Abstract
A steadily growing number of studies have shown that microRNAs have key roles in the regulation of cellular processes and that their dysregulation is essential to keep the malignant phenotype of cancer cells. The distorted and unique expression profile of microRNAs in different types and subsets of tumor coupled with their presence in biological fluids make of microRNAs an attractive source of sensitive biomarkers. Here, we will discuss how microRNA profiles are altered in cancer, highlighting their potential as sensitive biomarkers for cancer risk stratification, outcome prediction and classification of histological subtypes. We will also evaluate the current knowledge on the use of microRNAs as circulating biomarkers, hoping that further studies will lead to the application of microRNA signature in prognostic and predictive markers that can improve patient health.
Introduction
In the last decade non-coding RNAs have emerged as a new class of key regulators involved in development, normal physiology, and many different types of disease. MicroRNAs (miRNAs) represent the major class of small endogenous non-coding RNAs and control almost one-third of all human genes [1–2]. MiRNAs are single stranded RNAs of 19–25 nucleotides in length that negatively regulate gene expression by base-pairing to partially complementary sites on the target messenger RNAs (mRNAs), usually in the 3’ untranslated region (UTR) [3]. Binding of a miRNA to the target mRNA typically leads to translational repression and exonucleolytic mRNA decay, although highly complementary targets can be cleaved endonucleolytically [4]. As data accumulated proposing fundamental roles for miRNAs in proliferation, differentiation, survival and apoptosis, it is not surprising that miRNAs were found to be important in tumorigenesis and considered promising therapeutic targets for novel cancer treatments [5]. After our initial discovery that miR-15/16 cluster is deleted or downregulated in patients with chronic lymphocytic leukemia [6••], myriad reports established that neoplastic tissues had a cancer-associated miRNA signature. This miRNA alteration allows the accurate classification of the different malignancies and the identification of the tissue of origin for poorly differentiated tumors. The cause of the widespread differential expression of miRNA genes between malignant and normal cells can be explained by different mechanisms including a) chromosomal alterations of the miRNA genes, b) DNA point mutations, c) epigenetic mechanisms or d) alterations in the machinery responsible for miRNA production (Figure 1) [7]. Similar to the coding genes, miRNAs can be either over- or under-expressed and they can act as tumor-suppressors or oncogenes based on the downstream target that the miRNA controls. miR-15 and miR-16 were the first described tumor-suppressor miRNAs and their loss, by releasing the inhibition upon tumor-promoting genes, such as BCL2, BMI1, CCND2 and CCND1, promotes cell growth and tumor progression [6••, 8–9•,10]. Alternatively, miR-21 is highly up-regulated in the majority of cancer tissues and by repressing pro-apoptotic genes, such as PTEN or PDCD4, stimulates proliferation and tumor initiation [11–12••]. The development of different high-throughput miRNA profiling technologies (Table 1) has allowed the characterization of the miRNA expression profile for several malignancies including chronic lymphocytic leukemia [13], breast cancer [14], lung cancer [15], thyroid papillary carcinoma [16], pancreatic tumors [17], glioblastoma [18], gastric cancer [19], prostate cancer [20], hepatocellular carcinoma [21]. Surprisingly, the use of miRNA expression is newly becoming highly preferred to the traditional gene expression profiles for a variety of reasons. First, the remarkable stability of miRNAs, due to their short length, has allowed scientists to perform analyses also in samples considered to be technically challenging, such as formalin fixed specimens. Additionally, highly sensitive and refined miRNA detection techniques provide high reliability in the use of miRNAs as a diagnostic tools. Finally, miRNA expression has demonstrated the ability to identify the tissue of origin for cancer that have already spread in multiple metastatic sites, thereby reducing patient’s psychological burden and overall procedure costs [22•, 23]. In the following sections, we will discuss some of the more important approaches that have been taken to identify cancer-associated miRNA expression profiles and their potential use in clinical application.
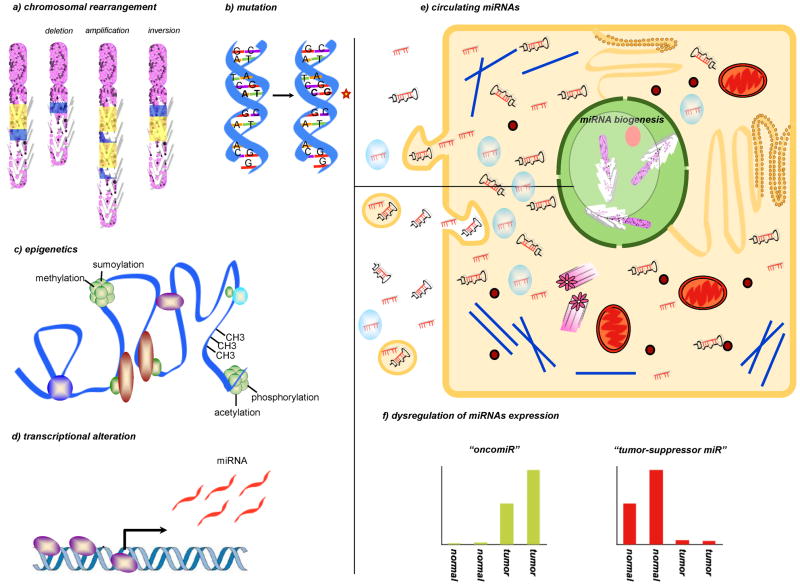
Figure 1. miRNA dysregulation in cancer.
Dysregulation of miRs expression in cancer compared to the normal tissues of origin is a general phenomenon that has been largely characterized in almost all neoplasia. Global repression of miRNAs expression in cancer cells is believed to induce an undifferentiated phenotype. Indeed the increase of specific miRs, the “oncomiRs”, confers aggressiveness and resistance to cell death. In the figure, we depicted the main processes involved in miRNA dysregulation, such as a) chromosomal alterations of the miRNA genes, b) DNA point mutations, c) epigenetic mechanisms or d) alterations in the machinery responsible for miRNA production. e) In addition to their intracellular functions, recent studies have demonstrated that miRNAs can be released or leaked from cancer cells and circulated in a remarkably stable form within blood. Many studies have demonstrated the circulating miRNA levels correlate significantly with cancer progression, therapeutic response, and patient survival. As RISC-associated, microvesicles-related or as a free miRNAs or pre-miRNAs, the function of circulatory miRNAs is largely unknown. Many questions are still missing an answer: are circulating miRNAs a result of leakage from cancer cells or active release? Besides bio-markers, can miRNA drive or activate molecules that help the body defend against cancer? f) Representation of the miRNA modulation in the neoplastic tissues compared to normal tissues of origin: an “oncomiR” upmodulation and a “tumor-suppressor miR” repression are shown in the two graphics.
Table 1.
microRNA profiling technologies
| technologies | advantages | disadvantages | Assay or platform/Vendor | cost |
|---|---|---|---|---|
| quantitative reverse transcription PCR (qRT-PCR) | Highly sensitivity and specificity. Low amount of RNA template and useful for absolute quantification | Useful only for known miRs. Relative low number of samples that can be processed per day. | TaqMan individual assays (ABI) miRCURY LNA qPCR (Exiqon) TaqMan OpenArray (ABI) TaqMan TLDA microfluidics card (ABI) Biomark HD system (Fluidigm) SmartChip human microRNA (Wafergen) miScript miRNA PCR array (SABiosciences/Qiagen) |
$$ |
| miRNA microarray | Relative low cost and high- throughput with respect to the number of samples that can be processed per day | Lower specificity than qRT-PCR or RNA sequencing. Cannot be use for new miRs | Geniom Biochip miRNA (CBC febit) GeneChip miRNA array (Affymetrix) GenoExplorer (Genosensor) MicroRNA microarray (Agilent) miRCURY LNA microRNA array (Exiqon) NCode miRNA array (Invitrogen) nCounter (Nanostring) OneArray (Phalanx Biotech) Sentrix array matrix and BeadChips (Illumina) μParaFlo biochip array (LC Biosciences) |
$ |
| RNA sequencing: high-throughput | Useful for the detection of novel miRs. High sensitivity in discriminating between very similar miRs | High cost. Large computational work for data analyses | HiSeq 2000 and Genome Analyzer IIX (Illumina) Solid (ABI) GS FLX+ 454 sequencing (Roche) |
$$$ |
| RNA sequencing: smaller scale | Useful for the detection of novel miRs. High sensitivity in discriminating between very similar miRs | High cost. Large computational work for data analyses | Ion Torrent (Invitrogen) MiSeq (Illumina) GS Junior(454) (Roche) |
$$$ |
miRNAs as a cancer biomarkers
A large number of miRNA genome-wide studies in different tumors have highlighted that selective groups of distinct miRNAs - miRNA fingerprints - are commonly dysregulated in specific types of human malignancies and often associated with diagnosis, staging, progression, prognosis and response to clinical therapies. These results have been confirmed over time in different cohorts of patients and provided the evidence that miRNA dysregulation in cancer is unlikely a random event (Table 2). In 2006, our laboratory published the first comprehensive profile of miRNAs in cancer by analyzing miRNA expression in 540 samples including 363 solid tumors from the six most common malignancies (breast, prostate, lung, stomach, pancreas, thyroid) and 177 normal tissues [24••]. A common “miRNoma” in cancer was identified consisting of 36 overexpressed and 21 downregulated miRNAs: among them, some of the well characterized cancer associated miRs, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155 (Table 2). Golub and coworkers also showed that miRNA profiles of tumor tissues paralleled the developmental origins of the malignant tissues, supporting the idea that miRNA expression patterns encode the developmental history of human cancers [25••]. For example, tumors of epithelial origin presented a different miRNA expression from haematopoietic malignancies. Furthermore, distinct patterns of miRNA expression can be observed within a single developmental lineage and reflect mechanisms of transformation. For example, miRNA profiles can subgroup acute lymphoblastic leukaemia (ALL) specimens into three major groups: one containing all t(9;22) BCR/ABL- and t(12;21) TEL/AML1-positive samples; a second group containing T-cell ALL samples; and a third group containing the MLL gene rearrangement. Thereafter, Rosenfeld and coworkers identified, by using miRNA microarray data of 253 samples, a transparent classifier based on 48 miRNAs that predicts with high confidence and accuracy the tissue from which cancers of unknown primary origin arose [22•]. Specifically, the classification accuracy reached 100% for most tissue classes analyzed, including 131 metastatic samples and an independent blinded test-set of 83 samples, demonstrating the great diagnostic value of miRNAs [22•].
Table 2.
microRNA dysregulated in cancer
| microRNA | genomic location | expression in cancer | function | mechanism of deregulation | targets |
|---|---|---|---|---|---|
| let-7a-2 | 11q24 | down in breast, lung, colon, ovarian and stomach cancer | tumor-suppressor | repressed by MYC | KRAS, HMGA2, MYC, DICER, BCL-XL, IMP-1, CDC34, IL6 |
| miR-15/-16 | 13q31 | down in CLL, prostate cancer and pituitary adenomas | tumor-suppressor | genomic loss, mutated, activated by p53 | BCL2, COX2, CHECK1, CCNE1, CCND1, CCND2, BMI-1, FGF2, FGFR1, VEGF, VEGFR2, CDC25a |
| miR-29 family | 7q32 1q30 |
down in AML, CLL, lung and breast cancer, lymphoma, hepatocarcinoma, rhabdomyosarcoma | tumor-suppressor | genomic loss, activated by p53, repressed by MYC | CDK6, MCL1, TCL1, DNMT1, DNMT3a, DNMT3b |
| miR-34 family | 1p36 11q23 |
Down in colon, lung, breast, kidney and bladder cancer | tumor-suppressor | repressed by MYC | SIRT1, BCL2, NOTCH, HMGA2, MYC, MET, AXL. NANOG, SOX2, MYCN, SNAIL |
| miR-26a | 3p22 | down in liver cancer | tumor-suppressor | repressed by MYC | CCND2, CCNE2 |
| miR-200 family | 1p36 12p13 |
down in aggressive breast cancer | tumor-suppressor | repressed by ZEB1/2; | ZEB1, ZEB2, BMI-1, SUZ-12, FN1, LEPR, CTNNB1, JAG1, MALM2, MALM3, p38alpha |
| miR-155 | 21q21 | up in high risk CLL, AML, breast, lung, colon cancer and lymphoma | oncogene | activated by NF-KB | SOCS1, BACH1, MEIS1, ETS1, FOXO3A, hMSH2, hMSH6, hMLH1, SMAD5, WEE1, SHIP1, CEBPB |
| miR-21 | 17q23 | up in lung, breast, pancreas stomach, prostate, CLL, AML, glioblastoma, myeloma | oncogene | activated by IL6, GF1alpha | PTEN, TPM1, PDCD4, SPRY1, TIMP3, RECK |
| miR-221/-222 | Xp11 | up in invasive ductal carcinoma, lung cancer, hepatocellular carcinoma,. papillary thyroid cancer | oncogene | activated by MET in lung cancer; repressed by ERalpha in breast; activated by PLZF in melanoma; activated by NF-Kb and cJun in prostate cancer and glioblastoma cells | p27(Kip1), p57(Kip2), PTEN, TIMP3, FOXO3A, ERalpha, KIT, TRSP1, DICER, APAF1, PUMA, PTPμ |
| miR-17/92 | 13q22 | up in lung, breast, colon, | oncogene | activated by E2F1 and MYC | PTEN, BIM, HIF1, PTPRO, p63, E2F2, E2F3, TSP-1, CTGF, p21(WAF1), JAK1, SMAD4, TGFbetaII, Page 21 of 22 MnSOD, GPX2, TRXR2 |
One of the most important aspect of the miRNA fingerprints in cancer is their discriminatory role for different cancer subtypes or correlation with specific neoplastic events, such as oncogenic activation. This characteristic provides additional information for the prognosis and treatment of cancer when combined with standard gene profiling. Numerous studies have shown that miRNAs can classify breast cancers into a specific tumor pathological phenotype (i.e., Estrogen Receptor and Progesterone Receptor status, proliferation, tumor stage, metastatic state, HER2 status) [14, 26], as well as the molecular subtype (Luminal A, Luminal B, Basal-like, HER2+ and Normal-like) [27]. To date, miR-200 family is positively associated with a well-differentiated breast cancer phenotype (luminal tumors) and it is very low expressed in the poorly differentiated claudin-low subtype of breast cancer [28]. In fact, it has been shown that miR-200 family members directly target ZEB1 and ZEB2, two transcription factors that control the epithelial-to-mesenchymal (EMT) transition, essential for cancer cells to become motile, invasive, and survive the metastatic journey [29••, 30]. While miR-200 family expression is high in the luminal breast cancer subtype, miR-221/222 cluster is overexpressed in triple negative breast cancers, particularly those that have undergone EMT [31,32]. Increasing miR-221 or -222 can affect various characteristics associated with EMT, including increased invasive capacity, and anoikis resistance [33]. Inhibition of miR-221/222 in basal-like cancer cells promoted mesenchymal to epithelial transition (MET) in part by directly targeting trichorhinophalangeal 1 (TRPS1), a transcriptional repressor of ZEB2 that can promote MET [33]. Furthermore, a recent publication from our laboratory showed that miR-221 is down-regulated in ductal carcinoma in situ and highly up-regulated in invasive ductal carcinoma [34], thus revealing a possible role for miR-222 and miR-221 cluster in the development of highly proliferative and aggressive breast cancer.
Another important feature of miRNA in cancer is their predictive role for the mutational status of important clinical biomarkers. The first association came from our laboratory in 2005, when we reported a 13 miRNA signature predictive of the ZAP70 and IgVH status in CLL patients [12]. Similarly, low expression of miR-193a, miR-338, and miR-565 was associated with melanomas carrying a BRAF missense mutation at the commonly involved residue V600E [35]. Interestingly, miR-193 was also reported to be downregulated in BRAFV600E mutated-thyroid cancer cell lines compared with normal thyroid tissue, suggesting that miR-193a may have an important and common role in BRAF-associated tumorigenesis. Likewise, up-regulation of miR-155 in acute myeloid leukemia was strongly but independently predictive of FLT3 gene mutations, which have been associated with patients that have increased risk of relapse [36]. miR155 is one of the most intensively studied miRNAs, not least of all because of the relatively early realization that it is abnormally expressed in specific forms of lymphomas, including Hodgkin lymphoma (HL), diffuse large B-cell (DLBCL) and primary mediastinal B cell lymphomas, but not primary Burkitt lymphoma [37–39]. miR-155 over-expression in mouse resulted in pre-B-cell expansion and bone marrow replacement, splenomegaly, and lymphopenia that preceded the development of lymphoblastic leukemia and lymphoma [40].
miRNAs have been also linked to the prediction of specific cytogenetic abnormalities that have prognostic implications and are present in the majority of hematological malignancies, such as CLL. Specifically, 32 miRNAs discriminated between CLL patients with the 17p and 11q deletions, who experience the aggressive form of the disease, and CLL patients with the 13q deletion or normal cytogenetic profiles, who experience the indolent form [41]. Another example of deregulated miRNA expression associated to cytogenetic abnormalities is represented by the miRNAs identified in multiple myeloma samples that are associated with the most common IgH translocations (t(4;14) and t(11;14)) and del(13q) [42]. Calin et al. have reported that more than 50% of miRNAs are located at genomic sites that are disrupted or amplified in various cancers [43]. For example, the 13q31 region, which is amplified in DLBCL [44], mantle cell [45], and follicular [46] B-cell lymphomas, harbors a cluster of seven miRNAs, the miR-17/92 cluster [47]. In agreement, this miRNA cluster was found to be increased ~10-fold in 65% of B-cell lymphoma samples [48]. When this cluster is overexpressed with the myc oncogene, it greatly accelerated c-myc-induced pre-B cell lymphomas in mice and reduced apoptosis [48]. Another example of relation between cytogenetic abnormalities and microRNAs is represented by the t(11;14)(q13;q32) translocation and miR-16 [10]. This translocation represents a genetic hallmark of mantle cell lymphoma that displaces the CCND1 gene on chromosome 11 downstream to the enhancer region of the IgH gene on chromosome 14 and causes its overexpression. Studies of patient samples have shown that truncations exist within the CCND1 mRNA 3′ UTR and the truncation leads to a worse prognosis. Since miR-16 represses CCND1, the truncation of the 3′ UTR reported in lymphoma cell lines prevents proper miR-16 repression of CCND1 mRNA, resulting in increased proliferation and shorter survival.
Since the large contribution of miRNAs in tumorigenesis, it is not surprising that miRNA expression can be also predictive of clinical survival and response to chemotherapy. To this regard, low miR-191 and high miR-193a were associated with a significant reduced survival in melanoma patients [49]. In CLL, high levels of miR-15/16 cluster are associated to a good prognosis, according with their tumor-suppressor activity demonstrated in vitro and in vivo [8, 13]. In lung cancer, a large miRNA genome-wide expression analyses showed that the levels of both miR-155 and let-7a-2 are associated with poor survival [15]. The potential prognostic value of let-7 in lung cancer was also shown in an independent study of Japanese patients where repression of let-7 correlated with significantly shorter survival time after curative surgery and stage of the disease [50]. Another study reported that liver cancer patients whose tumors had low miR-26 expression had shorter overall survival but a better response to interferon therapy [51]. In gastric cancer a robust 7-miRNAs signature predicts outcome [52]. In various cancer, miR-21 is an index of poor outcome [53–55] and it is also an important predictor of response to chemotherapy: high miR-21 expression levels are predictive of response to gemcitabine in pancreatic cancer patients [56]. Of note, Medina et al. demonstrated that miR-21 is a bona fide oncogenic miRNA [12••]. Induction of miR-21 in mouse resulted in spontaneous development of lymphoma that most closely resembled precursor B-cell lymphoblastic lymphoma/leukemia. Remarkably, repression of the miR-21 levels in mice with significant tumor loads resulted in complete disappearance of the tumors within 1 week.
miRNAs as new serum biomarkers
Current diagnostic techniques, whether based on more traditional approaches, such as histological evaluation, or based on a more personalized approach, such as Oncotype DX (Genomic Health, Inc., CA, USA) or Mammaprint (Agendia, Ammsterdam, The Netherland), both essentially rely on the direct sampling of the tumor material. Although direct measurements of biomarkers within tissues have greatly improved diagnosis and survivability, the invasive and unpleasant nature of the diagnostic procedures limits their application for most clinical conditions. In this regard, many researchers worldwide have made numerous efforts in the identification of new biomarkers in body fluids (serum, plasma and urine) due to their ease of collection and the fact that they reflect a particular physiological or pathological state. Many studies in fact have shown that specific cancer characteristics, both genetics and epigenetics, are detectable in the plasma and serum of cancer patients and can be useful in diagnostic to monitor ongoing pathological processes, such as alpha-fetoprotein (AFP) for liver cancer [57], prostate-specific antigen (PSA) for prostate cancer [58], carcinoembryonic antigen (CEA) and (CA125) for breast and ovarian cancer, respectively [59–60]. In 2007, small RNAs, including miRNAs, were identified in total RNAs preparation derived from biological fluid [61••]. The authors showed that exosomes from mast cells, natural vesicles secreted by a variety of cells, contain mature miRNAs which can be transferred to another cell and can be functional in this new recipient. Shortly afterwards, Lawrie et al. published the first report of miRs modulation in serum of B-cell lymphoma patients and that high miR-21 serum levels were associated with increased relapsed-free survival but not with overall survival [62]. In 2008, Tewari group showed that miRs are present in human plasma as a stable molecules and provided direct evidence that tumor-derived miRNAs can enter the circulation even when originating from an epithelial cancer type [63]. Recently, the global relationship between tissue miRNAs and circulating miRNAs in normal population has been investigated and showed that the composition of liver miRNAs correlates most closely with the miRNAs found in blood, suggesting that under normal conditions the liver may have the most contact with circulating blood [64]. Conversely, placenta, testis, and brain exhibit the weakest correlations between tissue miRNA populations and those found in blood [64]. Although reports indicate that circulating miRNAs are secreted or leaked from normal or tumor tissues, the mechanisms of the miRNA release and their function into circulation remain largely unknown. Even if many questions still remain unanswered, an immediate interest emerged for the investigation of miRs as non-invasive biomarkers in circulating blood, and a large list of new blood-derived miRs profiles emerged (Table 3). The most striking finding is the general upmodulation of miR-21 and/or miR-155 as a putative diagnostic and prognostic markers in several different malignancies. To date, high levels of circulating miR-21 were predictive of the estrogen receptor negative status while high levels of miR-155 were associated to a positive progesterone receptor status in breast cancer [65–66]. A comprehensive analysis of miRNA profiles in serum has been performed by Chen and coworkers [67]. The authors showed that miRNAs are abundant in the majority of body fluids (serum, plasma, amniotic fluid, urine, tears, ascetic fluid) and, by using Solexa deep sequencing, they identified 190 known miRs in the serum of healthy donors. They also determined that patients with lung cancer, diabetes and colorectal cancer have a significantly different expression of miRs in their serum compared to healthy subject. Specifically, miR-25 and miR-223, highly expressed in lung tumors, were also enriched in patient serum and their expression predicted lung cancer in an independent set of 75 healthy donors and 152 cancer patients. Recently, our group provided the first evidence for the roles of circulating miRNAs in cancer by demonstrating that tumor-secreted miRs are able to interact with the Toll-like receptors of immune cells to stimulate the production of prometastatic inflammatory cytokines and inducing the pro-tumor inflammatory processes [68]. In this scenario, circulating miRNAs can act as signals for receptor activation, a function that is completely independent of their conventional role in post-transcriptional gene regulation.
Table 3.
microRNA profile in blood
| Cancer type | biological fluid | modulated miRs | prognostic value |
|---|---|---|---|
| HNSCC | plasma | miR-184 | - |
| Breast cancer | serum | miR-34, -10b, -155 | all three miRs showed a correlation with metastasis |
| serum | miR-155 associated to PR status | ||
| serum | miR-21,-106a,-155; miR-126, -199a, -335 | ||
| whole blood | miR-195 | miR-21, -10b associated to ER-; let-7a reduced in lymph node metastasis | |
| NSCLC | serum | mir-25, -223 | - |
| exosome from plasma | miR-17, -21, -106a, -146, -155, -191, -192, -203, -205, - 210, -212, -214 | - | |
| serum | miR-486, -30d; miR-1, -499 | correlated with overall survival | |
| serum | miR-10b, -155 | miR-10b is correlated with lymph node metastasis | |
| Colorectal cancer | plasma | miR-221 | associated with poor overall survival |
| Hepatocellular carcinoma | serum | miR-25, -375, let-7f | - |
| serum | miR-21, -122, -223 | - | |
| Pancreatic carcinoma | plasma | miR-21 | - |
| plasma | miR-21, -155, -196a | - | |
| Prostate cancer | serum | miR-141 | - |
| serum | miR-20b, -874, -1274, -1207, -93, -106a, miR-24, -223, - 26b, -30c | miR-24 is correlated to metastasis | |
| serum | miR-21 | miR-21 is associated to docetaxel resistance |
Conclusion
There is no doubt that miRNAs are involved in tumorigenesis and regulate the development and progression of human malignancies. Although many progresses have been made in the understanding of miRNA involvement in cancer, many questions still need to be answered before translating miRNA profiling into clinical practice. One of the biggest challenges for the future years will be the creation of cancer specific miRNA signatures that can be highly reproducible and independently predictive of clinico-biological features of the tumor to improve diagnosis and treatment.
Acknowledgments
We are grateful for research support from The Ohio State University Targeted Investment in Excellence Award. We thanks Dr. Briskin Daniel for providing writing assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. Mirnas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT, Olson EN. Mirnas in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Huntzinger E, Izaurralde E. Gene silencing by miRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 5.Garofalo M, Croce CM. miRNAs: master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 6••.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. This study shows that miR15 and miR16 are located within a 30-kb region of loss in CLL, and that both genes are deleted or down-regulated in the majority ( approximately 68%) of CLL cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - miRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. This study demonstrates that deletion in mice of the 13q14-minimal deleted region (MDR), which encodes the DLEU2/miR-15a/16-1 cluster, causes development of indolent B cell-autonomous, clonal lymphoproliferative disorders, recapitulating the spectrum of CLL-associated phenotypes observed in humans. [DOI] [PubMed] [Google Scholar]
- 10.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA. Truncation of CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112:822–829. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 12••.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of miRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. By using Cre and Tet-off technologies to generate mice conditionally expressing miR-21, the authors show that miR-21 over-expression leads to a pre-B malignant lymphoid-like phenotype, demonstrating that mir-21 is a genuine oncogene. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A Mirna signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. Mirna gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique miRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of miRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. Mirna expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 18.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of miRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Tang Z, Sun Y, Zhang Y, Wang X, Shen Z, Liu F, Qin X. miRNA expression profile in primary gastric cancers and paired lymph node metastases indicates that miR-10a plays a role in metastasis from primary gastric cancer to lymph nodes. Exp Ther Med. 2012;3:351–356. doi: 10.3892/etm.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. Mirna expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 21.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of miRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 22•.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. Mirnas accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. This study demonstrates the effectiveness of miRNA signatures as biomarkers for tracing the tissue of origin of cancers of unknown primary origin. [DOI] [PubMed] [Google Scholar]
- 23.Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafà R, Magri E, Lunardi M, Munerato G, Querzoli G, Maestri I, et al. Mirna profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225:43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A miRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. Identification of a solid cancer miRNA signature composed by a large portion of overexpressed miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. First successful classification of poorly differentiated tumors using miRNA expression profiles, whereas messenger RNA profiles were highly inaccurate when applied to the same samples. [DOI] [PubMed] [Google Scholar]
- 26.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput miRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. Mirna expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bockmeyer CL, Christgen M, Müller M, Fischer S, Ahrens P, Länger F, Kreipe H, Lehmann U. Mirna profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat. 2011;130:735–745. doi: 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 29.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 30.Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 miRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia. 2012;17:65–77. doi: 10.1007/s10911-012-9244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. Mirna expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–517. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 32.Shah MY, Calin GA. Mirnas miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3:56. doi: 10.1186/gm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 34.Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of miRNA. Proc Natl Acad Sci U S A. 2012;109:3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. Mirna expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 36.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, et al. Distinctive miRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2008;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 39.Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–53. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 40.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD, Volinia S, Ferracin M, Palatini J, Balatti V, et al. Karyotype-specific miRNA signature in chronic lymphocytic leukemia. Blood. 2009;114:3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi J, Ballabio E, Chen XH, Kušec R, Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F, et al. Mirna expression in multiple myeloma is associated with genetic subtype, isotype and survival. CH Biol Direct. 2011;6:23. doi: 10.1186/1745-6150-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao PH, Houldsworth J, Dyomina K, Parsa NZ, Cigudosa JC, Louie DC, Popplewell L, Offit K, Jhanwar SC, Chaganti RS. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234–240. [PubMed] [Google Scholar]
- 45.De Leeuw RJ, Davies JJ, Rosenwald A, Bebb G, Gascoyne RD, Dyer MJ, Staudt LM, Martinez-Climent JA, Lam WL. Comprehensive whole genome array CGH profiling of mantle cell lymphoma model genomes. Hum Mol Genet. 2004;13:1827–1837. doi: 10.1093/hmg/ddh195. [DOI] [PubMed] [Google Scholar]
- 46.Neat MJ, Foot N, Jenner M, Goff L, Ashcroft K, Burford D, Dunham A, Norton A, Lister TA, Fitzgibbon J. Localisation of a novel region of recurrent amplification in follicular lymphoma to an 6. 8 Mb region of 13q32–33. Genes Chromosomes Cancer. 2001;32:236–243. doi: 10.1002/gcc.1187. [DOI] [PubMed] [Google Scholar]
- 47.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 48.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. Mirna expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 50.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 miRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 51.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. Mirna expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-miRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 53.Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, Fabbri M, Lerner S, Barron LL, Rassenti LZ, et al. miRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. Mirna-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, et al. Mirna expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Giannini EG, Erroi V, Trevisani F. Effectiveness of α-fetoprotein for hepatocellular carcinoma surveillance: the return of the living-dead? Expert Rev Gastroenterol Hepatol. 2012;6:441–444. doi: 10.1586/egh.12.30. [DOI] [PubMed] [Google Scholar]
- 58.Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. J Clin Oncol. 2003;21:383–391. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 59.Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 60.Díaz-Padilla I, Razak AR, Minig L, Bernardini MQ, Del Campo María. Prognostic and predictive value of CA-125 in the primary treatment of epithelial ovarian cancer: potentials and pitfalls. J Clin Transl Oncol. 2012;14:15–20. doi: 10.1007/s12094-012-0756-8. [DOI] [PubMed] [Google Scholar]
- 61••.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and miRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. This study shows that exosomes contain both mRNA and miRNA, which can be delivered to another cell, and can be functional in this new location. They propose that this RNAs are called “exosomal shuttle RNA” (esRNA) [DOI] [PubMed] [Google Scholar]
- 62.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated miRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating miRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen G, Wang J, Cui Q. Could circulating microRNAs contribute to cancer therapy? Trends Mol Med. 2012 doi: 10.1016/j.molmed.2012.10.006. pii: S1471-4914(12)00201-8. [DOI] [PubMed] [Google Scholar]
- 65.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating miRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 66.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating miRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of miRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 68••.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. Mirnas bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. First evidence that tumor-secreted miRs are able to interact with the Toll-like receptors of immune cells to stimulate the production of prometastatic inflammatory cytokines. [DOI] [PMC free article] [PubMed] [Google Scholar]