJ, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 39.Kiss T, Bortolin ML, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell. Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 41.de Almeida SF, Carmo-Fonseca M. Cotranscriptional RNA checkpoints. Epigenomics. 2010;2:449–455. doi: 10.2217/epi.10.21. [DOI] [PubMed] [Google Scholar]
- 42.O'Keeffe B, Fong Y, Chen D, Zhou S, Zhou Q. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem. 2000;275:279–287. doi: 10.1074/jbc.275.1.279. [DOI] [PubMed] [Google Scholar]
- 43.Chambers JC, Kurilla MG, Keene JD. Association between the 7 S RNA and the lupus La protein varies among cell types. J. Biol. Chem. 1983;258:11438–11441. [PubMed] [Google Scholar]
- 44.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim. Biophys. Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shumyatsky G, Shimba S, Reddy R. Capping signals correspond to the 5′ end in four eukaryotic small RNAs containing gamma-monomethylphosphate cap structure. Gene Expr. 1994;4:29–41. [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R, Gupta S, Reddy R. Capping of mammalian U6 small nuclear RNA in vitro is directed by a conserved stem-loop and AUAUAC sequence: conversion of a noncapped RNA into a capped RNA. Mol. Cell. Biol. 1990;10:939–946. doi: 10.1128/mcb.10.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Singh R, Reddy R. Capping of U6 small nuclear RNA in vitro can be uncoupled from transcription. J. Biol. Chem. 1990;265:9491–9495. [PubMed] [Google Scholar]
- 49.Shimba S, Reddy R. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J. Biol. Chem. 1994;269:12419–12423. [PubMed] [Google Scholar]
- 50.Shimba S, Buckley B, Reddy R, Kiss T, Filipowicz W. Cap structure of U3 small nucleolar RNA in animal and plant cells is different. Gamma-monomethyl phosphate cap structure in plant RNA. J. Biol. Chem. 1992;267:13772–13777. [PubMed] [Google Scholar]
- 51.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl Acad. Sci. USA. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 54.Kiss T, Solymosy F. Molecular analysis of a U3 RNA gene locus in tomato: transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 1990;18:1941–1949. doi: 10.1093/nar/18.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiss T, Marshallsay C, Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell. 1991;65:517–526. doi: 10.1016/0092-8674(91)90469-f. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharya R, Perumal K, Sinha K, Maraia R, Reddy R. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 2002;10:243–253. doi: 10.3727/000000002783992398. [DOI] [PMC free article] [PubMed] [Google Scholar]