Abstract
Teeth are unique to vertebrates and have played a central role in their evolution. The molecular pathways and morphogenetic processes involved in tooth development have been the focus of intense investigation over the past few decades, and the tooth is an important model system for many areas of research. Developmental biologists have exploited the clear distinction between the epithelium and the underlying mesenchyme during tooth development to elucidate reciprocal epithelial/mesenchymal interactions during organogenesis. The preservation of teeth in the fossil record makes these small organs essential for the work of paleontologists, anthropologists, and evolutionary biologists. In addition, with the recent identification and characterization of dental stem cells, teeth have become of interest to the field of regenerative medicine. Here, we review the major research areas and studies in the development and evolution of teeth, including morphogenesis, genetics and signaling, evolution of tooth development, and dental stem cells. Brief discussions of microRNAs and human disease as they apply to teeth are also included.
I. MORPHOGENESIS AND DEVELOPMENT
The formation of a head with complex jaws and networked sensory organs was a central innovation in the evolution of vertebrates, allowing the shift to an active predatory lifestyle.1 The earliest vertebrates were jawless fish (agnathans); the jaw-bearing gnathostomes arose later and have been more successful evolutionarily. An important event in head evolution was the emergence of specialized dentition. To function properly in grasping and crushing food, teeth must be of adequate hardness, proper shape, and anchored to underlying bone (Fig. 1). Tooth number, shape, and size vary significantly among species, because of natural selection in response to the environmental pressures provided by various types of food.
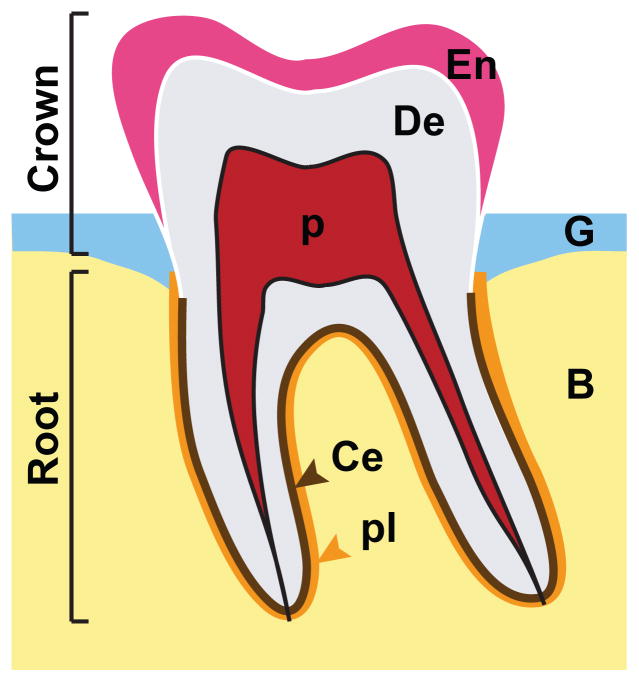
Figure 1. Cartoon depiction of a first lower molar from a human adult.
The crown (part of the tooth covered by enamel) and the root are shown. The tooth and its supporting structure, the periodontium, contain all four mineralized tissues in the body, bone (B), cementum (Ce), dentin (De), and enamel (En). The tooth is attached to the underlying bone via periodontal ligaments (pl) in humans. The pulp chamber (p) houses the blood vessels and nerves (not shown) as well as the putative odontoblast stem cells. The gingiva (G) is the oral mucosa that overlies alveolar bone (B).
Teeth, or tooth-like structures called odontodes or denticles, are present in all vertebrate groups, although they have been lost in some lineages. Most fish and reptiles, and many amphibians, possess dentitions that contain a large number of teeth (polyodont) of similar shape (homodont) that undergo continuous replacement (polyphyodont2). These teeth are comprised of dentin and enamel or an enamel-like structure, are rootless, and are attached directly to bone by ankylosis or fibrous tissue. In contrast, mammalian teeth are rooted and are connected to the jaws through interactions between the periodontal ligaments and alveolar sockets (Fig. 1).
Egg-laying monotremes, the most basal living mammals, possess a rudimentary unpaired egg tooth, similar to reptiles and birds, for use during hatching.3 Adult monotremes have horny plates as opposed to teeth. Therian mammals, which include all living mammals except monotremes, typically are heterodonts, meaning that the teeth have different shapes. Four types of teeth are present in mammals: incisors, canines, premolars and molars. In contrast to non-mammalian vertebrates, only two generations of teeth (diphyodonty) are present in mammals. The ancestral dental pattern for eutherian placental mammals in each quadrant is three incisors, one canine, four premolars, and three molars, and the premolars and molars are typically multicuspid.4 In most extant mammals, tooth number is reduced relative to this ancestral pattern (e.g., Fig. 2).
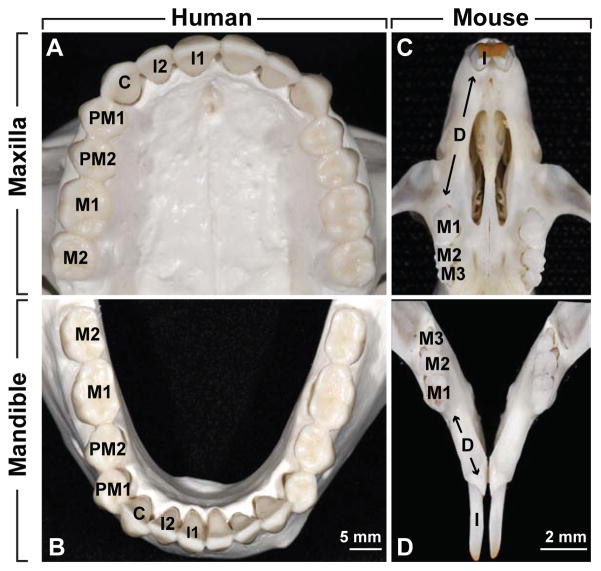
Figure 2. Human and mouse dentitions.
The maxillary (A, C) and mandibular (B, D) dental arches show the reduced dentitions in adult human (A, B) and mouse (C, D). Both species are derived from a common mammalian ancestor that is thought to have had 6 incisors, 2 canines, 8 premolars, and 6 molars in each dental arch. The third molar or wisdom tooth (M3) is absent in the human specimen. I, incisor; I1, central incisor; I2, lateral incisor; C, canine; PM1, first premolar; PM2, second premolar; M1, first molar, M2, second molar; M3, third molar; D, diastema. Images are courtesy of Dr. Kyle Burke Jones (UCSF).
The dentition is highly specialized in mice, which are the most commonly used model to study tooth development. In each quadrant, a single incisor is separated from three molars by a toothless region called the diastema (Fig. 2C, D). Rodent incisors are unusual because they grow continuously throughout the life of the animal, a property attributed to the presence of populations of adult stem cells. Such stem cell-fueled continuous growth of rodent teeth is discussed in more detail below. Also, rodents possess no replacement teeth, unlike humans, who have two sets of teeth.
A. STAGES OF TOOTH DEVELOPMENT
The tissues required for tooth development originate from two principal sources. The epithelium is derived from oral ectoderm and potentially pharyngeal endoderm,5, 6 whereas the mesenchyme is derived from cranial neural crest cells. Neural crest cells arise from the margins of the neuro-epithelium and migrate laterally and ventrally to fill the facial prominences with mesenchyme.7 The neural crest-derived mesenchyme (hereafter referred to as mesenchyme) eventually forms the facial and jaw skeletons, as well as most of the soft and hard tissues in teeth, including dentin, dental pulp, alveolar bone, and periodontal ligament;8 these tooth-specific tissues are discussed in greater detail below.
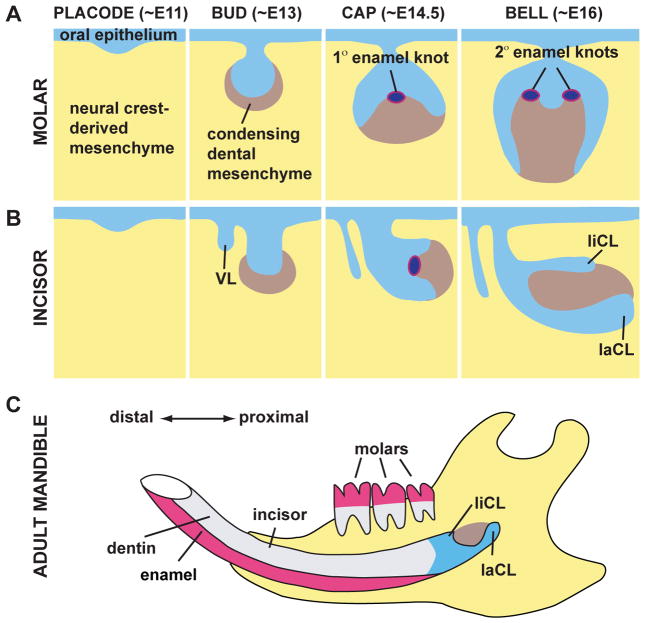
In mice, initiation of tooth development occurs between embryonic day (E) 8.5–10, when the sites of tooth formation are first apparent based on the expression of several genes. The first morphological sign of odontogenesis is a thickening of the oral epithelium at E11 in the mouse (week 7 of gestation in humans) (Fig. 3). During the subsequent bud stage at E12.5-E13.5, cells of the thickened oral epithelium proliferate and form a dental lamina that invaginates into the mesenchyme. Mesenchymal cells condense underneath the forming epithelial bud to generate the dental papilla. During the cap stage at E14, the epithelial bud extends further and begins to surround the dental papilla. Cells of the dental mesenchyme located adjacent to the dental papilla or outside the epithelial organ form the dental follicle or sac. Also, the primary enamel knot, a transient signaling center that regulates tooth shape, is present in the dental epithelium. During the bell stage beginning at E16, the tooth germ increases further in size, and the final shape of the tooth crown becomes increasingly apparent. In the forming molars, secondary enamel knots, which are the signaling centers in areas of epithelial folding whose initiation is controlled by the primary enamel knot, determine the sites of tooth cusp formation.9 Finally, tooth-specific cell types, such as ameloblasts and odontoblasts, begin to differentiate.
Figure 3. Tooth development.
The various stages of mouse molar (A) and incisor (B) development, and the adult mouse mandible (C) are depicted in sagittal views. The oral epithelium thickens at the placode stage and invaginates into the neural crest-derived mesenchyme. Mesenchymal condensation occurs at the bud stage and the enamel knot, a central signaling area, first appears at the cap stage. The extracellular matrices of dentin and enamel are secreted with the differentiation of ameloblasts and odontoblasts during the bell stage. The matrix will eventually mineralize forming the tooth crown and is followed by tooth eruption. Similar developmental events occur in incisors and molars with notable differences being the presence of a vestibular lamina (VL), as well as the labial and lingual cervical loops (laCL and liCL, respectively), during incisor development, and the presence of secondary enamel knots, the future site of cusps, during molar development.
The enamel-producing ameloblasts are generated from epithelial cells adjacent to the dental papilla called the inner enamel epithelium (IEE), and these cells secrete enamel matrix that eventually mineralizes. Dentin-producing odontoblasts differentiate from the outermost layer of the dental papilla and gradually migrate to the center of the dental papilla as they secrete dentin matrix. Root formation coincides with tooth eruption after formation of the crown, which is the part of the tooth covered by enamel.10 Cementum, periodontal ligament, and alveolar bone are all derived from the dental follicle, which has a mesenchymal origin, and are involved in anchoring the teeth to the jaws.11, 12 Rodent incisors do not have a typical crown or root but rather possess a crown-like labial (near the lip) surface covered by enamel and a root-like lingual (near the tongue) surface where enamel is absent.13 The first teeth to erupt in mice, the mandibular incisors, become visible at around postnatal day (P) 9, followed shortly by the maxillary incisors. Eruption of molar teeth begins at P15 with the first mandibular molars.
B. EPITHELIAL-MESENCHYMAL INTERACTIONS: THE TOOTH AS A MODEL FOR DEVELOPMENTAL BIOLOGISTS
Reciprocal interactions between the epithelium and the underlying mesenchyme regulate tooth morphogenesis, and studies of these interactions have made the tooth an important model for developmental biologists. The ability to form teeth, also called the odontogenic potential, was shown by classical tissue recombination experiments to reside in the epithelium at the placode stage. In these studies, oral epithelium of murine embryos between E9 and E11.5 induced tooth formation in non-dental mesenchyme.14, 15 After E11.5, the dental mesenchyme was able to induce non-dental epithelia to participate in odontogenesis,16 whereas the ability by the epithelium to induce tooth formation appeared to be lost at this stage. Thus, these early experiments suggested that, during early tooth development, the odontogenic potential shifts from the epithelium to the mesenchyme. During the cap stage and beyond, the dental mesenchyme regulates tooth shape formation and can induce formation of ameloblasts and enamel matrix secretion in non-dental epithelium.16, 17
C. ADULT TEETH AND TOOTH-SPECIFIC MINERALIZED TISSUES
In terms of hard tissues, the tooth is comprised of enamel, dentin and cementum, and it is anchored to the alveolar bone by periodontal ligaments; thus, the tooth and supporting structure, called the periodontium, contain all of the four types of mineralized tissues found in the vertebrate body (Fig. 1). The two main parts of an adult tooth are the crown and the root. The crown is covered by enamel, which is a highly mineralized, acellular substance secreted by cells derived from dental epithelium. Enamel is the hardest structure found in the body, and it consists primarily of hydroxyapatite, a crystalline calcium phosphate, which is also in the major component of dentin, cementum, and bone. The organization of mineral in enamel is unique, as this material is formed of rods of hydroxyapatite crystals running from the dentin-enamel junction to the surface of the tooth. Underlying dentin supports both the enamel layer of the tooth crown and the cementum layer of the tooth root. Dentin is less mineralized and less brittle than enamel and is necessary for the support of enamel in the crown and cementum in the roots. The individual collagen fibrils of the periodontal ligaments originate from the cementum and cementum-dentin junction and attach directly to the alveolar bone of the jaws (Fig. 1).18 The dental pulp is a mass of vascularized connective tissue enclosed by dentin in the central part of the tooth. The apical foramen, an opening in the area of the root apex, allows the supply of the dental pulp with blood vessels and nerves.
II. GENETICS AND SIGNALING
A number of signaling pathways work in concert to orchestrate tooth development, and this section summarizes some of the major pathways. Signaling cascades involved in development, response to intercellular signals and environment, cell cycle control, and pathogenesis require transcription factors that interact with DNA to regulate gene expression; some of the transcription factors involved in tooth development are summarized. Newly discovered functions for microRNAs are also briefly discussed. Expression data of the molecular factors discussed here at various stages of tooth development can be found at www.bite-it.helsinki.fi.
A. FGFs
Fgf8 and Fgf9 are amongst the earliest genes to be expressed in the oral epithelium. The conditional inactivation of Fgf8 in ectoderm caused defects in structures derived from the first pharyngeal arch including teeth, jaws, lateral skull wall, and middle ear, as well as part of the tongue and other soft tissues.19 Although molars and the proximal mandible were absent, the distal-most structures such as lower incisors were present. These results suggested that a large proximal derivative of the first pharyngeal arch primordium is specified by FGF8, but a small distal region depends on other signaling molecules for its outgrowth and patterning.19 In mice over-expressing a dominant negative form of Fgfr2b, tooth development did not progress beyond the bud stage.20 Fgf4 and Fgf9, which are expressed in the enamel knot, are thought to stimulate proliferation in adjacent epithelial and mesenchymal tissues.9, 21 Deletion of Fgf3 and Fgf10 in mice results in smaller teeth with aberrant cusp morphology22, but Fgf3 and Fgf10 do not appear to be required individually for ameloblast differentiation.22, 23 The inactivation of sprouty genes, which are inhibitors of FGF signaling, results in the formation of supernumerary teeth24 and the generation of ectopic enamel on the lingual surface of the incisor.25 FGF signaling is also important in zebrafish tooth morphogenesis, and decreases in FGF signaling have been proposed to lead to the loss of oral teeth.26, 27
B. BMPs
BMPs function at multiple stages during odontogenesis. BMP4, in particular, is an important mediator of signaling between epithelial and mesenchymal tissues.28 During initiation of tooth formation, BMP signaling in the oral epithelium antagonizes FGF signaling, which is thought to determine the sites of tooth formation.29–31 Mesenchymal BMP4 regulates Shh expression30 and is critical for the transition from tooth bud to cap stage and for induction of the enamel knot in the epithelium.32, 33 The inactivation of activin or Bmpr1a in either epithelium or mesenchyme results in the arrest of tooth development after the bud stage.34–36 During the cap stage, BMP4 induces the cyclin dependent kinase inhibitor, p21, and the expression of Bmp4 and p21 is associated with differentiation as well as apoptosis of the primary enamel knot.33 Therefore, BMP signaling regulates patterning of the cusps and ultimately, the shape of the tooth crown. Fst1, which acts as an antagonist of BMP and INHBA (formerly known as activin) signaling, and Sostdc1, an inhibitor of both BMP and WNT signaling, are important regulators of enamel knot formation.37, 38 BMP signaling is also known to function during root formation39 and during differentiation of odontoblasts and ameloblasts.38, 40, 41
C. SHH
During initiation of tooth formation, Shh is expressed in specific regions of the epithelium of the molar and incisal placodes.42 At this early stage, the mitogenic activity of SHH is thought to stimulate proliferation in the placode epithelium, which enables invagination into the underlying mesenchyme and formation of the epithelial bud.43, 44 Analysis of expression of the receptor Ptch1 and the transcription factor Gli1, which are both downstream targets of SHH signaling, showed that SHH signals to the mesenchyme as well as the epithelium.44, 45 Expression of Shh is retained at the tip of the epithelial bud, becomes down-regulated towards the end of the bud stage, is re-induced in the enamel knot, and remains expressed in the epithelium throughout ameloblast differentiation. The regulation of these interesting expression patterns is not well understood and remains the subject of much interest.
SHH from the enamel knot regulates crown formation by stimulating proliferation in epithelial and mesenchymal tissues adjacent to the signaling center. Conditional inactivation of Shh or the signal transducer smoothened from the epithelium, or inhibition of signaling using an antibody against SHH, demonstrated that SHH signaling regulates tooth separation, size and morphology as well as cytological organization of matrix secreting cells.46–48 Primary cilia exert a negative regulatory effect on SHH activity and function to repress tooth formation.49 In zebrafish, SHH signaling is required continuously throughout tooth development from initiation to morphogenesis.50 Despite gene duplication and differences in the location of where teeth form between mice and zebrafish, the role of SHH signaling in tooth development appears to be conserved between these two species.50
D. WNTs
The modulation of WNT signaling leads to variations in tooth number. Supernumerary teeth arise with the up-regulation of WNT signaling. Multiple ectopic teeth in the molar region were observed after constitutive activation of the transcriptional effector CTNNB1 (beta catenin),51, 52 and mis-expression in epithelial tissues of Lef1, the binding partner of CTNNB1, led to multiple teeth in the incisor region.53 Overexpression of the zinc finger protein-encoding gene, Sp6, led to an increase in WNT signaling and mice with up to 50 teeth.54 Inactivation of WNT antagonists such as Apc55, 56 and Sostdc137, 57, 58 also led to increases in the number of teeth. Several of these studies demonstrated that the dental epithelium undergoes multiple invaginations leading to the formation of extra enamel knots and ultimately, supernumerary teeth.51, 54, 56, 59 Conversely, there is evidence in humans that decreases in WNT signaling lead to tooth loss.60, 61
The mechanism by which WNT signaling regulates tooth number is still unclear. Surprisingly, Msx1, which is required for normal tooth development, was dispensable for WNT-mediated supernumerary tooth formation, whereas Fgf8 was identified as a direct target of WNT signaling.56 WNT signaling regulates Shh48, 62 and Bmp4 expression,63 and it affects multiple stages of tooth development such as bud to cap transition, formation of the enamel knot, molar tooth size, and dentinogenesis.41, 52, 54, 62, 64 Conditional inactivation in the dental mesenchyme of Smad4, which encodes a BMP signal transducer, led to an up-regulation of CTNNB1 and down-regulation of WNT antagonist genes such as Dkk1 and Sfrp1.41 In these mice, enamel appeared normal whereas dentin formation was compromised, an observation that challenges the traditional notion that ameloblast differentiation is dependent upon odontoblast differentiation.65
E. Notch
Components of the Notch signaling pathway, which include four transmembrane Notch receptors (Notch1-4) and 5 transmembrane ligands (Jag1, Jag2, Dll1, Dll3, and Dll4), are expressed during tooth development and affect several aspects of tooth formation. Notch signaling was demonstrated to regulate tooth morphogenesis and ameloblast differentiation.66 Specifically, inactivation of the Notch-interacting domain of JAG2 in mice caused abnormal molar shapes, additional cusps, and inhibition of ameloblast differentiation and enamel matrix deposition.66
The stratum intermedium (SI), a layer of cells that is subjacent to the ameloblast layer during enamel formation and whose function is still unclear, expresses Notch1 and its downstream target, Hes1, whereas the IEE and ameloblasts express Jag1.67 In HAT-7 cells, a dental epithelial cell line, treatment with exogenous JAG1 led to the differentiation of SI cells, and this effect was neutralized with an anti-JAG1 antibody, pointing to the importance of Notch signaling in the SI.67
F. EDA
Ectodysplasin-A (Eda) is a member of the tumor necrosis factor (TNF) superfamily of signaling molecules. The EDA-A1 isoform, its receptor EDAR, and the adapter protein EDARADD act in a linear fashion and activate canonical NFKB signaling as well as other pathways.68 The EDA-A2 isoform signals through another receptor, EDA2R (formerly known as XEDAR), rather than EDAR, and activates similar pathways but appears to play a less important role in development.69 Mice with mutations in Eda, Edar or Edaradd (initially found as the spontaneous mutants tabby, downless, and crinkled, respectively) all have a decrease in the number of teeth with abnormal cusp morphology.68
During early odontogenesis, EDA signaling is crucial for determining the size of the tooth field and the number of teeth generated. Specifically, mutations in Eda or Edar resulted in formation of smaller teeth and frequently the absence of third molars.70 In contrast, increased levels of Eda expression, or expression of a constitutively active form of Edar, led to the formation of a supernumerary tooth in the diastema region.71, 72 In zebrafish, mutations in eda and edar led to defects in ectodermal structures such as scales and glands and partial or complete loss of pharyngeal teeth.73
EDA signaling also affects tooth shape. Mutations in any of the three pathway components in mice result in molars with reduced cusp number and rounded cusps. Eda is expressed in oral and dental epithelium throughout tooth formation, whereas Edar and Edaradd are expressed in the enamel knot. The enamel knots in tabby or crinkled mutants were smaller,74, 75 whereas loss of Edar in downless mice led to the formation of an elongated-rope like enamel knot.76 Interestingly, overexpression of Edar but not of Eda resulted in formation of extra cusps.71, 72 It is clear that the loss of function of Eda versus Edar has distinct effects on tooth size and morphology. This may be due to activation of the EDA2R pathway, which is influenced by Eda but not Edar expression, to interaction between EDAR and a yet unidentified protein, and/or to a ligand-independent activity for EDAR.77
G. Transcription Factors
The initial patterning as well as the coordinated interplay of signals at each step of tooth development is greatly dependent on the actions of transcription factors. Here, some of the general concepts and recent advances in our understanding of the roles of transcription factors in tooth development are discussed.
At E8.5, prior to any morphological signs of tooth development, Pitx2 is expressed in the stomatodeal epithelium, the precursor to oral and dental epithelium, and it is considered to be the earliest transcription factor expressed during tooth development.30, 78 Pax9 expression has been shown to specify the mesenchymal regions at the prospective sites of all teeth at E10.29 The direct regulation by PITX2 of Dlx2, a gene that is expressed at E9.5, is attenuated by a physical interaction between DLX2 and PITX2.79 At later stages, DLX2 and FOXJ1, a transcription factor expressed in the oral epithelium that plays a fundamental role in embryonic development, activate transcription of amelogenin, a tooth-specific protein required in enamel formation and mineralization.80
During initiation of tooth development, epithelial FGF8 and BMP4 induce the expression of numerous transcription factor genes including Barx1, Dlx1, Dlx2, Msx1, Msx2, Pax9, Pitx1, and Pitx2.29–31 The expression in prospective mesenchyme of many non-HOX homeobox-containing genes, such as Barx1, Dlx1, Dlx2, Dlx3, Dlx4, Dlx5, Dlx6, Lhx6, Lhx7, Msx1, and Msx2,81–83 led to the proposal of the odontogenic homeobox code model, which postulates that expression of specific combinations of homeobox gene directs the formation of specific tooth types.84
Msx1 and Pax9 act in the dental mesenchyme to maintain expression of Bmp4, which is crucial for establishing the enamel knot.32, 33 Absence of either of these transcription factors led to an arrest in tooth development at the bud stage, similar to that reported in Lef1-null embryos.64 Recently, it was shown that during early tooth formation, mesenchymal condensation (i.e. compression of mesenchyme) alone could regulate expression of Msx1 and Pax9, as well as Bmp4.85
H. microRNAs
There is an emerging role for microRNAs (miRNAs) in the development and evolution of teeth. Small RNAs, and miRNAs in particular, have important effects on development and disease through modulation of specific signaling pathways.86 miRNAs are endogenously expressed, short (~21 nucleotides), non-coding RNA molecules that affect protein synthesis by posttranscriptional mechanisms.87 The involvement of miRNAs in various ectodermal derivatives has been demonstrated in skin,88, 89 hair,90 and teeth.91–94 Pitx2-Cre;Dicer deleted mice showed a multiplication of enamel-free incisors, demonstrating the importance of miRNAs in ameloblast differentiation as well as their role in the regulation of ameloblast stem cells;91 Dicer is an RNAse III enzyme required for conversion of pre-miRNAs to mature miRNAs.95, 96 Krt14-Cre;Dicer deleted mice showed milder changes in tooth shape, epithelial homeostasis, and enamel formation.93 The differences in phenotype between the two mutant mice are likely due to the early expression of Pitx2 in stomatodeal epithelium compared to Krt14 expression. The expression of miRNAs in distinct regions of the mouse incisor and pulp was profiled using microarray experiments, laying the groundwork for future investigations.93, 94 These initial studies indicate that there is much work ahead in understanding the roles of miRNAs during tooth development.
III. EVOLUTION OF TOOTH DEVELOPMENT
A fundamental question in evolutionary developmental biology is how genetic changes contribute to morphologic variations that are subjected to natural selection. Teeth or tooth-like structures such as odontodes and denticles are invaluable in the study of evolutionary developmental biology for several reasons. First, teeth are ancient structures that are found in multiple locations in the vertebrate body such as the posterior pharynx of extinct jawless fish and extant fish, the dermal surface of sharks and rays, the oral cavity of rodents and humans, and lining the oro-pharyngeal cavity of fish in association with gill arches.97, 98 Second, there is great variation in the shape, size, number, and rows of teeth, and these variations are relatively easy to characterize. Third, teeth are readily fossilized vertebrate structures with excellent preservation of morphology due to the hardness of enamel, and thus they provide a large number of specimens for comparative genomic, anatomic, and phylogenetic studies. This section provides an overview of the main ideas and current research in the evolution of tooth development and highlights some of the multi-disciplinary approaches that can be utilized to answer important questions in the evolution and development of teeth.
A. THE ORIGIN OF TEETH IN VERTEBRATES
Teeth are an ancient and key vertebrate innovation, and their origin is a hotly debated question. The first occurrence of tooth-like structures is believed to be in the posterior pharynx of jawless fishes more than 500 million years ago.98, 99 With the evolution of jawed vertebrates, teeth developed on oral jaws and helped to establish the dominance of gnathostomes on land and in water.
It is still unclear whether oral teeth evolved with jaws for predation and mastication or first appeared as external dental armor as protection from predation. At least two opposing theories have been put forth regarding the evolution of oral teeth. The ‘outside-in’ theory posits that teeth evolved from ectoderm-derived, skin denticles that folded and integrated into the mouth.100 The ‘inside-out’ theory suggests that teeth originated from endoderm, with the formation of pharyngeal teeth in jawless vertebrates and moved anteriorly to the oral cavity with the evolution of jaws.101 However, recent studies suggest that neither theory may be entirely correct.102
Fate-mapping approaches using transgenic axolotls showed that teeth formed normally regardless of whether the oral epithelium was derived from ectoderm or endoderm.6 Experiments utilizing chicken embryos, which have lost the ability to form teeth,103 have demonstrated the dominant role of mesenchyme in the initiation of tooth development. Specifically, transplantation of mouse neural crest cells into developing chicken embryos showed the formation of tooth germ-like structures.104
Some extant fish, such as certain cichlids, possess both oral and pharyngeal teeth (Fig. 4). Pharyngeal teeth develop on discrete pharyngeal jaws in hox-positive, endoderm-derived sites, whereas oral teeth develop in hox-negative, ectoderm-derived regions.99 Pharyngeal teeth of jawless vertebrates appear to utilize an ancient gene network that predates the origin of oral jaws, oral teeth, and ectodermal appendages.99 During mouse development, expression of various genes such as claudin6, Foxa2, alpha-fetoprotein, Esrp1 (formerly known as Rbm35a), and Sox2 is observed in the presumptive molar region but not in the incisor region.5 In Chuk- (formerly known as Ikka) null mice, there was abnormal epithelial evagination in incisors but not in molars.105 These and other studies suggest differences in the epithelium from which incisor and molar teeth develop. However, despite distinct developmental environments, which suggest different molecular mechanisms that result in heterodont dentition, both oral and pharyngeal teeth also show striking similarities in their gene regulatory networks.99
Figure 4. Simplified evolutionary progression of dentitions and jaws.
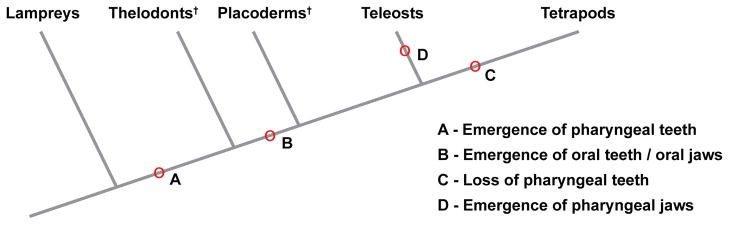
Point A indicates the origin of pharyngeal teeth in extinct (†) jawless fish. Oral teeth and jaws are thought to have arisen at point B. The pharyngeal teeth were lost in common ancestors to tetrapods at point C. In some extant teleosts such as cichlids, both oral and pharyngeal teeth are present and pharyngeal jaws are thought to have arisen at point D. Adapted from Fraser et al.99
Taken together, the studies using axolotl, cichlids, chicken, and mice demonstrate that teeth can form despite different epithelial origins and demonstrate the important role of mesenchyme in the initiation of tooth development, challenging the primacy of oral ectoderm in this role.14 These studies also demonstrate the conservation of gene regulatory networks across lineages with origins in different germ layers and the role of deep homology106 in the evolution and development of teeth. Thus, teeth appear to have evolved both ‘inside and out’, wherever and whenever the odontogenic-specific gene network of the mesenchyme was present.97
B. EVOLUTION OF TOOTH SHAPE, SIZE, NUMBER, AND ROWS
Both humans and rodents evolved from a common mammalian ancestor that is thought to have had a full complement of teeth comprising three incisors, one canine, four premolars, and three molars in each dental quadrant that replaced its teeth a single time.4 During mammalian evolution, teeth were lost along the lineages that gave rise to both rodents and humans (Fig. 2). Humans have all four of the major classes of teeth but have lost members of several of these classes; for example, we only have two incisors and two premolars (Fig. 2A, B). Rodent ancestors underwent a further reduction in dental formula, such that mice have only one incisor and three molars per quadrant, and no replacement teeth. Rodent teeth are considered to be deciduous teeth that do not undergo replacement,107–109 but the potential for replacement teeth in mice appears to have been retained.109–111
In addition to modifications in the number of teeth, the morphology of mammalian teeth is enormously diverse. These modifications involve variations in cusp shape and crest organization, and in the case of a number of species, the evolution of stem cell-fueled continuous growth, as discussed below. Comparative studies of tooth morphology have been greatly advanced by improvements in three-dimensional (3D) imaging techniques such as high-resolution micro-computed tomography. Some recent studies regarding the regulation of tooth shape, size, number, and rows are discussed below.
The various tooth shapes observed in heterodont animals are believed to have evolved from ancestral conical teeth, perhaps similar to canines, through the addition of cones and grooves.112 Relatively little is known regarding the molecular mechanisms underlying such changes, and therefore they are the subject of much current interest. Decreasing BMP signaling in the incisor region can lead an incisor to acquire a molar-like phenotype.113 However, it has recently been proposed that the molar-like phenotype was a result of the splitting of the incisor placode rather than a change in tooth identity.114 Lrp4-null mice displayed enamel grooves on the labial surface of incisors that exhibited similar molecular characteristics as molar cusps, suggesting that WNT signaling may be involved in cusp development.112
Two recent studies have provided important information about the developmental regulation of the relative size and number of molars. By using mouse molar cultures, it was proposed that a combination of activators and inhibitors governs the relative relationship between size and number of teeth.115 Detailed studies of tooth shape indicated that the complexity of the cusps directly reflects the animal’s diet across many mammalian species.115, 116 These studies pointed to higher order, generalizable principles that govern tooth shape and size.
Several studies have shown that alterations in signaling pathways can lead to variation in tooth number, and such studies point to mechanisms that may have determined tooth number during mammalian evolution. An example of dramatic tooth loss was highlighted in the cypriniform fish, a group including zebrafish. Zebrafish possess pharyngeal teeth, and fossil evidence suggests that zebrafish lost their oral teeth 50 million years ago (Fig. 4).117 This was associated with the loss of dlx2a and dlx2b expression in the oral epithelium. Because DLX genes are required for tooth development in mice,118 changes in trans-acting regulators of DLX genes that may be downstream of FGF signaling have been proposed as candidates responsible for the loss of cypriniform oral teeth.27 Interestingly, a region in the upstream regulatory element of dlx2b was retained that drives specific expression in the oral epithelium, and the retention of this cis-regulatory element is posited to be due to its requirement in other tissues, as the DLX genes have pleiotropic effects.119 These studies suggest that teeth lost from specific regions may be relatively easy to reacquire during evolution119 and they are exciting because they challenge Dollo’s Law of the Irreversibility of Evolution, which states that an organism can never exactly return to a previous evolutionary state120 because a lost structure cannot reappear in evolution.
A number of mouse mutants with changes in tooth number or pattern have provided tantalizing hints about the evolution of dentition. Supernumerary teeth present in the diastema region of mice (Fig. 2C, D), a species with reduced dentition, may represent the revival of teeth present in ancestral species. The following experiments in mice led to supernumerary teeth in the diastema region of the mandible: epithelial overexpression of Eda under the control of the Krt14 promoter;71 inactivation of the receptor tyrosine kinase antagonists, Spry2 and Spry4;24 production of a hypomorphic allele of the gene encoding Polaris, a protein involved in SHH signaling;121 a null mutation of the SHH antagonist, Gas1;49 and inactivation of the BMP/WNT pathway inhibitor gene, Sostdc1.37, 57, 58, 122 Diastema teeth in Krt14-Eda mutants and in Spry2-, Spry4-, polaris-, and Gas1-null mice have a size and shape characteristic of premolars, a tooth type that was lost in mice around 50–100 million years ago. Interestingly, diastema teeth in Sostdc1-null mice showed a molar-like phenotype, as well as enlarged enamel knots and altered cusp patterns.37, 57, 58, 122 In the diastema region, it was previously observed that the tooth primordium was present but failed to further develop because it does not maintain Shh expression.123–126 These studies demonstrate how loss of teeth from specific regions may be relatively easy to reacquire during evolution.
Mice carrying mutations in Sostdc1,58, 127 Lrp4,122 or inheriting the Di (duplicate incisor) trait128 have supernumerary upper or lower incisors that are located lingual to the normal incisor. Decreases in sprouty gene dosages also led to increasing numbers of incisors.129 The detailed study of Sostdc1 mutants indicated that the supernumerary incisors corresponded to replacement teeth.127 Splitting of the incisor placode has been observed in Sostdc1/Fst1 double-null mice, which have bifid incisors.114 The potential mechanisms by which supernumerary incisors arise in mice include: failure of integration of the ancestral dental primordia;109 development of replacement teeth;127 splitting of a large placode into smaller elements;114 or development of supernumerary tooth germs.130 These examples of supernumerary teeth may also reflect ancestral rodent dentition, in which a larger number of incisors was found, and highlight potential mechanisms by which humans and mice have evolved their reduced dentition.
Mammals possess a single row of teeth in the upper and lower jaws, unlike the multiple rows observed in some non-mammalian species such as fish and snakes. Teeth are replaced only once in most mammals, whereas in many non-mammalian species, teeth are continuously replaced. Additionally, in rodents, there are no replacement teeth, but there is continuous, stem cell-fueled growth of incisors, as well as of molars in species such as voles.131 Interestingly, supernumerary teeth developed lingual to the first molars in mice with inactivation of Osr2 (odd-skipped related-2), a gene homologous to the Drosophila transcriptional repressor odd-skipped.110 Osr2 limits the odontogenic field by suppressing the BMP4-MSX1 signaling cascade.110 The development of supernumerary teeth in Osr2 mutants may represent a second row of teeth similar to the multiple rows observed in some fish, and it may represent a reawakening of replacement teeth in mice.
C. COMPARATIVE TOOTH MORPHOLOGY AND MAMMALIAN EVOLUTION
Due to the highly mineralized nature of enamel, there is excellent preservation of detailed dental features in teeth from extant and extinct species. Using this vast repository of specimens, detailed 3D images can be constructed to compare subtle differences in tooth morphology. This information can be applied in interesting ways to further our understanding on the evolution of tooth development.
Comparative morphologic studies of mutant mice and various extinct and extant species have shed light on the role of specific genes in the evolution and development of tooth morphology. One such study showed that varying dosages of the Fgf3 gene caused morphological changes in teeth of mutant mice and in human patients (Fig. 5).132 Using comparisons between mice and humans carrying Fgf3 mutations with primitive rodent and primate fossils, it was observed that decreases in Fgf3 dosage led to tooth phenotypes that resembled the progressive reappearance of ancestral morphologies (Fig. 5).132 Progressive decreases in sprouty dosage caused increasing numbers of incisors, mimicking the dentition of rodent ancestors.129 Multidisciplinary approaches that integrate development and evolution can thus help to correlate subtle dental modifications with genetic mutations in a variety of mammalian lineages.
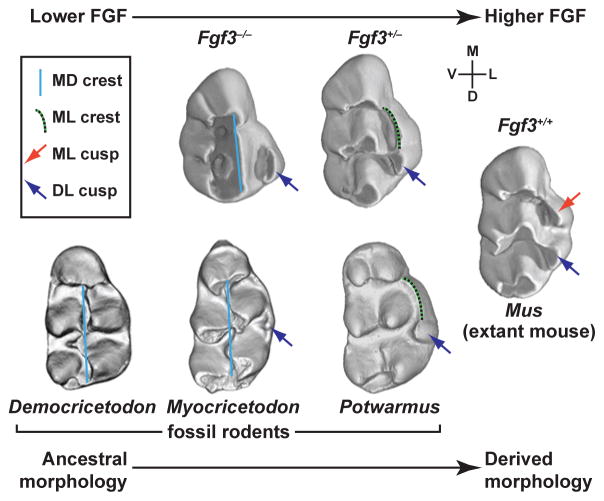
Figure 5. Dental morphology of Fgf3 mutant mice and fossil rodents.
As Fgf3 dosage is decreased in mice, the mesio-lingual (ML) cusp of the upper first molar is transformed into the ML crest (Fgf3+/−) and is eventually lost (Fgf3−/−), whereas the mesio-distal (MD) crest appears (Fgf3−/−). Comparisons of mutant and wild-type mice with fossil rodents such as Democricetodon, Myocricetodon, and Potswarmus show several features: during the transition from ancestral to derived morphologies, there is a loss of the MD crest, an emergence of the DL cusp, and an emergence of the ML crest that is transformed into the ML cusp in mice. The arrow indicating the relative levels of FGF signaling applies only to the allelic series of Fgf3 mutant mice, as the expression levels of Fgf3 in muroid ancestors is unknown. The following abbreviations are used for orientation: M, mesial; D, distal; V, vestibular; L, lingual. Figure adapted from Charles et al.132
A large amount of information can be extracted from the analysis of fossilized teeth. For example, a record of growth can be attained from the enamel and dentin that allows the reconstruction of the developmental history and the timing of crown and root formation. Measurements of daily enamel cross-striations can be used to infer information about the timing and rate of enamel/crown formation; accentuated neonatal lines in the enamel of deciduous and permanent molars may denote the time of birth; incremental markings in the dentin indicate the timing of root completion; and the quality of the enamel-dentin junction, where the crown meets the root, provides a window to tooth development and the actions of the enamel knot.133 Using such techniques, tooth development in Neanderthals was shown to closely resemble that of human populations, underscoring the similarities between humans and Neanderthals.133
In another study utilizing fossilized teeth to understand the evolution of species, the worn cusp apices of teeth (mesowear) from North American horses for the past 55.5 million years was analyzed. Hypsodonty (high-crowned teeth) was correlated with mesowear, thereby strengthening the argument that the evolution of hypsodonty relative to brachydonty (short-crowned teeth) was adapted for abrasive diets associated with the spread of grasslands in North America.134 In brachydont species such as humans, the tooth crown is entirely above the level of the jaw bone upon initial eruption, whereas in hypsodont species, some of the tooth crown is retained below the level of the jaw bone.135 In rodents, hypsodonty is posited to be an intermediate stage on the evolutionary path towards hypselodonty (ever-growing teeth).135
Thus, by utilizing model and non-model organisms to analyze genetic and signaling pathways, along with detailed 3D reconstructions of teeth in extant and extinct species and in combination with ecological data, some of the exciting multi-disciplinary studies discussed above are at the forefront of research in the evolution of tooth development.
IV. DENTAL STEM CELLS
The regenerative ability of many adult tissues is dependent on tissue-specific stem cell populations that maintain stable numbers by self-renewal and that possess the capacity to differentiate into distinct cell lineages. Regeneration and renewal in adult mammals has been studied in several organs, including the blood, gut, brain, skin, and hair. Here, we describe the advent of the continuously growing mouse incisor as an adult stem cell model system. Although the study of incisor stem cells is a relatively new field, advances have recently been made in the identification of these cells, the understanding of their function, and the characterization of molecular mechanisms that regulate their behavior.
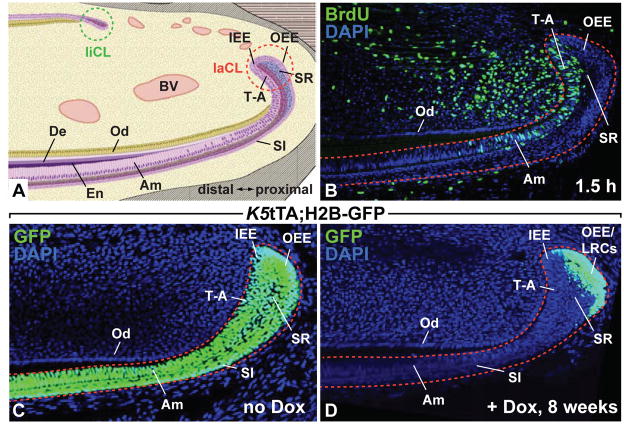
In mice, both molars and incisors go through similar developmental stages at early stages of odontogenesis, but incisors continue to grow throughout postnatal life, whereas molars cease growth in the perinatal period. The ability of the incisor to grow continuously is dependent on the presence of epithelial and mesenchymal stem cells that have the capacity to self-renew and differentiate into all of the cell types of the adult tooth, including ameloblasts, odontoblasts, and the SI. Importantly, in the wild-type rodent incisor, the labial CL, but not the lingual CL, contains stem cells that give rise to ameloblasts and the SI (Fig. 6). Labeling experiments demonstrated that cells in the dental epithelium move in a proximal to distal direction.136 In the labial CL, the stem cell progeny contribute to a population of transit amplifying (T-A) cells (Fig. 6B). T-A cells undergo several rounds of cell division before they move distally and differentiate into ameloblasts. Early labeling experiments examining the rate of ameloblast and odontoblast migration in mice and rats gave the first clues that turnover of these specialized cells was rapid, underlining the need for progenitor pools to resupply differentiated cell populations.136, 137 Mouse incisor epithelia appear to function as a “conveyor belt”, moving cells from a proximal, undifferentiated source to regularly repopulate the tooth with specialized cell types. Initial studies using explant cultures of the CL region from 2 day-old mice showed that new epithelial structures could be generated in culture, indicating that the labial CL housed the dental stem cells that give rise to ameloblasts and the SI.138
Figure 6. The adult mouse incisor.
The lower incisor is shown in sagittal view (A–D). (A) The diagram indicates the two stem cell compartments in the lingual (liCL) and labial (laCL) cervical loops. Also shown are the inner enamel epithelium (IEE), from which the transit-amplifying (T-A) cells and ameloblasts (Am) arise, the outer enamel epithelium (OEE) that house the enamel stem cells in the laCL, stellate reticulum (SR), stratum intermedium (SI), odontoblasts (Od), dentin (De), enamel (En), and blood vessels (BV). (B) Adult mice were injected with BrdU for 1.5 h. BrdU-positive cells indicate rapidly proliferating cells in the T-A region. (C–D) Images from incisors of Krt5-tTA; H2B-GFP mice. In the absence of doxycycline (no Dox; C), GFP is present in all the cells expressing Krt5, which includes the OEE, IEE, SR, SI, and Am. In the presence of doxycycline (+ Dox; D) for 8 weeks, H2B-GFP expression was turned off, leading to the retention of GFP in the slowly proliferating label-retaining cells (LRCs) of the OEE. The LRCs are putative dental epithelial stem cells.
Identification of organ-specific adult stem cell populations can be challenging, because stem cells often reside in heterogeneous niches intermingled with support cells. A useful character of stem cells that has aided in their identification in vivo is the relatively slow cell-division kinetics of many stem cells relative to surrounding tissue.139 Slow-cycling cell populations have largely been identified through label retention experiments, traditionally utilizing BrdU incorporation, because cells that divide slowly do not dilute the BrdU label as quickly as their rapidly dividing neighbors. Using this technique, BrdU label-retaining cells (LRCs) were identified in the labial CL of cultured perinatal incisors and in adult incisors in situ.138,119 Another approach to label retention is the use of transgenic mice harboring a tetracycline-sensitive, histone H2B-GFP cassette under the control of a tissue specific trans-activator (Fig. 6C, D).140 Expression of H2B-GFP is initially activated in all cells of the tissue of interest followed by a “chase” period when the transgene is repressed by exposure of the animal to doxycycline, such that rapidly dividing cells dilute the label. This technique was used to identify LRCs in the outer enamel epithelium (OEE) of the adult labial CL.141 The LRCs of the dental epithelium expressed Gli1, a target of SHH signaling, and lineage tracing experiments demonstrated that the Gli1-expressing cells were indeed stem cells.141 More recently, identification of LRCs in non-mammalian vertebrates has been pursued.142, 143
Understanding the regulation of adult stem cell populations is key to future utilization of such cells for clinical therapies. How stem cells are maintained at the appropriate number, what signals regulate their differentiation, and how they are established within the context of the developing organism are important questions in stem cell research. Many signaling molecules and pathways are implicated in development and homeostasis of the incisor, including WNTs, BMPS and FGFs. One theme that has emerged from several recent studies is the convergence of distinct FGF signaling pathways that maintain the size and shape of the CL through regulation of cell division and death. Expression analyses first indicated that components of the FGF pathways may play major roles in the mouse incisor.22, 138 Specifically, Fgf3 and Fgf10 are expressed in the mesenchyme immediately adjacent to and surrounding the labial CL, whereas Fgfr1b and Fgfr2b are enriched in the epithelium. Incisors of Fgf3 null mice at P0 are indistinguishable from those in wild-type mice, yet Fgf3−/−;Fgf10+/− compound mutants reveal a severely hypoplastic labial CL, indicating that precise levels of FGF signaling are responsible for regulating the size and shape of the stem cell niche.22 Consistent with this result, attenuation of signaling via FGFR2B in the embryo using a tetracycline-inducible dominant negative system gave similar results in E18.5 embryos.144
It appears that several additional signaling pathways either directly or indirectly converge on FGF signaling via regulation of Fgf3 to control proliferation in developing incisors. Mis-expression of Fst throughout the dental epithelium under the control of the Krt14 promoter resulted in complete down-regulation of mesenchymal Fgf3 expression with reduction in epithelial proliferation and severely hypoplastic labial CLs.22 Conversely, loss of Fst in the epithelium of incisors led to up-regulated Fgf3 expression in mesenchyme adjacent to the lingual CL, causing increased proliferation and expansion of the lingual CL.22 Deletion of Tgfbr1 (formerly known as Alk5) in the mesenchyme led to down-regulation of Fgf3, Fgf9, Fgf10, and a reduced labial CL size, likely due to proliferation defects.145. Notably, fewer LRCs survived in this mutant, and the defect could be reversed by the addition of exogenous FGF10. Thus, several lines of evidence indicate that FGF signaling is involved in the maintenance of incisor stem cell number.
The sprouty genes encode negative feedback regulators of FGF signaling that are expressed in both the lingual and labial epithelia as well as in the mesenchyme adjacent to the labial CL. Loss of sprouty function in the incisor resulted in up-regulation of FGF gene expression in the lingual epithelium and mesenchyme and the presence of ameloblasts in the lingual epithelium.25 This study highlighted the importance of balanced FGF signaling in the incisor to maintain asymmetric production of ameloblasts on the labial side, while preventing ameloblast formation and activity on the lingual side.
In addition to FGF signaling, incisor stem cells require the activity of the Notch pathway to ensure development and survival. Three Notch receptors are expressed in the CL regions in the developing incisor. Notch1 and Notch2 are expressed in the epithelium and mesenchyme, whereas Notch3 is restricted to the mesenchyme.138 The Notch ligand encoded by Jag2 is strongly expressed in the epithelium, and Jag2-null mice show incisors with defects in cellular morphology.66 Inhibition of Notch signaling with DAPT led to a reduction in the size of the labial CL in explant experiments.146
Comparative analysis of two different rodent species provides a potential mechanism for the evolution of ever-growing teeth. The sibling vole (Microtus rossiaemeridionalis), in contrast to mice, possesses ever-growing incisors and molars.131 During development, CLs in mouse molars undergo a transition to the root fate and cease producing enamel. Vole molars do not undergo such a transition, and enamel is continuously generated for the life of the animal.131 Interestingly, mouse and vole molars are practically identical in morphology and distribution of developmental markers until E17, when the molars are in the late bell stage. Given the roles of FGF and Notch signaling in maintenance of the stem cell niche in the mouse incisor, these pathways were compared at stages during which mouse molars initiate root formation.131 The development of roots in the mouse coincides with loss of epithelial Notch and mesenchymal FGF signaling, whereas vole molars continue to express key signaling components and partially bypass the root fate. This idea is substantiated by evidence showing that the maintenance of Notch signaling in the cervical loops of mouse molars grown in vitro resulted in continuous crown development in lieu of root formation.147 Although correlative, these studies suggest that vole molars have evolved mechanisms that maintain the necessary morphology for continuous growth fueled by molar cervical loops, which may be analogous to the stem cell niche of the mouse incisor.
CONCLUDING REMARKS
Important advances have been made in our understanding of the development and evolution of teeth. The tooth provides a valuable model for the elucidation of major biological questions, and the identification and initial characterization of dental stem cells have been exciting recent developments. Many tooth-specific defects in model and non-model organisms mirror conditions present in humans and provide a means to study their genetics, development, and pathology. For a comprehensive review on the diseases of the tooth, please refer to the review by XX et al. However, much work remains to be done, and the utilization of cellular, molecular, and genetic approaches, as well as anthropological and paleontological techniques, will enable continued progress. The rapid increases in our understanding of dental development in extant and extinct vertebrate species using techniques including 3D imaging, genetic manipulations, omics analyses, and genome-wide association studies make this an exciting time to study the development and evolution of teeth.
Acknowledgments
We apologize to those colleagues whose work we were unable to cite due to space constraints. We would like to thank our colleagues in the UCSF Craniofacial and Mesenchymal Biology Program and Jukka Jernvall, Irma Thesleff, Renata Peterkova, Herve Lesot, Vagan Mushegyan, Cyril Charles, Laurent Viriot, Ann Huysseune, and Gail Martin for helpful discussions. The authors are funded in part by the National Institutes of Health (DP2-OD007191 and R01-DE021420 to O.D.K., and K99-DE022059 to A.H.J.).
Abbreviations used
- Am
ameloblasts
- BV
blood vessels
- B
bone
- C
canine
- Ce
cementum
- CL
cervical loop
- De
dentin
- E
embryonic day
- D
distal
- G
gingiva
- GFP
green fluorescent protein
- I
incisor
- IEE
inner enamel epithelium
- LRCs
label-retaining cells
- laCL
labial cervical loop, L, lingual
- liCL
lingual cervical loop
- M
mesial
- M
molar
- Od
odontoblasts
- OEE
outer enamel epithelium
- pl
periodontal ligament
- P
postnatal day
- PM
premolar
- p
pulp chamber
- SI
stratum intermedium
- SR
stellate reticulum
- T-A
transit-amplifying
- V
vestibular
- VL
vestibular lamina
- 3D
3-dimensional
Footnotes
SUPPORTING INFORMATION
Gene and protein names in the mouse and human genome databases are available from the NCBI website, http://www.ensembl.org/index.html.
References
- 1.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 2.Huysseune A, Witten PE. Developmental mechanisms underlying tooth patterning in continuously replacing osteichthyan dentitions. J Exp Zool B Mol Dev Evol. 2006;306:204–215. doi: 10.1002/jez.b.21091. [DOI] [PubMed] [Google Scholar]
- 3.Hill JP. Development of the Monotremata. Part VII. The development and structure of the egg-tooth and the caruncle in the monotremes and on the occurrence of vestiges of the egg-tooth and caruncle in marsupials. Trans Zool Soc Lond. 1949;26:503–544. [Google Scholar]
- 4.Line SR. Variation of tooth number in mammalian dentition: connecting genetics, development, and evolution. Evol Dev. 2003;5:295–304. doi: 10.1046/j.1525-142x.2003.03036.x. [DOI] [PubMed] [Google Scholar]
- 5.Ohazama A, Haworth KE, Ota MS, Khonsari RH, Sharpe PT. Ectoderm, endoderm, and the evolution of heterodont dentitions. Genesis. 2010;48:382–389. doi: 10.1002/dvg.20634. [DOI] [PubMed] [Google Scholar]
- 6.Soukup V, Epperlein HH, Horacek I, Cerny R. Dual epithelial origin of vertebrate oral teeth. Nature. 2008;455:795–798. doi: 10.1038/nature07304. [DOI] [PubMed] [Google Scholar]
- 7.Noden DM, Schneider RA. Neural crest cells and the community of plan for craniofacial development: historical debates and current perspectives. Adv Exp Med Biol. 2006;589:1–23. doi: 10.1007/978-0-387-46954-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 9.Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- 10.Avery JK. Development of teeth: crown formation. 3. Stuttgart: Thieme; 2002. [Google Scholar]
- 11.Ten Cate A, Mills C, Solomon G. The development of the periodontium. A transplantation and autoradiographic study. Anat Rec. 1971;170:365–380. doi: 10.1002/ar.1091700312. [DOI] [PubMed] [Google Scholar]
- 12.Ten Cate AR, Mills C. The development of the periodontium: the origin of alveolar bone. Anat Rec. 1972;170:365–380. doi: 10.1002/ar.1091730106. [DOI] [PubMed] [Google Scholar]
- 13.Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol Dev. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- 14.Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- 15.Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103 (Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- 16.Kollar EJ, Baird GR. Tissue interactions in embryonic mouse tooth germs. II. The inductive role of the dental papilla. J Embryol Exp Morphol. 1970;24:173–186. [PubMed] [Google Scholar]
- 17.Kollar EJ, Baird GR. The influence of the dental papilla on the development of tooth shape in embryonic mouse tooth germs. J Embryol Exp Morphol. 1969;21:131–148. [PubMed] [Google Scholar]
- 18.Ho SP, Marshall SJ, Ryder MI, Marshall GW. The tooth attachment mechanism defined by structure, chemical composition and mechanical properties of collagen fibers in the periodontium. Biomaterials. 2007;28:5238–5245. doi: 10.1016/j.biomaterials.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 21.Kettunen P, Karavanova I, Thesleff I. Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4; and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev Genet. 1998;22:374–385. doi: 10.1002/(SICI)1520-6408(1998)22:4<374::AID-DVG7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- 24.Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Stock DW, Jackman WR, Trapani J. Developmental genetic mechanisms of evolutionary tooth loss in cypriniform fishes. Development. 2006;133:3127–3137. doi: 10.1242/dev.02459. [DOI] [PubMed] [Google Scholar]
- 28.Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 29.Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 30.St Amand TR, Zhang Y, Semina EV, Zhao X, Hu Y, Nguyen L, Murray JC, Chen Y. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- 31.Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 33.Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- 34.Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 35.Ferguson CA, Tucker AS, Christensen L, Lau AL, Matzuk MM, Sharpe PT. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes Dev. 1998;12:2636–2649. doi: 10.1101/gad.12.16.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen Y. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev Biol. 2011;349:451–461. doi: 10.1016/j.ydbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassai Y, Munne P, Hotta Y, Penttila E, Kavanagh K, Ohbayashi N, Takada S, Thesleff I, Jernvall J, Itoh N. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- 38.Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Yamashiro T, Tummers M, Thesleff I. Expression of bone morphogenetic proteins and Msx genes during root formation. J Dent Res. 2003;82:172–176. doi: 10.1177/154405910308200305. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Huang X, Xu X, Mayo J, Bringas P, Jr, Jiang R, Wang S, Chai Y. SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development. 2011;138:1977–1989. doi: 10.1242/dev.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 43.Cobourne MT, Hardcastle Z, Sharpe PT. Sonic hedgehog regulates epithelial proliferation and cell survival in the developing tooth germ. J Dent Res. 2001;80:1974–1979. doi: 10.1177/00220345010800110501. [DOI] [PubMed] [Google Scholar]
- 44.Hardcastle Z, Mo R, Hui CC, Sharpe PT. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- 45.Dassule HR, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 46.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 47.Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- 48.Cho SW, Kwak S, Woolley TE, Lee MJ, Kim EJ, Baker RE, Kim HJ, Shin JS, Tickle C, Maini PK, et al. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development. 2011;138:1807–1816. doi: 10.1242/dev.056051. [DOI] [PubMed] [Google Scholar]
- 49.Ohazama A, Haycraft CJ, Seppala M, Blackburn J, Ghafoor S, Cobourne M, Martinelli DC, Fan CM, Peterkova R, Lesot H, et al. Primary cilia regulate Shh activity in the control of molar tooth number. Development. 2009;136:897–903. doi: 10.1242/dev.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackman WR, Yoo JJ, Stock DW. Hedgehog signaling is required at multiple stages of zebrafish tooth development. BMC Dev Biol. 2010;10:119. doi: 10.1186/1471-213X-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura T, de Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J Biol Chem. 2008;283:4825–4833. doi: 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- 55.Kuraguchi M, Wang XP, Bronson RT, Rothenberg R, Ohene-Baah NY, Lund JJ, Kucherlapati M, Maas RL, Kucherlapati R. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XP, O’Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn Y, Sanderson BW, Klein OD, Krumlauf R. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 2010;137:3221–3231. doi: 10.1242/dev.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murashima-Suginami A, Takahashi K, Kawabata T, Sakata T, Tsukamoto H, Sugai M, Yanagita M, Shimizu A, Sakurai T, Slavkin HC, et al. Rudiment incisors survive and erupt as supernumerary teeth as a result of USAG-1 abrogation. Biochem Biophys Res Commun. 2007;359:549–555. doi: 10.1016/j.bbrc.2007.05.148. [DOI] [PubMed] [Google Scholar]
- 59.Liu F, Dangaria S, Andl T, Zhang Y, Wright AC, Damek-Poprawa M, Piccolo S, Nagy A, Taketo MM, Diekwisch TG, et al. beta-Catenin initiates tooth neogenesis in adult rodent incisors. J Dent Res. 2010;89:909–914. doi: 10.1177/0022034510370090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bohring A, Stamm T, Spaich C, Haase C, Spree K, Hehr U, Hoffmann M, Ledig S, Sel S, Wieacker P, et al. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am J Hum Genet. 2009;85:97–105. doi: 10.1016/j.ajhg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarkar L, Cobourne M, Naylor S, Smalley M, Dale T, Sharpe PT. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc Natl Acad Sci U S A. 2000;97:4520–4524. doi: 10.1073/pnas.97.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujimori S, Novak H, Weissenbock M, Jussila M, Goncalves A, Zeller R, Galloway J, Thesleff I, Hartmann C. Wnt/beta-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev Biol. 2010;348:97–106. doi: 10.1016/j.ydbio.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- 65.Bei M. Molecular genetics of ameloblast cell lineage. J Exp Zool. 2009;312B:437–444. doi: 10.1002/jez.b.21261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025–3035. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, Wakisaka S. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006;340:611–616. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 68.Courtney JM, Blackburn J, Sharpe PT. The Ectodysplasin and NFkappaB signalling pathways in odontogenesis. Arch Oral Biol. 2005;50:159–163. doi: 10.1016/j.archoralbio.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 69.Mikkola ML. TNF superfamily in skin appendage development. Cytokine Growth Factor Rev. 2008;19:219–230. doi: 10.1016/j.cytogfr.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Sofaer JA. The teeth of the “sleek” mouse. Arch Oral Biol. 1977;22:299–301. doi: 10.1016/0003-9969(77)90117-0. [DOI] [PubMed] [Google Scholar]
- 71.Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, Pakkasjarvi L, Jaatinen R, Thesleff I. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- 72.Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev Biol. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 73.Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nusslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 2008;4:e1000206. doi: 10.1371/journal.pgen.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pispa J, Jung HS, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, Kere J, Thesleff I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- 75.Ohazama A, Courtney JM, Tucker AS, Naito A, Tanaka S, Inoue J, Sharpe PT. Traf6 is essential for murine tooth cusp morphogenesis. Dev Dyn. 2004;229:131–135. doi: 10.1002/dvdy.10400. [DOI] [PubMed] [Google Scholar]
- 76.AS, Headon DJ, Schneider P, Ferguson BM, Overbeek P, Tschopp J, Sharpe PT. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- 77.Charles C, Pantalacci S, Tafforeau P, Headon D, Laudet V, Viriot L. Distinct impacts of Eda and Edar loss of function on the mouse dentition. PLoS One. 2009;4:e4985. doi: 10.1371/journal.pone.0004985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitsiadis TA, Mucchielli ML, Raffo S, Proust JP, Koopman P, Goridis C. Expression of the transcription factors Otlx2, Barx1 and Sox9 during mouse odontogenesis. Eur J Oral Sci. 1998;106 (Suppl 1):112–116. doi: 10.1111/j.1600-0722.1998.tb02161.x. [DOI] [PubMed] [Google Scholar]
- 79.Venugopalan SR, Li X, Amen MA, Florez S, Gutierrez D, Cao H, Wang J, Amendt BA. Hierarchical interactions of homeodomain and forkhead transcription factors in regulating odontogenic gene expression. J Biol Chem. 2011;286:21372–21383. doi: 10.1074/jbc.M111.252031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 81.Tissier-Seta JP, Mucchielli ML, Mark M, Mattei MG, Goridis C, Brunet JF. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech Dev. 1995;51:3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- 82.Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- 83.Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 84.Sharpe PT. Homeobox genes and orofacial development. Connect Tissue Res. 1995;32:17–25. doi: 10.3109/03008209509013701. [DOI] [PubMed] [Google Scholar]
- 85.Mammoto T, Mammoto A, Torisawa YS, Tat T, Gibbs A, Derda R, Mannix R, de Bruijn M, Yung CW, Huh D, et al. Mechanochemical Control of Mesenchymal Condensation and Embryonic Tooth Organ Formation. Dev Cell. 2011 doi: 10.1016/j.devcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 88.Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 89.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cao H, Wang J, Li X, Florez S, Huang Z, Venugopalan SR, Elangovan S, Skobe Z, Margolis HC, Martin JF, et al. MicroRNAs play a critical role in tooth development. J Dent Res. 2010;89:779–784. doi: 10.1177/0022034510369304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jevnaker AM, Osmundsen H. MicroRNA expression profiling of the developing murine molar tooth germ and the developing murine submandibular salivary gland. Arch Oral Biol. 2008;53:629–645. doi: 10.1016/j.archoralbio.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Michon F, Tummers M, Kyyronen M, Frilander MJ, Thesleff I. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev Biol. 2010;340:355–368. doi: 10.1016/j.ydbio.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 94.Jheon AH, Li CY, Wen T, Michon F, Klein OD. Expression of MicroRNAs in the Stem Cell Niche of the Adult Mouse Incisor. PLoS One. 2011;6:e24536. doi: 10.1371/journal.pone.0024536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 96.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 97.Fraser GJ, Cerny R, Soukup V, Bronner-Fraser M, Streelman JT. The odontode explosion: the origin of tooth-like structures in vertebrates. Bioessays. 2010;32:808–817. doi: 10.1002/bies.200900151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sire JY, Huysseune A. Formation of dermal skeletal and dental tissues in fish: a comparative and evolutionary approach. Biol Rev Camb Philos Soc. 2003;78:219–249. doi: 10.1017/s1464793102006073. [DOI] [PubMed] [Google Scholar]
- 99.Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7:e31. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reif W-E. Evolution of dermal skeleton and dentition in vertebrates. Evol Dev. 1982;15:287–368. [Google Scholar]
- 101.Smith MM. Vertebrate dentitions at the origin of jaws: when and how pattern evolved. Evol Dev. 2003;5:394–413. doi: 10.1046/j.1525-142x.2003.03047.x. [DOI] [PubMed] [Google Scholar]
- 102.Huysseune A, Sire JY, Witten PE. Evolutionary and developmental origins of the vertebrate dentition. J Anat. 2009;214:465–476. doi: 10.1111/j.1469-7580.2009.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sire JY, Delgado SC, Girondot M. Hen’s teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mitsiadis TA, Cheraud Y, Sharpe P, Fontaine-Perus J. Development of teeth in chick embryos after mouse neural crest transplantations. Proc Natl Acad Sci U S A. 2003;100:6541–6545. doi: 10.1073/pnas.1137104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohazama A, Hu Y, Schmidt-Ullrich R, Cao Y, Scheidereit C, Karin M, Sharpe PT. A dual role for Ikk alpha in tooth development. Dev Cell. 2004;6:219–227. doi: 10.1016/s1534-5807(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 106.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 107.Moss-Salentijn L. Studies on dentin. 2. Vestigial lacteal incisor teeth of the rat. Acta Anat (Basel) 1975;92:329–350. [PubMed] [Google Scholar]
- 108.Moss-Salentijn L. Vestigial teeth in rabbit, rat and mouse; their relationship to the problem of lacteal dentitions. London: Academic Press; 1978. [Google Scholar]
- 109.Hovorakova M, Prochazka J, Lesot H, Smrckova L, Churava S, Boran T, Kozmik Z, Klein O, Peterkova R, Peterka M. Shh expression in a rudimentary tooth offers new insights into development of the mouse incisor. J Exp Zool B Mol Dev Evol. 2011;316:347–358. doi: 10.1002/jez.b.21408. [DOI] [PubMed] [Google Scholar]
- 110.Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang XP, Aberg T, James MJ, Levanon D, Groner Y, Thesleff I. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138–143. doi: 10.1177/154405910508400206. [DOI] [PubMed] [Google Scholar]
- 112.Ohazama A, Blackburn J, Porntaveetus T, Ota MS, Choi HY, Johnson EB, Myers P, Oommen S, Eto K, Kessler JA, et al. A role for suppressed incisor cuspal morphogenesis in the evolution of mammalian heterodont dentition. Proc Natl Acad Sci U S A. 2010;107:92–97. doi: 10.1073/pnas.0907236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 114.Munne PM, Felszeghy S, Jussila M, Suomalainen M, Thesleff I, Jernvall J. Splitting placodes: effects of bone morphogenetic protein and Activin on the patterning and identity of mouse incisors. Evol Dev. 2010;12:383–392. doi: 10.1111/j.1525-142X.2010.00425.x. [DOI] [PubMed] [Google Scholar]
- 115.Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449:427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 116.Evans AR, Wilson GP, Fortelius M, Jernvall J. High-level similarity of dentitions in carnivorans and rodents. Nature. 2007;445:78–81. doi: 10.1038/nature05433. [DOI] [PubMed] [Google Scholar]
- 117.Nelson JS. Fishes of the World. 4. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- 118.Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- 119.Jackman WR, Stock DW. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc Natl Acad Sci U S A. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dollo L. Les lois de l’évolution. Bull Soc Belge Geol Pal Hydr. 1893;7:164–166. [Google Scholar]
- 121.Zhang Q, Murcia NS, Chittenden LR, Richards WG, Michaud EJ, Woychik RP, Yoder BK. Loss of the Tg737 protein results in skeletal patterning defects. Dev Dyn. 2003;227:78–90. doi: 10.1002/dvdy.10289. [DOI] [PubMed] [Google Scholar]
- 122.Ohazama A, Johnson EB, Ota MS, Choi HY, Porntaveetus T, Oommen S, Itoh N, Eto K, Gritli-Linde A, Herz J, et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peterkova R, Peterka M, Viriot L, Lesot H. Development of the vestigial tooth primordia as part of mouse odontogenesis. Connect Tissue Res. 2002;43:120–128. doi: 10.1080/03008200290000745. [DOI] [PubMed] [Google Scholar]
- 124.Tureckova J, Lesot H, Vonesch JL, Peterka M, Peterkova R, Ruch JV. Apoptosis is involved in the disappearance of the diastemal dental primordia in mouse embryo. Int J Dev Biol. 1996;40:483–489. [PubMed] [Google Scholar]
- 125.Cobourne MT, Miletich I, Sharpe PT. Restriction of sonic hedgehog signalling during early tooth development. Development. 2004;131:2875–2885. doi: 10.1242/dev.01163. [DOI] [PubMed] [Google Scholar]
- 126.Prochazka J, Pantalacci S, Churava S, Rothova M, Lambert A, Lesot H, Klein O, Peterka M, Laudet V, Peterkova R. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci U S A. 2010;107:15497–15502. doi: 10.1073/pnas.1002784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Munne PM, Tummers M, Jarvinen E, Thesleff I, Jernvall J. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 2009;136:393–402. doi: 10.1242/dev.025064. [DOI] [PubMed] [Google Scholar]
- 128.Danforth CH. The Occurrence and Genetic Behavior of Duplicate Lower Incisors in the Mouse. Genetics. 1958;43:139–148. doi: 10.1093/genetics/43.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Charles C, Hovorakova M, Ahn Y, Lyons D, Marangoni P, Churava S, Biehs B, Jheon A, Lesot H, Balooch G, et al. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development. 2011;138:4063–4073. doi: 10.1242/dev.069195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sofaer JA. The genetics and expression of a dental morphological variant in the mouse. Arch Oral Biol. 1969;14:1213–1223. doi: 10.1016/0003-9969(69)90159-9. [DOI] [PubMed] [Google Scholar]
- 131.Tummers M, Thesleff I. Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development. 2003;130:1049–1057. doi: 10.1242/dev.00332. [DOI] [PubMed] [Google Scholar]
- 132.Charles C, Lazzari V, Tafforeau P, Schimmang T, Tekin M, Klein O, Viriot L. Modulation of Fgf3 dosage in mouse and men mirrors evolution of mammalian dentition. Proc Natl Acad Sci U S A. 2009;106:22364–22368. doi: 10.1073/pnas.0910086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Macchiarelli R, Bondioli L, Debenath A, Mazurier A, Tournepiche JF, Birch W, Dean MC. How Neanderthal molar teeth grew. Nature. 2006;444:748–751. doi: 10.1038/nature05314. [DOI] [PubMed] [Google Scholar]
- 134.Mihlbachler MC, Rivals F, Solounias N, Semprebon GM. Dietary change and evolution of horses in North America. Science. 2011;331:1178–1181. doi: 10.1126/science.1196166. [DOI] [PubMed] [Google Scholar]
- 135.Damuth J, Janis CM. On the relationship between hypsodonty and feeding ecology in ungulate mammals, and its utility in palaeoecology. Biol Rev Camb Philos Soc. 2011;86:733–758. doi: 10.1111/j.1469-185X.2011.00176.x. [DOI] [PubMed] [Google Scholar]
- 136.Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec. 1975;183:523–561. doi: 10.1002/ar.1091830405. [DOI] [PubMed] [Google Scholar]
- 137.Hwang WS, Tonna EA. Autoradiographic Analysis of Labeling Indices and Migration Rates of Cellular Component of Mouse Incisors Using Tritiated Thymidine (H3tdr) J Dent Res. 1965;44:42–53. doi: 10.1177/00220345650440012901. [DOI] [PubMed] [Google Scholar]
- 138.Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Handrigan GR, Leung KJ, Richman JM. Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development. 2010;137:3545–3549. doi: 10.1242/dev.052415. [DOI] [PubMed] [Google Scholar]
- 143.Huysseune A, Thesleff I. Continuous tooth replacement: the possible involvement of epithelial stem cells. Bioessays. 2004;26:665–671. doi: 10.1002/bies.20039. [DOI] [PubMed] [Google Scholar]
- 144.Parsa S, Kuremoto K, Seidel K, Tabatabai R, Mackenzie B, Yamaza T, Akiyama K, Branch J, Koh CJ, Al Alam D, et al. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development. 2010;137:3743–3752. doi: 10.1242/dev.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor beta signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol. 2011;31:2079–2089. doi: 10.1128/MCB.01439-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010;80:241–248. doi: 10.1016/j.diff.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 147.Tummers M, Yamashiro T, Thesleff I. Modulation of epithelial cell fate of the root in vitro. J Dent Res. 2007;86:1063–1067. doi: 10.1177/154405910708601108. [DOI] [PubMed] [Google Scholar]