Abstract
The LIM homeobox containing genes of the LIM-3 group, Lhx3 and Lhx4, are critical for normal development. Both genes are involved in the formation of the pituitary and the motoneuron system and loss of either gene causes perinatal lethality. Previous studies had shown that Lhx3 is overexpressed in hyperplastic placentas of mouse interspecies hybrids. To determine the role of LHX3 in the mouse placenta, we performed expression and function analyses. Our results show that Lhx3 exhibits specific spatial and temporal expression in the mouse placenta, however, deletion of Lhx3 does not produce a placental phenotype. To test whether this is due to functional substitution by Lhx4, we performed a phenotype analysis of Lhx3−/−;Lhx4−/−double-mutant placentas. A subset of Lhx3−/−;Lhx4−/− placentas exhibited abnormal structure of the labyrinth, however, absence of both LIM-3 genes did not interfere with placental transport nor consistently with expression of target genes such as Gnrhr. Thus, LHX3 and LHX4 appear to be dispensable for placental development and function.
Keywords: LIM-homeobox gene, Lhx3, Lhx4, mouse placenta
Introduction
A functional placenta is an absolute prerequisite for development to term in all eutherian mammals, including man and defects that severely compromise placental function will result in fetal lethality (Rossant & Cross, 2001, Cross et al., 2003). Significantly impaired placental transport function that does not jeopardize survival of the developing fetus will still cause fetal undernutrition and intrauterine growth restriction (IUGR), which is again associated with high incidence of perinatal lethality (Cross, 2006). However, even newborns that survive this critical perinatal period will suffer from late to very late effects of IUGR on brain development (Lane et al., 2001), energy metabolism (Simmons et al., 2001, Tsirka et al., 2001), cardiac function (Gagnon, 2003, Leipala et al., 2003, Vonnahme et al., 2003, Xu et al., 2006), and other organ systems (Vonnahme et al. 2003). These findings stress the pivotal role of the placenta in normal fetal development. In recent years, numerous genes have been identified in the mouse that, when mutated, cause abnormal placental phenotypes. This clearly demonstrates that these genes are important in normal placentation (Rossant & Cross, 2001, Hemberger & Cross, 2001, Cross et al., 2003). However, the functional roles of many other genes expressed in the placenta have not been revealed to date.
We had previously reported global mRNA expression profiles of murine hyperplastic placentas (Singh et al., 2004) generated by interspecific hybridization between different mouse species (interspecies hybrid placental dysplasia; IHPD) (Zechner et al., 1996, Zechner et al., 2002), reproductive cloning (Tanaka et al., 2001) and mutation of the X-linked gene Esx1 (Li & Behringer, 1998). In that study, we had identified a considerable number of genes that are differentially expressed between normal and hyperplastic placentas, thus forming a pool of potential placentation genes. Altered expression of some of these genes could reflect secondary changes due to disturbed gene expression cascades or shifts in tissue proportions. Indeed, deletion of some genes identified in our previous study (Singh et al., 2004) did indeed not cause any placental phenotypes (Singh et al., 2005, Singh et al., 2006a). On the other hand, deletion of other such genes was associated with placental phenotypes, thus providing evidence for their functional roles in placental development (Singh et al., 2006b, Singh et al., 2007). Thus, analysis of more genes from this set of data is likely to increase our knowledge about gene function in mouse placentation.
A gene that was shown to be upregulated in hyperplastic IHPD placentas, Lhx3, encodes a LIM homeodomain transcription factor (Singh et al., 2004). Together with Lhx4, Lhx3 forms the LIM-3 group within the LIM homeobox gene family, which is characterized by a conserved homeodomain that it is distinctive from that of other homeodomain containing families (Hobert & Westphal, 2000). As shown by gene targeting in the mouse, the LIM-3 group genes are important in pituitary and motor neuron development (Sheng et al., 1996, Sheng et al., 1997, Sharma et al., 1998, Mullen et al., 2007, Raetzman et al., 2002, Ellsworth et al., In press). Interestingly, Lhx3 and Lhx4 have both redundant and complementary functions in these developmental processes (Sheng et al., 1997; Sharma et al., 1998). Thus, formation of the definite Rathke’s pouch is controlled in a redundant manner by both Lhx3 and Lhx4 as shown by the analysis of Lhx3−/−;Lhx4−/− mouse embryos. However, the presence of one wild-type allele, such as in Lhx3+/−;Lhx4−/− or Lhx3−/−;Lhx4+/− is sufficient for formation of a definitive pouch (Sheng et al., 1997). For the next developmental step, formation of a proper pituitary structure and specification of pituitary lineages, Lhx3 expression is an absolute requirement (Sheng et al., 1997). Thus, the presence of one Lhx3 wild-type allele is sufficient for pituitary development, but Lhx4 wild-type alleles are not able to substitute for Lhx3 (Sheng et al., 1997). In a subsequent step, both Lhx3 and Lhx4 together control proliferation and differentiation of pituitary-specific cell lineages (Sheng et al., 1997). In contrast to this, during the differentiation of motor neurons Lhx3 and Lhx4 act in a plain redundant manner, that is, either Lhx3 or Lhx4 alone are competent to specify motor neuron identity (Sharma et al., 1998).
Up-regulation of Lhx3 in abnormal placentation raised the possibility that this transcription factor could be important in placental development. To test this possibility, we performed Lhx3 in situ hybridization to characterize the spatio-temporal expression pattern of this gene. We furthermore analyzed Lhx3−/− placentas and, when these failed to exhibit any obvious phenotypes, Lhx3−/−;Lhx4−/− double mutant placentas. Unexpectedly, even though Lhx3 and Lhx4 are both expressed in the spongiotrophoblasts, double-homozygous mutant placentas exhibited a specific but not fully penetrant phenotype characterized by defective labyrinth structure. This finding suggests that other, to date unidentified genes, can substitute for both LIM-3 transcription factors in mouse placental development and function.
Materials and methods
Mice and Tissues
All experiments with mice were conducted according to the guidelines issued by Uppsala University. For isolation of wild-type placentas, C57BL/6 (B6) × B6 matings were performed. Pregnant females were killed by cervical dislocation, with the day of vaginal plug being counted as day 1. Lhx3 and Lhx4 mutant mice (Sheng et al., 1997, Sheng et al., 1996) were kindly given to us by Dr. Sally A. Camper, University of Michigan. Both strains were propagated in the original B6 strain background by mating heterozygous males with wild-type females, however, neither strain was systematically backcrossed to produce inbred B6 genetic background. Fetuses and placentas were weighed. Placentas were halved, and one half was frozen on dry ice for RNA extraction, while the other half was fixed in Serra’s fixative overnight at 4–8°C and later processed for paraffin histology. Fetal tissue was frozen prior to DNA extraction for genotyping. To generate Lhx3 and Lhx4 double mutant mice, het × het matings were performed between female Lhx3+/− and male Lhx4+/− to produce double heterozygous mutant mice. Subsequently, females and males Lhx3+/−, Lhx4+/− double heterozygous mice were mated to obtain Lhx3/Lhx4 double mutants. Females of the AT24 congenic strain (Hemberger et al., 1999, Elliott et al., 2001), kindly provided by Dr. J. Forejt, Prague, were mated with Lhx3+/− ;Lhx4+/- double heterozygous males to generate heterozygous AT24/+;Lhx3+/−;Lhx4+/− conceptuses. AT24 contains a M. spretus derived proximal X chromosome. AT24 mice exhibit a mild but consistent placental hyperplasia that mimics IHPD not only phenotypically (Hemberger et al., 1999), but also in terms of gene expression (Singh et al., 2005). To determine the role of Gnrhr−/− in placental development, Gnrhr+/− heterozgous mice were mated to generate Gnrhr+/+ and Gnrhr−/− conceptuses for comparison (Pask et al., 2005). Placentas were dissected from the uteri of pregnant females on E18 and E19. The extraembryonic membranes were removed and fetal and placental weights were measured prior to fixation of the placentas in 4% paraformaldehyde. Placental sections were phenotypically assessed independently by at least three investigators who were not aware of the genotypes of the conceptuses.
Histology and immunohistochemistry
Isolectin B4 (Vector Labs) and anti-human alpha smooth muscle actin (aSMA; Dako) staining and histology were performed as described before (Singh et al., 2007). Semiquantitative morphometry was performed on isolectin B4 stained sections as described by Salas et al. (Salas et al., 2004) using Adobe PhotoshopCS. Values from two randomly selected sections each from two placentas of each genotype were averaged. Sections near the placental midline were used to exclude shifting of placental compartments in lateral sections. The area occupied by the chorionic plate was excluded.
mRNA in situ Hybridization
Radioactive and non-radioactive in situ hybridizations were performed as described previously (Meunier et al., 2003, Anson-Cartwright et al., 2000). As placental cell type specific markers, Tpbpa (Lescisin et al., 1988), Prl3d1 (Guillemot et al., 1994), Prl3b1 (Monkley et al., 1996), Eomes (Ciruna and Rossant, 1999; Russ et al., 2000) and Tfeb (Steingrimsson et al., 1998) were applied. A linearized Lhx3 clone (accession number: AI893926), derived from the cDNA library described in a previous study (Singh et al. 2004), was used as in vitro transcription template. The Lhx4 probe was generated by RT-PCR. The purified PCR product was cloned into pGEMT Easy vector (Promega). Digoxigenin (Roche) labeled sense and antisense riboprobes were synthesized by in vitro transcription using SP6 or T7 RNA polymerases. Primers used to generate the Lhx4 in situ probe were: forward, 5′-GTCACCCTCCTTGTCCTCCTG-3′; reverse, 5′-GGTTCTGGTGAGAGGGATGA-3′.
DNA Extraction and Genotyping PCRs
Genomic DNA was extracted from fetal tissues by using the Promega Wizard genomic DNA extraction kit. To genotype Lhx3 mutant mice, two different primer pairs were used in separate reactions. The wild-type allele was amplified using the primers 5′-GTG ACT GCC ATG CTG CTA GA-3′ and 5′-GGA TCT CTG GAG TCC TGC TG-3′, whereas the neomycin cassette inserted in the knockout allele was amplified using the primer pair 5′-CGG CAT CAG AGC AGA TTG TA- 3′ and 5′-GCT TCC TCT TGC AAA ACC AC - 3′. Reaction conditions were 94°C for 3 min (94°C for 35 sec, 56°C for 30 sec, 72°C for 45 sec) × 35, 72°C for 7 min, for the wild-type allele and 94°C for 3 min (94°C for 30 sec, 59°C for 35 sec, 72°C for 45 sec) × 35, 72°C for 7 min for the mutant allele. To genotype Lhx4 mutant mice, multiplex primers 5′–CTC GGA AGC TCA GCT CAC TGT CAC TAA CCC- 3′, 5′–ATC AGG ATG ATC TGG ACG AAG AGC ATC AGG- 3′ and 5′–CAG AAA ATC CGA CTG TGA CCA CAG CTT GAA- 3′ were used. Reaction conditions were 94°C for 3 min (94°C for 35 sec, 61°C for 30 sec, 72°C for 45 sec) × 35 cycles, 72°C for 5 min. Fetal sex was determined by a Y chromosome-specific PCR using the primer pair 5′-CAT TTA TGG TGT GGT CCC GTG - 3′ and 5′-GTG TGC AGC TCT ACT CCA G-3′. Conditions of the reaction were 94°C for 3 min (94°C for 30 sec, 60°C for 30 sec, 72°C for 40 sec) × 35, 72°C for 7 min. All genotyping PCRs were performed in PTC-100 PCR machines (MJ Research, Watertown, MA). PCR products were resolved on 1.5% agarose gels. The genotype of Gnrhr mice was assessed as previously described (Pask et al., 2005). Exon 1 of the Gnrhr gene was amplified by PCR using the following primers: forward, 5′ - CTC CAC TCT TGA AGC CTG TCC–3′; reverse, 5′ - TCA CCA TGT TCA CAC AAA TTC–3′. Subsequently, the PCR product was digested with DdeI and resolved on 3% Nusieve agarose gel (Cambrex Bioproducts, Baltimore, MD).
RNA extraction, RT-PCR and qRT-PCR
RNA was extracted using Trizol (Invitrogen). RQ1 DNAse (Promega) was used to treat RNA, which was then reverse transcribed using M-MLV reverse transcriptase and random primers (Promega) as described earlier (Singh et al., 2004). QuantiTect SYBR green PCR mix (Qiagen) and RotorGene RG3000 (Corbett Research) was used for qRT-PCRs and each sample was analysed in duplicate. Melt curve was analysed to ensure specificity of the PCR. Delta-delta Ct method was used for analysis and the ratio of gene expression for each sample was calculated by normalizing the comparative quantitation values to those of Gapdh and Tubb3. Primer sequences used in qRT-PCRs are listed in Suppl. Table 1.
Lipid extraction and analysis
Total lipid was extracted and purified from altogether 8 fetuses and their placentas (for detailed information see suppl. Table 2), using methanol/chloroform reagent according to a standardized protocol (Bligh & Dyer, 1959, Wuhrer et al., 2000). Lipids were treated with 500 ml of 1 M HCl and 10 M H2O in methanol for 16 h at 100°C according to Gaver and Sweeley (Gaver & Sweeley, 1965). Fatty acids, released as their methyl esters, were recovered by a three-fold phase partition using n-hexane. Fatty acid methyl esters were analyzed by gas chromatography/mass spectrometry using a VF 5ms capillary column (60 m, 0.25 mm inner diameter, 0.1 mm film thickness; Varian, Darmstadt Germany) and 2.5 ml / min helium as carrier gas. Temperature was maintained at 50°C for 2 min, raised to 130°C with 40°C/min and to 300°C with 6°C/min and was finally maintained at 300°C for 5 min. Fatty acid compounds were registered by mass spectrometry in the positive ion mode after chemical ionization with methanol using an PolarisQ instrument (ThermoQuest Analytical Systems).
Statistical Analysis
Statistical analysis was carried out by t-test using GraphPad Prism 3.0 (San Diego, CA). A value of p < 0.05 was considered significant. Data are presented as mean ± SD.
Results
Spatio-temporal expression of Lhx3 and Lhx4 mRNA in the placenta
Lhx3 expression was assayed on E8, E10, E12, E14 and E18 wild type placentas. No expression of Lhx3 was detected at E8 (Fig. 1A). At E10, the expression was in the ectoplacental cone and the developing spongiotrophoblast (Fig. 1B, arrow). From E12 onwards, the expression was predominantly in parts of the spongiotrophoblast (Fig. 1C, arrow) and in many small foci in the labyrinth (suppl. Fig 1). Use of several markers for placental cell types, such as Tpbpa, Prl3d1, Prl3b1, Eomes and Tfeb, did not allow identification of the cell types present in these foci. Similar expression was seen at E14 and E16 (D, E, arrows). Between E12 and E16, only a limited overlap in the spatial expression patterns of Tpbpa and Lhx3 could be observed, showing that not all spongiotrophoblasts expressed Lhx3 (Fig. 1E-L). At E14 and E16, weak expression was seen in the glycogen cells situated in the decidua (arrows in E, F). Lhx3 expression declined after E16 and extremely weak expression was detected in the spongiotrophoblast of E18 placentas (F). Decidua glycogen cells however expressed Lhx3 at comparable levels in E16 and E18 placentas (arrows in F and E). Expression was not observed in giant cells and in the glycogen cells present in spongiotrophoblast or labyrinth.
Fig 1.
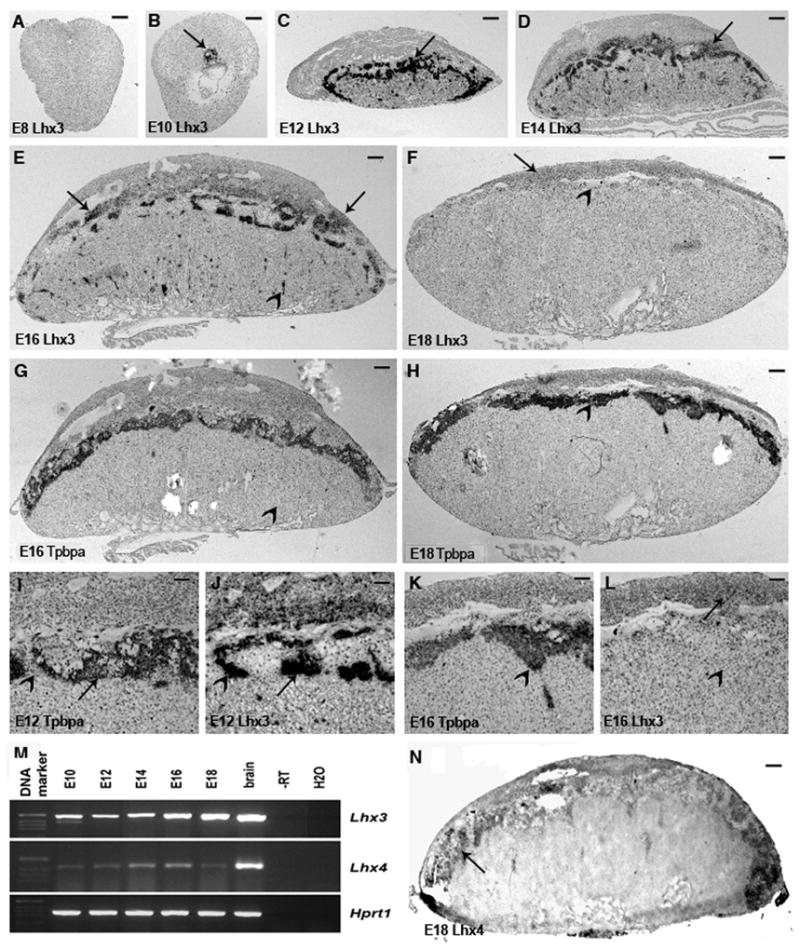
In situ expression analysis of Lhx3 and Lhx4: E8 conceptus shows no expression of Lhx3 (A). At E10 (B), the expression is seen in the ectoplacetal cone and the invasive trophoblasts (arrow), whereas the embryo is devoid of any expression. In E12 placentas (C), predominant expression is detected in many parts of spongiotrophoblast (arrow) and multiple foci in the labyrinth. Expression in the rare patches of the decidua is not clearly seen at this magnification. Similar expression pattern is seen in E14 placentas (D), but expression is reduced in spongiotrophoblast and increased in the decidua (arrow). At E16 (E), decidual expression is even stronger (arrows), but further reduced in spongiotrophoblast and in the labyrinth (arrowhead). Finally at E18 (F), weak expression is seen only in the decidua (arrows) with no expression in spongiotrophoblast (arrowhead), which earlier was a major site for Lhx3 expression. Overviews of Tpbpa expression on sections adjacent to those shown in (E) and (F) show that Lhx3 is expressed only in a subset of spongiotrophoblasts at E16 and confirms that it is not expressed in this tissue at E18. Higher magnification views of Lhx3 and Tpbpa hybridizations on adjacent sections of an E12 placenta (I and J) show that in some parts of spongiotrophoblast expression is spatially separated, whereas in other nearby parts, they overlap. Decidual expression of Lhx3 in E12 placenta can be seen (J). At E18 Lhx3 and Tpbpa are not co-expressed anymore (K and L). (M) RT-PCR analysis of Lhx3 and Lhx4 expression during placental development (N) Lhx4 expression in E18 placentas, arrows show positive signal (in spongiotrophoblast). Scale bar = μm in (A) and (B), 300 μm in (C), 400 μm in (D), 500 μm in (E) till (H), 125 μm in (I) and (J) and 100 μm in (K) and (L), 250μm in (N).
Lhx4 in situ hybridization was carried out only on E18 wild type placentas. Lhx4 expression was detected specifically in the spongiotrophoblast region (Fig 1N). Temporal expression pattern of both Lhx3 and Lhx4 was determined by RT-PCR. In contrast to the Lhx3 in situ analysis, RT-PCR showed no major changes in expression levels for both genes between E10 and E10 of gestation. Expression of other LIM-homeobox genes, Lhx1, Lhx2, Lhx4, Lhx5, Lhx6, Lhx8, and Lhx9, was also assessed by RT-PCR only. By this method, transcripts from all genes were detected in the E18 mouse placenta (not shown).
Lhx3 mutation causes no changes in placental weight and morphology
From the Lhx3 het × het matings, four litters with a total of 42 E18 conceptuses were obtained. These consisted of 25 heterozygous (14 females and 11 males), 10 homozygous mutant (5 females and 5 males) and 7 wild type (5 females and 2 males) conceptuses. Weights of Lhx3+/+, Lhx3+/−, and Lhx3−/− placentas were 88.7 ± 10.0 mg, 87.4 ± 11.5 mg, and 89.2 ± 13.6 mg, respectively. Thus there was no consistent effect of the Lhx3 mutation on the wet placental weight and this effect was independent of the fetal sex (not shown).
Isolectin B4 stained sections were studied for changes in placental morphology. Light microscopy did not reveal any significant differences. Furthermore, 2 isolectin B4 stained sections from 2 placentas of each genotype and sex combination, altogether 24 sections, were analyzed by semi-quantitative morphometry for subtle differences in the section-areas occupied by decidua, spongiotrophoblast and labyrinth. As with placental weight, no significant difference was found between different genotypes (not shown). Furthermore, in situ hybridization revealed that the spatial expression patterns of the placental cell type-specific markers Tpbpa, Prl3d1, Prl3b1, Eomes and Tfeb were not altered in Lhx3 mutant placentas (not shown). Together these results indicated that loss of LHX3 does not cause any obvious placental phenotypes.
Deletion of both Lhx3 and Lhx4 causes an inconsistent labyrinthine phenotype
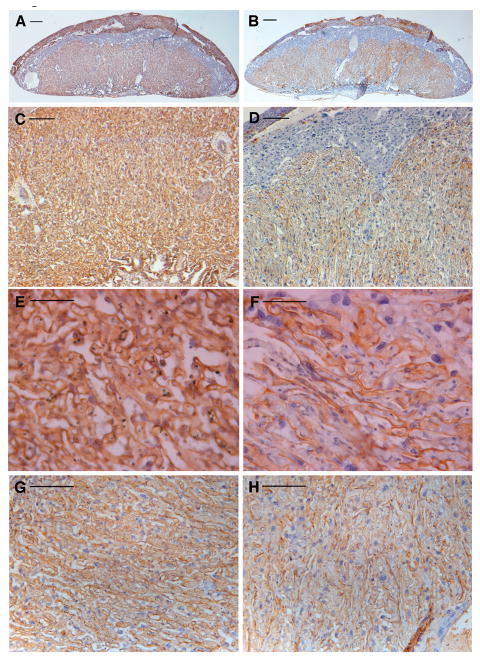
As it is known that in pituitary and motoneuron development LHX3 and LHX4 may substitute for each other, the possibility existed that expression of Lhx4 could compensate for loss of LHX3. This possibility was supported by the similar expression of Lhx3 and Lhx4 in the mouse placenta, however, as assessed by RT-PCR, Lhx4 transcript levels did not vary between Lhx3+/+ and Lhx3−/− placentas (not shown). Thus, Lhx3+/−;Lhx4+/− double het × het matings were performed. A total of 17 E18 litters with 176 conceptuses were obtained and among these 7 Lhx3−/−;Lhx4−/− conceptuses were identified (4%). No significant differences in placental weight were observed between the different genotypes (Table 1). Furthermore, histological analysis of placentas showed no discernible difference between any of the genotypes, with exception of some Lhx3−/−;Lhx4−/− placentas (see below). This suggests that presence of a single LIM-3 allele, as in Lhx3−/−;Lhx4+/− or in Lhx3+/−;Lhx4−/−, is fully compatible with normal placental development. However, in three E18 litters (litter 1, 3 and 5; Suppl. Table 2), double mutant fetuses were smaller as compared to other fetuses in the same litter (Table 1). Comparative histological analysis of isolectin B4 stained sections from 5 Lhx3−/−;Lhx4−/− and control placentas from the same litters showed that 3 double-mutant placentas exhibited a specific abnormal phenotype, which did not correlate with reduced fetal size (Suppl. Table 2). This phenotype was characterized by loose organization of labyrinthine architecture, with reduced density of isolectin B4 positive trophoblast basement membranes (Fig. 2A-F). This finding was supported by ASMA staining, which revealed similar disorganization of pericytes in the 3 abnormal Lhx3−/−;Lhx4−/− placentas (Fig. 2G, H). Pericytes function in capillary looping in the labyrinth (Ohlsson et al., 1999). However, in the remaining 2 double-mutant placentas, no difference could be detected compared to control placentas. Semi-quantitative morphometrical analysis, which was performed on three double mutant and six control placental sections stained with isolectin B4, showed no significant alterations in placental tissue distribution.
Table 1.
Placental and fetal weights of Lhx3−/−;Lhx4−/− double mutants and other genotypes
| Litter number and litter size (N) | Average placental weight (mg) | Lhx3−/−;Lhx4−/− placental weight (mg) | Average fetal weight (mg) | Lhx3−/−;Lhx4−/− fetal weight (mg) |
|---|---|---|---|---|
| 1 (N = 9) | 83.9±9.0 | 89.5 | 1251.0±67.9 | 1006.4 |
| 2 (N = 10) | 93.7±10.1 | 88.1 | 924.6±52.2 | 886.4 |
| 3 (N = 10) | 82.6±9.3 | 76.4 | 930.7±36.7 | 754.7 |
| 4 (N = 6) | 124.8±11.7 | 129.5 | 882.4±105.5 | 903.8 |
| 5 (N = 10) | 100.7±15.3 | 88.1 | 947.8±70.8 | 842.3 |
| 6 (N = 11) | 87.4 ± 6.8 | 90.0 | 876.7 ± 61.7 | 813.3 |
| 7 (N = 10) | 111.1 ± 17.6 | 90.0 | 743.33 ± 49.2 | 770.0 |
Values are given as means ± SD. Average placental and fetal weights do not include the Lhx3−/−;Lhx4−/− weights.
Fig 2.
Morphological analysis of a Lhx3−/−;Lhx4−/− and control placentas
A, C, E and G are control placentas, B, D, F and H show Lhx3−/−;Lhx4−/− placentas. A and B: overview of isolectin B4 staining; C, D, E and F: isolectin B4 staining in labyrinth; G and H: ASMA staining. Arrows show positive staining.
Scale bar: 1mm.
Normalization of Lhx3 and Lhx4 expression levels does not rescue the placental phenotypes of AT24 mice
Like the hyperplastic IHPD placentas, AT24 placentas over express Lhx3 (not shown). In order to establish whether over expression of LIM-3 group genes is a cause or a consequence of placental over growth in IHPD, we tested the effect of reduction of LHX3 and LHX4 levels in AT24 placentas. From 2 E18 AT24/AT24 × Lhx3+/−;Lhx4+/− pregnancies, 18 conceptuses were isolated. Four conceptuses were AT24/+;Lhx3+/+;Lhx4+/+ and 3 were AT24/+;Lhx3+/−;Lhx4+/−. The remaining conceptuses were either Lhx3+/− or Lhx4+/−. Mean placental weights were 165.8 ± 10.1 mg for AT24/+;Lhx3+/−;Lhx4+/− and 162.5 ± 21.0 mg for AT24/+;Lhx3+/+;Lhx4+/+. Thus reduction of LIM-3 gene expression by half has no influence on the AT24 placental phenotype.
Placental transport function analysis in Lhx3 and Lhx4 double mutant mice
The abnormal morphology of the labyrinth in a sub-set of Lhx3−/−;Lhx4−/- placentas raised the possibility that placental transport functions could be compromised in these Lhx3−/−;Lhx4−/− placentas. Therefore, one parameter of placental transport was assessed by determining the accumulation of the essential fatty acids (EFAs) linoleic acid (18:2), arachidonic acid (20:4), and docosahexaenoic acid (22:6), and the non-essential fatty acids (NEFAs) palmytic acid (16:0) and stearic acid (18:0). EFAs are obtained through the mother’s diet and are therefore transported to the fetal organism via the placenta. NEFAs are synthesized in the fetus. With this approach, compromised placental transport function is indicated by distorted ratios of EFAs and NEFAs in favor of the latter (Wu et al., 2003). Our lipid measurements provided no evidence for defective lipid transport function of Lhx3−/−;Lhx4−/− placentas. Thus, in control E18 fetuses the EFAs made up 22.0 ± 9.4 % (N = 6) of all FAs. In Lhx3−/−;Lhx4−/− fetuses this value was 21.2 ± 4.4 (N = 2; P > 0.05). In the placentas from the same conceptuses, these values were 23.4 ± 4.9% and 20.6 ± 11.3%, for control and Lhx3−/−;Lhx4−/− placentas, respectively (P > 0.05). However, this finding does not exclude the possibility that other transport systems are affected in Lhx3−/−;Lhx4−/−placentas.
Expression of Lhx3/Lhx4 target genes in Lhx3 and Lhx4 double mutant mice
One of the known target genes of Lhx3 that is expressed in the placenta is gonadotropin-releasing hormone receptor (Gnrhr) (McGillivray et al., 2005, Granger et al., 2006). To determine if absence of the LIM-3 transcription factors alters expression of genes involved in the gonadotrope pathway, levels of Gnrhr and gonadotropin releasing hormone (Gnrh) were determined in 4 Lhx3−/−;Lhx4−/− placentas and 10 control litter-mate placentas (Suppl. Table 2) by qRT-PCR. Furthermore, expression of Dlx3 was assessed, as the product of this gene is involved in the regulation of the subunit gene of chorionic gonadotropin(Roberson et al., 2001) and its deletion causes placental failure, which is however associated with down-regulation of Esx1 (Morasso et al., 1999), a gene shown to be relevant for labyrinthine morphogenesis (Li and Behringer, 1998). These experiments provided no indication for differential expression of Dlx3 and Gnrh (not shown) in Lhx3−/−;Lhx4−/− and control placentas. However, for Gnrhr up-regulation was seen in the phenotypically normal Lhx3−/−;Lhx4−/− placentas of 2 litters, whereas in 2 litters where the Lhx3−/−;Lhx4−/− placentas were abnormal, Gnrhr expression was not modified by genotype (Fig. 3B; Suppl. Table 2). This finding provided some indication that Gnrhr could possibly function in placental development. To test this possibility, isolectin B4 stained sections of E18 and E19 Gnrhr+/+ and Gnrhr−/− littermate placentas were analyzed. At both developmental stages, 2 mutant and 2 wild-type placentas were compared. With this light-microscopy approach no phenotypic differences could be observed between Gnrhr+/+ and Gnrhr−/− placentas.
Fig 3.
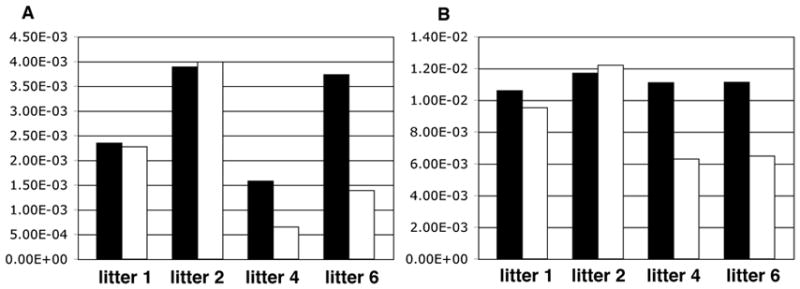
Gnrhr expression level in Lhx3−/−;Lhx4−/− and control placentas
A: Relative ratio of Gnrhr/Tubb3 in Lhx3−/−;Lhx4−/− and control placentas; B: Relative ratio of Gnrhr/Gapdh Lhx3−/−;Lhx4−/− and control placentas. Black column: gene expression level in Lhx3−/−;Lhx4−/− placentas. White column: gene expression level in control placentas. For detailed information on mutant and control placentas see Suppl. Table 2.
Discussion
We had previously identified a large number of genes, which exhibited changed expression in the placental over growth models (Singh et al., 2004) interspecific hybridization (Zechner et al., 1996), cloning by somatic cell nuclear transfer (Tanaka et al., 2001), and mutation of Esx1 (Li and Behringer, 1998). For a considerable number of these genes there were no previous reports that demonstrated or suggested that they had functions in placental development. Therefore, in the present study we initially focused on one gene, Lhx3, whose expression was increased in IHPD placentas, as assessed by microarray, Northern and in situ hybridization analyses (Singh et al., 2004). Our study shows that Lhx3 is expressed in the mouse placenta in a cell-type specific manner, in that only three cell types, spongiotrophoblasts, decidual glycogen cells, and some cells in the labyrinth zone express the gene. In the spongiotrophoblast proper, which excludes the glycogen cells also present in the junctional zone, not all cells expressed Lhx3. Similarly, Lhx3 expression in glycogen cells was strictly dependent on their location. The significance of the differential Lhx3 expression in these two cell types is presently not clear.
The function of LHX3 as transcription factor and the perinatal lethality of the Lhx3 deletion suggested that absence of Lhx3 might also result in abnormal placental development and the finding that deletion of Lhx3 alone was not sufficient to cause any alterations in placental histology and morphology was unexpected. However, it had previously been shown that in some developmental processes, such as motoneuron formation (Sharma et al., 1998), Lhx4 is fully competent to substitute for Lhx3, which suggested that the same could apply to placental development. This possibility was also supported by the finding that on E18 Lhx4 is expressed in the spongiotrophoblast, that is, in the tissue layer where high Lhx3 transcripts levels were detected between E12 and E16. While deletion of both LIM-3 factors resulted in a mild placental phenotype, this was not fully penetrant and in addition affected the labyrinth, whereas both Lhx3 and Lhx4 are expressed mainly in the junctional zone. The ectopic phenotype may possibly be explained by the fact that LHX3 and LHX4 are involved in transcriptional regulation of several hormones, such as glycoprotein α-subunit, prolactin, and follicle stimulating hormone-β, which do not act in a autocrine manner (McGillivray et al., 2005). It is difficult to explain the incomplete penetrance, as it seems to be established that no other LIM homeodomain containing transcription factors can substitute for LHX3 and LHX4 (Hobert and Westphal, 2000), which excludes functional redundancy. However, it is important to note that several other LIM-homeobox genes are expressed in the placenta, including Lhx5, which belongs to the related LIN-11 group (Hobert and Westphal, 2000). A simple explanation could be that neither of the two strains has been bred to full homozygosity for a B6 background, so there is the possibility that genetic heterogeneity underlies the incomplete penetrance. Alternatively, it has been shown that co-expression of Lhx3, Nr5a1 (steroidogenic factor 1) and the LIM-homeodomain containing transcription factor Isl1 enhances expression of Gnrhr expression in mouse gonadotrope T3-1 and L T2 cells (Granger et al., 2006). In non-gonadotrope COS cells, which normally do not express Gnrhr, co-expression of these expression factors is also competent to induce Gnrhr and interestingly Nr5a1 and Isl1 alone induce expression, albeit at lower levels than when Lhx3 was cotransfected. Co-expression of either Lhx5 or Lhx9 did not increase Gnrhr activation above that produced by Nr5a1 and Isl1 (Granger et al., 2006). Performance of Lhx4 was not tested in that study. Thus the possibility remains that in the mouse placenta deletion of Lhx3 and Lhx4 can be rescued to some degree by expression of Nr5a1 and Isl1, or Isl2, which is also competent to induce Gnrhr (Granger et al., 2006). However, to the best of our knowledge it is not known if these genes are expressed in the mouse placenta. In addition, it is not clear if the transcription factors encoded by these genes are at all involved in transcriptional regulation of Gnrhr in the placenta (Cheng et al., 2001) While we have analyzed only some genes involved in the gonadotrope pathway, as these are known to be expressed in the mouse placenta (Khodr & Siler-Khodr, 1980, Tan & Rousseau, 1982, Miyake et al., 1982, Cheng et al., 2000), it is possible that the inconsistent placental phenotype of Lhx3−/−;Lhx4−/− double-mutant conceptuses is in fact caused by compromised regulation of other target genes such as Foxl2 (Ellsworth et al., 2006). However, the finding that up-regulation of Gnrhr coincides with phenotypical normality of Lhx3−/−;Lhx4−/−placentas may suggest that the placental phenotypes were indeed caused by altered GNRHR levels. Over expression of Lhx3/Lhx4 in Jeg3 trophoblast tumor cells or trophoblast stem cells in vitro or in transgenic mice in vivo could probably clarify the functions of Lhx3 and Lhx4 in placental development, as over expression does not allow rescue through redundancy (Heintz, 2001).
This study was initiated because of our previous finding that Lhx3 is up-regulated in enlarged IHPD placentas (Singh et al., 2004). The fact that reduced Lhx3/Lhx4 expression has no discernible effect on the phenotype of AT24 placentas demonstrates that over expression of Lhx3 does not underlie placental overgrowth. However, as demonstrated previously (Singh et al., 2004) Lhx3 expression in IHPD placentas is ectopic and does not reflect the expression in normal placentas, which shows that Lhx3 over expression was not just a consequence of increased spongiotrophoblast. Again, over expression of Lhx3 in trophoblasts in vitro or in vivo could possibly provide information of the LIM-3 transcription factors in normal and abnormal placental development.
Supplementary Material
Suppl. Fig. 1. High magnification (40x) of Lhx3 (A) and Tpbpa (B) mRNA in situ hybridization on consecutive sections of a E14 wt placenta. The arrows in (A) indicate Lhx3 positive areas in the labyrinth, which are negative for Tpbpa (B) and the other placental cell-type specific markers used (not shown).
Acknowledgments
The authors are grateful to Drs. S. Camper, R. Elliott, and J. Forejt for valuable help and again to Dr. S. Camper for critically reading the manuscript. This work was supported by grants from the Swedish Research Council (Vetenskapsrådet) and the Wallenberg Consortium North to RF; from the Deutsche Forschungsgemeinschaft (SFB 535) to RG; and from the U.S. National Institutes of Health HD30284 to RRB and R01HD34283 and R37HD30428 to Dr. S. Camper. MH was supported by a Career Development Award from the Medical Research Council, UK.
References
- Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet. 2000;25:311–4. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Cheng KW, Nathwani PS, Leung PC. Regulation of human gonadotropin-releasing hormone receptor gene expression in placental cells. Endocrinology. 2000;141:2340–9. doi: 10.1210/endo.141.7.7543. [DOI] [PubMed] [Google Scholar]
- Cross JC. Placental function in development and disease. Reprod Fertil Dev. 2006;18 :71–6. doi: 10.1071/rd05121. [DOI] [PubMed] [Google Scholar]
- Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–30. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Elliott RW, Miller DR, Pearsall RS, Hohman C, Zhang Y, Poslinski D, Tabaczynski DA, Chapman VM. Genetic analysis of testis weight and fertility in an interspecies hybrid congenic strain for Chromosome X. Mamm Genome. 2001;12:45–51. doi: 10.1007/s003350010234. [DOI] [PubMed] [Google Scholar]
- Ellsworth BS, Butts DL, Camper SA. Mechanisms Underlying Pituitary Hypoplasia and Failed Cell Specification in Lhx3 Deficient Mice. Developmental Biology. doi: 10.1016/j.ydbio.2007.10.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, Osamura RY, Camper SA. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- Gagnon R. Placental insufficiency and its consequences. Eur J Obstet Gynecol Reprod Biol. 2003;110(Suppl 1):S99–107. doi: 10.1016/s0301-2115(03)00179-9. [DOI] [PubMed] [Google Scholar]
- Gaver RC, Sweeley CC. Methods For Methanolysis Of Sphingolipids And Direct Determination Of Long-Chain Bases By Gas Chromatography. J Am Oil Chem Soc. 1965;42:294–8. doi: 10.1007/BF02540132. [DOI] [PubMed] [Google Scholar]
- Granger A, Bleux C, Kottler ML, Rhodes SJ, Counis R, Laverriere JN. The LIM-homeodomain proteins Isl-1 and Lhx3 act with steroidogenic factor 1 to enhance gonadotrope-specific activity of the gonadotropin-releasing hormone receptor gene promoter. Mol Endocrinol. 2006;20:2093–108. doi: 10.1210/me.2005-0184. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–6. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–70. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol Metab. 2001;12:162–8. doi: 10.1016/s1043-2760(01)00375-7. [DOI] [PubMed] [Google Scholar]
- Hemberger MC, Pearsall RS, Zechner U, Orth A, Otto S, Ruschendorf F, Fundele R, Elliott R. Genetic dissection of X-linked interspecific hybrid placental dysplasia in congenic mouse strains. Genetics. 1999;153:383–90. doi: 10.1093/genetics/153.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16 :75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Khodr GS, Siler-Khodr TM. Placental luteinizing hormone-releasing factor and its synthesis. Science. 1980;207:315–7. doi: 10.1126/science.6985750. [DOI] [PubMed] [Google Scholar]
- Lane RH, Ramirez RJ, Tsirka AE, Kloesz JL, McLaughlin MK, Gruetzmacher EM, Devaskar SU. Uteroplacental insufficiency lowers the threshold towards hypoxia-induced cerebral apoptosis in growth-retarded fetal rats. Brain Res. 2001;895:186–93. doi: 10.1016/s0006-8993(01)02074-1. [DOI] [PubMed] [Google Scholar]
- Leipala JA, Boldt T, Turpeinen U, Vuolteenaho O, Fellman V. Cardiac hypertrophy and altered hemodynamic adaptation in growth-restricted preterm infants. Pediatr Res. 2003;53:989–93. doi: 10.1203/01.PDR.0000061564.86797.78. [DOI] [PubMed] [Google Scholar]
- Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 1988;2:1639–46. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet. 1998;20:309–11. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–5. doi: 10.1210/en.2004-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Peters T, Luttges A, Curfs J, Fundele R. Preferential expression of the G90 gene in post-mitotic cells during mouse embryonic development. Anat Embryol (Berl) 2003;207:109–17. doi: 10.1007/s00429-003-0330-9. [DOI] [PubMed] [Google Scholar]
- Miyake A, Sakumoto T, Aono T, Kawamura Y, Maeda T, Kurachi K. Changes in luteinizing hormone-releasing hormone in human placenta throughout pregnancy. Obstet Gynecol. 1982;60:444–9. [PubMed] [Google Scholar]
- Monkley SJ, Delaney SJ, Pennisi DJ, Christiansen JH, Wainwright BJ. Targeted disruption of the Wnt2 gene results in placentation defects. Development. 1996;122:3343–53. doi: 10.1242/dev.122.11.3343. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A. 1999;96:162–7. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, Bhangoo AP, Ten S, Weigel J, Pfaffle RW, Rhodes SJ. Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol. 2007;265–266:190–5. doi: 10.1016/j.mce.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Falck P, Hellstrom M, Lindahl P, Bostrom H, Franklin G, Ahrlund-Richter L, Pollard J, Soriano P, Betsholtz C. PDGFB regulates the development of the labyrinthine layer of the mouse fetal placenta. Dev Biol. 1999;212:124–36. doi: 10.1006/dbio.1999.9306. [DOI] [PubMed] [Google Scholar]
- Pask AJ, Kanasaki H, Kaiser UB, Conn PM, Janovick JA, Stockton DW, Hess DL, Justice MJ, Behringer RR. A novel mouse model of hypogonadotrophic hypogonadism: N-ethyl-N-nitrosourea-induced gonadotropin-releasing hormone receptor gene mutation. Mol Endocrinol. 2005;19:972–81. doi: 10.1210/me.2004-0192. [DOI] [PubMed] [Google Scholar]
- Raetzman LT, Ward R, Camper SA. Lhx4 and Prop1 are required for cell survival and expansion of the pituitary primordia. Development. 2002;129:4229–39. doi: 10.1242/dev.129.18.4229. [DOI] [PubMed] [Google Scholar]
- Roberson MS, Meermann S, Morasso MI, Mulvaney-Musa JM, Zhang T. A role for the homeobox protein Distal-less 3 in the activation of the glycoprotein hormone alpha subunit gene in choriocarcinoma cells. J Biol Chem. 2001;276:10016–24. doi: 10.1074/jbc.M007481200. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–48. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;2:95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Salas M, John R, Saxena A, Barton S, Frank D, Fitzpatrick G, Higgins MJ, Tycko B. Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev. 2004;121:1199–210. doi: 10.1016/j.mod.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–28. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H. Multistep control of pituitary organogenesis. Science. 1997;278:1809–12. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Zhadanov AB, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science. 1996;272:1004–7. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- Simmons RA, Templeton LJ, Gertz SJ. Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes. 2001;50:2279–86. doi: 10.2337/diabetes.50.10.2279. [DOI] [PubMed] [Google Scholar]
- Singh U, Fohn LE, Wakayama T, Ohgane J, Steinhoff C, Lipkowitz B, Schulz R, Orth A, Ropers HH, Behringer RR, Tanaka S, Shiota K, Yanagimachi R, Nuber UA, Fundele R. Different molecular mechanisms underlie placental overgrowth phenotypes caused by interspecies hybridization, cloning, and Esx1 mutation. Dev Dyn. 2004;230:149–64. doi: 10.1002/dvdy.20024. [DOI] [PubMed] [Google Scholar]
- Singh U, Sun T, Larsson T, Elliott RW, Kostka G, Fundele RH. Expression and functional analysis of fibulin-1 (Fbln1) during normal and abnormal placental development of the mouse. Placenta. 2006a;27:1014–21. doi: 10.1016/j.placenta.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Singh U, Sun T, Looman C, Heuchel R, Elliott R, Freichel M, Meissner M, Flockerzi V, Fundele R. Expression and function of the gene encoding the voltage-dependent calcium channel beta3-subunit in the mouse placenta. Placenta. 2007;28:412–20. doi: 10.1016/j.placenta.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Singh U, Sun T, Shi W, Schulz R, Nuber UA, Varanou A, Hemberger MC, Elliott RW, Ohta H, Wakayama T, Fundele R. Expression and functional analysis of genes deregulated in mouse placental overgrowth models: Car2 and Ncam1. Dev Dyn. 2005;234:1034–45. doi: 10.1002/dvdy.20597. [DOI] [PubMed] [Google Scholar]
- Singh U, Yu Y, Kalinina E, Konno T, Sun T, Ohta H, Wakayama T, Soares MJ, Hemberger M, Fundele RH. Carboxypeptidase E in the mouse placenta. Differentiation. 2006b;74:648–60. doi: 10.1111/j.1432-0436.2006.00093.x. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–16. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- Tan L, Rousseau P. The chemical identity of the immunoreactive LHRH-like peptide biosynthesized in the human placenta. Biochem Biophys Res Commun. 1982;109:1061–71. doi: 10.1016/0006-291x(82)92047-2. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod. 2001;65:1813–21. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- Tsirka AE, Gruetzmacher EM, Kelley DE, Ritov VH, Devaskar SU, Lane RH. Myocardial gene expression of glucose transporter 1 and glucose transporter 4 in response to uteroplacental insufficiency in the rat. J Endocrinol. 2001;169:373–80. doi: 10.1677/joe.0.1690373. [DOI] [PubMed] [Google Scholar]
- Vonnahme KA, Hess BW, Hansen TR, McCormick RJ, Rule DC, Moss GE, Murdoch WJ, Nijland MJ, Skinner DC, Nathanielsz PW, Ford SP. Maternal undernutrition from early- to mid-gestation leads to growth retardation, cardiac ventricular hypertrophy, and increased liver weight in the fetal sheep. Biol Reprod. 2003;69:133–40. doi: 10.1095/biolreprod.102.012120. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–7. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Dennis RD, Doenhoff MJ, Geyer R. Stage-associated expression of ceramide structures in glycosphingolipids from the human trematode parasite Schistosoma mansoni. Biochim Biophys Acta. 2000;1524:155–61. doi: 10.1016/s0304-4165(00)00152-5. [DOI] [PubMed] [Google Scholar]
- Xu Y, Williams SJ, O’Brien D, Davidge ST. Hypoxia or nutrient restriction during pregnancy in rats leads to progressive cardiac remodeling and impairs postischemic recovery in adult male offspring. Faseb J. 2006;20:1251–3. doi: 10.1096/fj.05-4917fje. [DOI] [PubMed] [Google Scholar]
- Zechner U, Hemberger M, Constancia M, Orth A, Dragatsis I, Luttges A, Hameister H, Fundele R. Proliferation and growth factor expression in abnormally enlarged placentas of mouse interspecific hybrids. Dev Dyn. 2002;224:125–34. doi: 10.1002/dvdy.10094. [DOI] [PubMed] [Google Scholar]
- Zechner U, Reule M, Orth A, Bonhomme F, Strack B, Guenet, Hameister H, Fundele R. An X-chromosome linked locus contributes to abnormal placental development in mouse interspecific hybrid. Nat Genet. 1996;12:398–403. doi: 10.1038/ng0496-398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1. High magnification (40x) of Lhx3 (A) and Tpbpa (B) mRNA in situ hybridization on consecutive sections of a E14 wt placenta. The arrows in (A) indicate Lhx3 positive areas in the labyrinth, which are negative for Tpbpa (B) and the other placental cell-type specific markers used (not shown).