Abstract
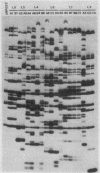
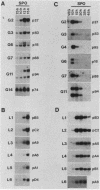
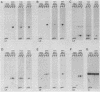
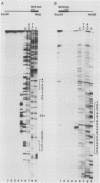
We previously described the use of a differential hybridization screen of a genomic DNA library of Saccharomyces cerevisiae to identify sporulation-specific (SPS) genes (A. Percival-Smith and J. Segall, Mol. Cell. Biol. 4:142-150, 1984). This initial screen identified 14 SPS genes that are first expressed 6 to 8 h after transfer of cells to sporulation medium. Accumulation of transcripts corresponding to these genes becomes maximal at 8 to 12 h of sporulation, the time at which meiotic events are nearing completion, and by 15 h of sporulation, transcript levels are beginning to decrease. In the present study two additional SPS genes, first expressed at 12 h of sporulation, were isolated. The steady-state level of transcripts corresponding to these two genes, termed SPS100 and SPS101, remains unchanged from 15 to 35 h, a time coincident with spore wall maturation. The nature of the putative 34.2-kilodalton protein encoded by the SPS100 gene is consistent with its being a component of the glycoprotein matrix of the spore wall; the protein contains a potential signal sequence and cleavage site and numerous sites for potential glycosylation. A MATa sps100/MAT alpha sps100 strain was found to be indistinguishable from the wild-type strain when assessed for efficiency of ascus formation and spore viability. However, a more detailed analysis of the mutant strain revealed that the SPS100 gene product serves a protective role during the early stages of spore wall formation. The time at which resistance to ether could first be detected in developing spores was delayed by 5 h in the mutant strain relative to the wild-type strain. This phenotype is presumably a reflection of a defect in spore wall maturation. This study has confirmed that temporally distinct classes of sporulation-specific genes are sequentially activated during the process of meiosis and spore formation and has shown that the SPS100 gene, identified on the basis of its developmental-specific expression pattern, contributes to spore development.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Clancy M. J., Buten-Magee B., Straight D. J., Kennedy A. L., Partridge R. M., Magee P. T. Isolation of genes expressed preferentially during sporulation in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 May;80(10):3000–3004. doi: 10.1073/pnas.80.10.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984 Oct;38(3):841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dawes I. W., Hardie I. D. Selective killing of vegetative cells in sporulated yeast cultures by exposure to diethyl ether. Mol Gen Genet. 1974;131(4):281–289. doi: 10.1007/BF00264859. [DOI] [PubMed] [Google Scholar]
- Donovan W., Zheng L. B., Sandman K., Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- Esposito M. S., Esposito R. E. Genes controlling meiosis and spore formation in yeast. Genetics. 1974 Sep;78(1):215–225. doi: 10.1093/genetics/78.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber A. T., Segall J. The SPS4 gene of Saccharomyces cerevisiae encodes a major sporulation-specific mRNA. Mol Cell Biol. 1986 Dec;6(12):4478–4485. doi: 10.1128/mcb.6.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlin-Ninfa E., Kaback D. B. Isolation and functional analysis of sporulation-induced transcribed sequences from Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2185–2197. doi: 10.1128/mcb.6.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holaway B. L., Lehman D. J., Primerano D. A., Magee P. T., Clancy M. J. Sporulation-regulated genes of Saccharomyces cerevisiae. Curr Genet. 1985;10(3):163–169. doi: 10.1007/BF00798745. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Magee P. T., Welch S. K., Friedman M., Hall B. D. Macromolecule synthesis and breakdown in relation to sporulation and meiosis in yeast. J Bacteriol. 1974 Aug;119(2):619–628. doi: 10.1128/jb.119.2.619-628.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics. 1980 Nov;96(3):567–588. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kraig E., Haber J. E. Messenger ribonucleic acid and protein metabolism during sporulation of Saccharomyces cerevisiae. J Bacteriol. 1980 Dec;144(3):1098–1112. doi: 10.1128/jb.144.3.1098-1112.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Changing patterns of gene expression during sporulation in yeast. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7323–7327. doi: 10.1073/pnas.81.23.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Subcellular differentiation in sporulating yeast cells. Cell. 1986 Jun 6;45(5):771–779. doi: 10.1016/0092-8674(86)90791-9. [DOI] [PubMed] [Google Scholar]
- Lynn R. R., Magee P. T. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J Cell Biol. 1970 Mar;44(3):688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell. 1976 Nov;9(3):431–438. doi: 10.1016/0092-8674(76)90088-x. [DOI] [PubMed] [Google Scholar]
- Mason J. M., Setlow P. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J Bacteriol. 1986 Jul;167(1):174–178. doi: 10.1128/jb.167.1.174-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2443–2451. doi: 10.1128/mcb.6.7.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Increased copy number of the 5' end of the SPS2 gene inhibits sporulation of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jul;7(7):2484–2490. doi: 10.1128/mcb.7.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Isolation of DNA sequences preferentially expressed during sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):142–150. doi: 10.1128/mcb.4.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petko L., Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986 Jun 20;45(6):885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. The new yeast genetics. 1983 Sep 29-Oct 5Nature. 305(5933):391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- Trew B. J., Friesen J. D., Moens P. B. Two-dimensional protein patterns during growth and sporulation in Saccharomyces cerevisiae. J Bacteriol. 1979 Apr;138(1):60–69. doi: 10.1128/jb.138.1.60-69.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M. The isolation and genetic analysis of sporulation-deficient mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191(1):17–21. doi: 10.1007/BF00330883. [DOI] [PubMed] [Google Scholar]
- Wang H. T., Frackman S., Kowalisyn J., Esposito R. E., Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987 Apr;7(4):1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir-Thompson E. M., Dawes I. W. Developmental changes in translatable RNA species associated with meiosis and spore formation in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Apr;4(4):695–702. doi: 10.1128/mcb.4.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. F., Ajam N., Dawes I. W. Nature and timing of some sporulation-specific protein changes in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Oct;1(10):910–918. doi: 10.1128/mcb.1.10.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]