Abstract
The sphingosine 1-phosphate receptor 1 (S1P1) and its ligand, sphingosine 1-phosphate (S1P), have now emerged as critical regulators of lymphocyte trafficking, vascular development and integrity, and immunity. S1P1 is targeted by the phosphorylation product of fingolimod, which has been approved for the treatment of multiple sclerosis. The recent progress in the structural biology of heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptors has now enabled the elucidation of the structure of S1P1. Analysis of the structure, along with structure activity and mutagenesis analysis, highlighted key interactions associated with the binding of S1P and agonists and suggested that the ligand may gain access to the binding pocket by lateral diffusion within the plasma membrane. The S1P1 crystal structure will be helpful for designing ligands that specifically target S1P1.
The pleiotropic sphingolipid mediator, sphingosine 1-phosphate (S1P), exerts many of its functions by binding to a family of five specific S1P receptors (S1PRs). Some flavor of these receptors is present on almost every cell in the human body, although the receptor S1P1 has attracted much attention because it controls many functions, including lymphocyte trafficking, immunity, and vascular integrity (1). Additional interest in this receptor has arisen with the development of the first Food and Drug Administration–approved S1PR-targeted therapeutic, fingolimod [also known as Gilenya or FTY720 (Novartis)] (2), for the treatment of multiple sclerosis. Fingolimod is a sphingosine analog that is phosphorylated in vivo to an S1P mimetic that is thought to act as a functional antagonist of S1P1, decreasing its cell surface abundance on lymphocytes, which prevents their egress from secondary lymphoid organs and suppresses immune responses. The report by Hanson et al. (3) that deciphered the crystal structure of S1P1 has provided important clues as to how the natural ligand S1P binds to S1P1 as well as how to develop even more effective and specific S1P1 agonists and antagonists.
To solve the structure of the S1P1 heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptor (GPCR), Hanson et al. implemented advanced x-ray microdiffraction data collection and data manipulation methods (3). Because of rapid crystal decay during data collection, complete data sets had to be constructed by merging partial data of many microcrystals. With traditional data processing and merging methods of isotropic resolution cutoffs, which systematically exclude data with poor statistics, they successfully obtained a 3.35 Å–resolution structure of the S1P1 receptor complexed with the selective antagonist (R)-3-amino-(3-hexylphenylamino)-4-oxobutylphosphonic acid [ML056, Protein Data Bank identification code (PDB ID) 3V2W]. By the pioneering use of a microdiffraction data assembly method, they were also able to include or exclude individual reflections on the basis of their correlation threshold to a peak fitting profile and effectively assemble a data set containing many of the reflections at high resolution that were not affected by radiation damage but that were excluded by the traditional processing methods. This allowed Hanson et al. to report the structure of the S1P1 receptor–ML056 complex at 2.8 Å resolution (PDB ID 3V2Y), which highlights details of the structure that were not clearly defined in the lower-resolution data assembly. Improved electron density was observed for the ML056 antagonist and for the residues that line the ligand binding cavity, revealing critical ligand-receptor interactions, and they reported that the higher quality maps show electron density for an N-terminal helix that caps the ligand binding site by packing tightly against extracellular loops (ECLs) 1 and 2.
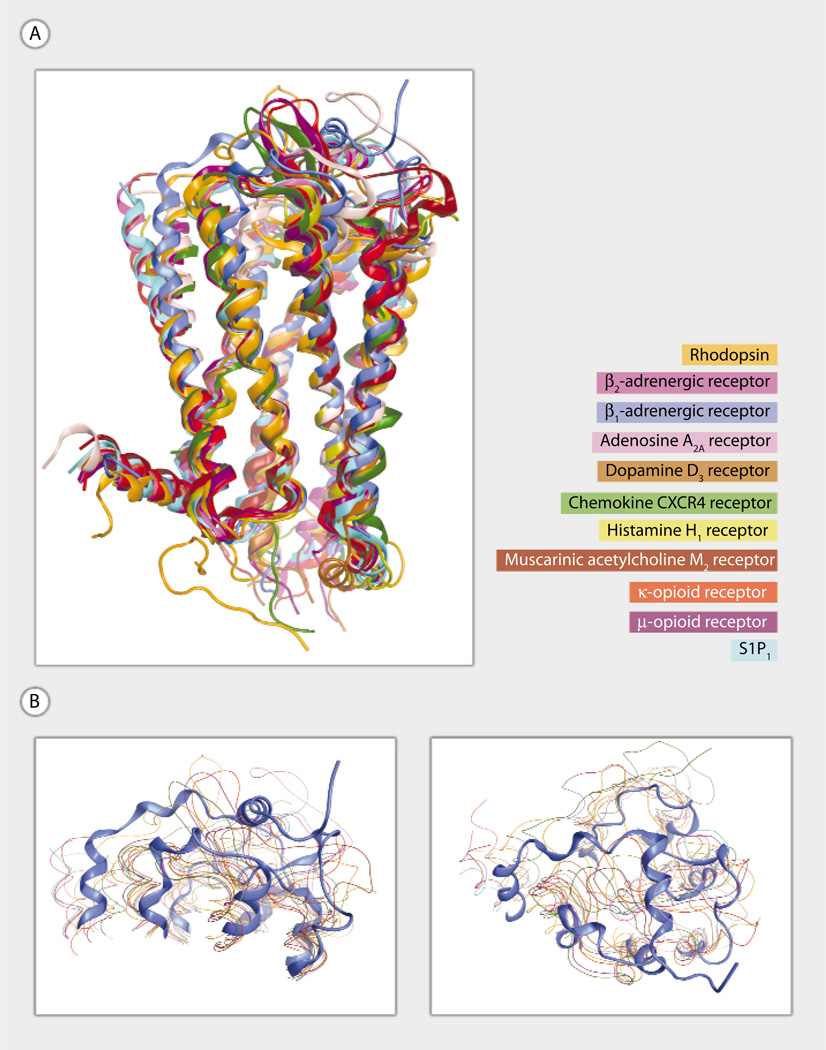
The S1P1 crystal structure exhibits the structurally conserved helical bundle characteristic of the 12 different GPCR family members for which crystallographic structures are available (4) (Fig. 1A). In contrast, the extracellular regions of the S1P1 receptor, including three ECLs (ECL1, ECL2, and ECL3) and the N terminus, adopt a previously undiscovered GPCR architecture (Fig. 1A). Key features include a helical portion of the N terminus that packs between ECL1 and ECL2 and intraloop disulfide bonds within both ECL2 and ECL3. The unique extracellular structure comes as no surprise based on several observations. First, only three pairs of receptors have exhibited a shared extracellular architecture so far. The β2 and β1 adrenoceptors, which share >60% sequence identity, exhibit a short helical stretch in the second ECL and an α-carbon root mean square positional deviation (RMSD) of 0.9 Å for residues in the first, second, and third ECLs. Visual comparison of the magenta and cyan ribbons in Fig. 1 demonstrates the high degree of structural similarity that is reflected by this low RMSD value. The κ and μ opioid receptors, which share over 55% sequence identity, both exhibit a β sheet in the second ECL but have lower overall structural similarity than the adrenergic pair because of differences in the lengths of the second and third ECLs. The M2 and M3 muscarinic acetylcholine receptors, which share >60% sequence identity, possess an outward bend at the extracellular end of the fourth transmembrane helical segment and exhibit an RMSD of 1.2 for residues in the ECL1, ECL2, and ECL3. Because S1P1 is the only member of the S1PR subfamily to be crystallized to date, a closely homologous extracellular structure was not expected. Second, S1P1 shares little sequence identity (<22%) with any of the other 11 crystallized sequences. The S1P1 structure, therefore, provides a window through which a new region of GPCR sequence space may be viewed. In particular, S1P1 is the closest homolog to other phospholipid receptors responsive to S1P (S1P2 to S1P5) or to lysophosphatidic acid (LPA1 to LPA3), previously known as the endothelial differentiation gene family, as well as to the two cannabinoid receptors (CB1 and CB2) that are responsive to N-arachidonoyl ethanolamide and the structurally related orphan receptors, GPR3, GPR6, and GPR12. The first ECL is the same length in these receptors except in the GPR3/6/12 group, which have three fewer amino acids in this loop. The entire set of related receptors also has the cysteine residues in the second ECL that in S1P1 form a disulfide bond. However, S1P4, LPA1 to LPA3, and CB2 have an additional cysteine residue that may lead to a different disulfide bond pattern. The length of the second ECL is well conserved, with only CB2 having two fewer amino acids. The third ECL shows greater differences, and the cysteine residues that form a disulfide bond in S1P1 are found only in S1P2, S1P3, and S1P5. The length of the third ECL is also conserved only among these receptors, with other receptors having both longer (S1P4) and shorter (LPA1 to LPA3, GPR3/6/12, and CB1 and CB2) loops. The limited conservation of the ECL lengths and disulfide bonds in S1P4 may be responsible for the substantially reduced affinity for S1P (5) relative to other S1PRs. Despite the noted differences, the crystal structure of S1P1 provides the best starting point to develop models for these lipid receptors.
Fig. 1.
(A and B) Superposition of 11 crystallized GPCR family members. Rhodopsin (PDB ID 1F88) (16), β2-adrenoceptor (PDB ID 2RH1) (12), β1-adrenoceptor (PDB ID 2VT4) (17), adenosine A2A (PDB ID 3EML) (18), dopamine D3 (PDB ID 3PBL) (19), chemokine CXCR4 (PDB ID 3OE0) (20), histamine H1 (PDB ID 3RZE) (21), muscarinic acetylcholine M2 (PDB ID 3UON) (22), κ-opioid (PDB ID 4DJH) (23), and μ-opioid (PDB ID 4DKL) (24) are shown using ribbon (A) and line (B) representations. S1P1 (PDB ID 3V2Y) (3) is shown using a ribbon representation in all panels. The muscarinic acetylcholine M3 (PDB ID 4DAJ) (25) structure is not shown. Ribbon representations are oriented with extracellular segments at the top of the figure and transmembrane domain 1 on the left (A). T4 lysozyme replacements for IL3 are not shown. (B) Two views of the extracellular segments are shown to emphasize the unique extracellular architecture of the S1P1 receptor.
The crystal structure of S1P1 not only provides insights into other GPCR structures but also provides answers to questions that have long puzzled phospholipid researchers, as well as a new set of puzzles for future research to solve. A long-standing question that arose from the first S1P1 model structure (6) relates to the entry path taken by the poorly water-soluble S1P from carrier proteins such as albumin or high-density lipoprotein (HDL) into the receptor binding site. Hanson et al. show compelling structural evidence that this entry path is likely to involve delivery of S1P to the outer leaflet of the cell membrane, followed by lateral diffusion into the binding pocket and entry between helical segments 1 and 7. One new puzzle presented by the crystal structure of S1P1 centers on the role of Arg292 in helix 7, position 34. Mutation of this arginine residue to either alanine or valine results in receptors that properly localize to the cell membrane but fail to respond to S1P (7). These mutations were selected on the basis of a modeled S1P1 complex with S1P, which predicted a direct, favorable ion-pairing interaction between Arg292 (7) and the phosphate headgroup of S1P. The crystal structure places the cationic side chain of Arg292 almost 12 Å from the phosphate headgroup of the crystallized antagonist, oriented into the gap between helical segments 1 and 7. This distance is so large that Arg292 does not make a direct binding interaction with the antagonist. We speculate whether the role of Arg292 differs substantially between antagonists and agonists, with a direct binding interaction only important for agonist binding, or whether Arg292 serves as a cationic lure to draw phospholipids from the outer leaflet of the cell membrane toward the receptor, thus directing lateral diffusion of candidate ligands in the bilayer toward the entryway into the receptor without retaining a direct interaction with bound ligands in the final complex.
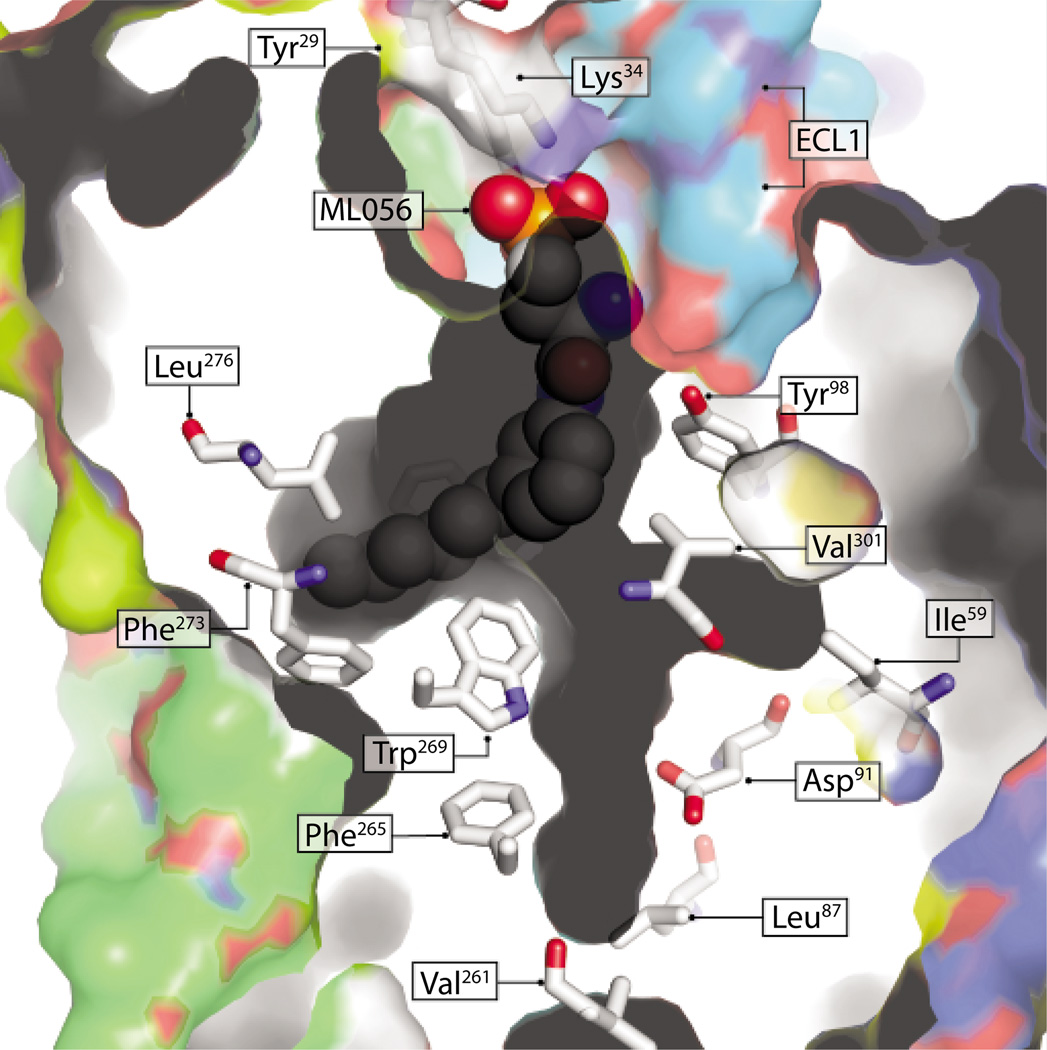
The S1P1-ML056 complex reveals important interactions that contribute to the binding and stabilization of hydrophobic and hydrophilic moieties of S1P1 ligands within the receptor cavity (Fig. 2). Like many GPCRs, S1P1 and other members of this GPCR family have a conserved Trp in helix 6, position 48 (Trp269 in S1P1), which along with other residues on this helix are critical for GPCR ligand-binding receptor activation (8). This aromatic side-chain rotamer toggle switch controls the ligand-induced conformational states of rhodopsin light/dark activation cycle (9) and is a primary determinant in receptor activation, but not for ligand binding in GPR119, GPR39, or the ghrelin, β2-adrenergic, or neurokinin-1 receptors (10). Through mutagenesis of Trp269 (3), Hanson et al. showed that this residue also contributes to the differential stabilization and binding of different subtypes of S1P ligands. For example, S1P and the agonist CYM-5442, which have substantially different molecular structures, interact with the receptor in similar and different ways. Both ligands require the binding energy contribution of the amide carbonyl of Gln101. In addition, S1P interactions with S1P1 require a hydrophobic residue at position 269 of the receptor. In contrast, CYM-5442 interactions specifically require an aromatic residue at this position because ligand binding in Trp269 mutants could not be rescued by insertion of an aromatic residue vicinal to this position. These observations, and others provided by the crystal structure of the S1P1-ML056 complex, provide a powerful basis for the design of S1PR subtype–specific ligands because CYM-5442 preferentially binds only to S1P1. The amphiphilic S1P1 binding site provides important interactions that stabilize ML056 atoms within the receptor cavity (Fig. 2). As in other GPCRs, S1P1 ligands act as agonists or antagonists on the basis of small variations in ligand structure. For example, differential effects on S1P1 activation can be seen between VPC22277 and VPC23019 ligands, which differ in the relative position of substituents on the aromatic ring (11). In the S1P1-ML056 complex, ML056 atoms occupy only a portion of the ligand cavity, possibly because of the meta-substituted aromatic ring (Fig. 2). It will be interesting to see whether the unoccupied regions of the cavity in this complex are occupied by agonists such as VPC22277, which exhibit more linear shapes because of the para-substituted aromatic ring. The crystal structure of S1P1 provides a critical starting point for the design of experiments to probe these questions.
Fig. 2.
Surface-rendering representation of S1P1 and its extended ligand cavity. Representative residues forming the cavity are shown in CPK coloring and stick illustration. Some residues critical for ligand binding, such as Gln101, Arg292, and Phe210, are not shown. Gray marks interior-facing protein surfaces, and colored faces illustrate solvent- or membrane-facing protein surfaces. The ML056 antagonist is colored CPK and illustrated as spheres. Residues Tyr29 and Lys34 are located on an N-terminal helix that is believed to occlude ligand cavity access, and ECL1 faces the membrane or solvent interface through which membrane imbedded ligands are predicted to enter the cavity (3). Residues Val261, Phe265, and Trp269 are in positions 40, 44, and 48, respectively, of helix 6; all of these positions are critical for GPCR activation or ligand binding (or both).
The crystal structure of S1P1 will be important for modeling and understanding agonistic functions of other members of the S1PR family, particularly S1P2, which has unique functions and ligand binding specificity and does not bind phosphorylated fingolimod. The structure will be helpful for designing ligands that will have specificity for S1P1 over other receptors for new therapies. The next step will be to crystallize the other members of the S1PR family, particularly those with the largest differences in ECL lengths and sequences, such as S1P4. Moreover, it will be useful in the future to obtain the structure of the S1P1 receptor complexed with its natural ligand, S1P, to verify the conclusions drawn by this study. This is especially important if the crystal structure is the inactive form of the receptor that was trapped by the antagonist. Lastly, following the pioneering report of the crystal structure of the β2-adrenergic receptor–Gs protein complex that provides a high-resolution view of transmembrane signaling by a GPCR (12–15), it will be important to elucidate a crystal structure of the agonist-bound S1P1 coupled to its G protein partner to unravel the molecular underpinning of G protein activation by this receptor.
Acknowledgments
Funding: Supported by NIH grants R37GM043880, RO1CA61774, RO1AI050094, and U19AI077435-018690 (S.S.); T32 HL094290 (S.L.); and R01HL0084003 (A.L.P.).
References and Notes
- 1.Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011;11:403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun J, Brinkmann V. A mechanistically novel, first oral therapy for multiple sclerosis: The development of fingolimod (FTY720, Gilenya) Discov. Med. 2011;12:213–228. [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, Sanna MG, Han GW, Kuhn P, Rosen H, Stevens RC. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale JJ, Doherty G, Toth L, Li Z, Mills SG, Hajdu R, Ann Keohane C, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Rosen H, Mandala S. The discovery of 3-(N-alkyl)aminopropylphosphonic acids as potent S1P receptor agonists. Bioorg. Med. Chem. Lett. 2004;14:3495–3499. doi: 10.1016/j.bmcl.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 6.Parrill AL, Baker DL, Wang D, Fischer DJ, Bautista DL, van Brocklyn J, Spiegel S, Tigyi G. In: Lysophospholipids and Eicosanoids in Biology and Pathophysiology. Goetzl EJ, Lynch KR, editors. vol. 905. New York: New York Academy of Sciences; 2000. pp. 330–339. [DOI] [PubMed] [Google Scholar]
- 7.Parrill AL, Wang D-A, Bautista DL, Van Brocklyn JR, Lorincz Z, Fischer DJ, Baker DL, Liliom K, Spiegel S, Tigyi G. Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. J. Biol. Chem. 2000;275:39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 8.Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: The transmembrane segments and second extracellular loop. Annu. Rev. Pharmacol. Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 9.Crocker E, Eilers M, Ahuja S, Hornak V, Hirshfeld A, Sheves M, Smith SO. Location of Trp265 in metarhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. J. Mol. Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 10.Holst B, Nygaard R, Valentin-Hansen L, Bach A, Engelstoft MS, Petersen PS, Frimurer TM, Schwartz TW. A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 2010;285:3973–3985. doi: 10.1074/jbc.M109.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis MD, Clemens JJ, Macdonald TL, Lynch KR. Sphingosine 1-phosphate analogs as receptor antagonists. J. Biol. Chem. 2005;280:9833–9841. doi: 10.1074/jbc.M412356200. [DOI] [PubMed] [Google Scholar]
- 12.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein–coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 17.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien EYT, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, Kobayashi T, Stevens RC, Iwata S. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012 doi: 10.1038/nature10939. 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012 doi: 10.1038/nature10954. 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]