Abstract
Peptidoglycan (PGN) recognition proteins (PGRPs) and gram-negative bacteria binding proteins (GNBPs) play an essential role in Toll/Imd signaling pathways in arthropods. The existence of homologous pathways involving PGRPs and GNBPs in other major invertebrate phyla such as the Mollusca remains unclear. In this paper, we report four full-length PGRP cDNAs and one full-length GNBP cDNA cloned from the snail Biomphalaria glabrata, the intermediate host of the human blood fluke Schistosoma mansoni, designated as BgPGRPs and BgGNBP, respectively. Three transcripts are generated from a long form PGRP gene (BgPGRP-LA) by alternative splicing and one from a short form PGRP gene (BgPGRP-SA). BgGNBP encodes a putative secreted protein. Northern blots demonstrated that expression of BgPGRP-SA and BgGNBP was down-regulated in B. glabrata at 6 h after exposure to three types of microbes. No significant changes in expression were observed in snails at 2 days post-exposure (dpe) to the trematodes Echinostoma paraensei or S. mansoni. However, up-regulation of BgPGRP-SA in M line snails at later time points of infection with E. paraensei (i.e., 12 and 17 dpe) was observed. Our study revealed that exposure to either microbes or trematodes did not alter the expression levels of BgPGRP-LAs, which were consistently low. This study provides new insights into the potential pathogen recognition capabilities of molluscs, indicates that further studies of the Toll/Imd pathways in this phylum are in order, and provides additional ways to judge the importance of this pathway in the evolution of internal defense across the animal phyla.
Keywords: Peptidoglycan recognition protein (PGRP), Gram-negative bacteria binding protein (GNBP), Toll/Imd pathways, Innate immunity, Biomphalaria glabrata, Schistosoma mansoni
Introduction
Unlike jawed vertebrates that have both adaptive and innate immune systems, invertebrates rely solely on innate immunity. The evident success of many invertebrate phyla with respect to abundance of both species and individuals clearly indicates that innate immune systems are both effective and sufficient to ensure success. The argument has been made that there is considerable conservatism in the mechanisms of innate immunity across the animal phyla, as most dramatically shown by the evidence suggesting similarities between insect Toll/Imd (immune deficiency) and mammalian TLR (Toll-like receptor)/TNFR (tumor-necrosis factor receptor) signaling pathways (Hoffmann 2003). Discovery of the Toll/Imd pathways in invertebrates (Lemaitre et al. 1996), followed by the subsequent discovery of similar pathways in mammals (Medzhitov et al. 1997), has revolutionized our understanding of innate immunity. Although there is a growing appreciation for the fact that immune systems among invertebrate phyla are complex and diversified and that innate immunity in these groups is by no means solely a function of Toll/Imd pathway activity (Loker et al. 2004; Zhang et al. 2004; Litman et al. 2005; Watson et al. 2005; Terwilliger et al. 2006), it is nonetheless instructive to examine the possible presence and function of these pathways in different invertebrate phyla.
Previous studies in invertebrates of the Toll/Imd pathways have concentrated on arthropods, particularly on the model organism Drosophila melanogaster. More recently, attention has also been directed toward mosquitoes and their role as malaria vectors (Meister et al. 2005; Frolet et al. 2006; Shin et al. 2006). With respect to the phylum Mollusca, the second largest phylum in the animal kingdom after the arthropods, studies of species of aquaculture importance imply that Toll/Imd pathways also exist in this phylum. Genes or expression sequence tags (ESTs) encoding intracellular molecules of relevance to the pathways (e.g., Rel/NF-kB, IKK-like genes, and Toll-like sequences) have been reported in the oyster, scallop, and abalone (Escoubas et al. 1999; Montagnani et al. 2004; Tanguy et al. 2004; Jiang and Wu 2006; Song et al. 2006). Moreover, several ESTs homologous to genes associated with Toll/Imd pathways have been identified in the squid Euprymna scolopes, a cephalopod mollusc (Goodson et al. 2005).
However, homologous genes or gene products have not been reported from the freshwater gastropod Biomphalaria glabrata (Walker 2006), the best-studied snail host involved in transmission of the human blood fluke, Schistosoma mansoni. This parasite is one of three main disease-causing schistosome species that in aggregate infect 200 million people worldwide (Gryseels et al. 2006). Better understanding the fundamental mechanism of snail internal defense and associated immune signaling pathways will provide new insights that could lead to innovative control strategies for blocking the transmission of schistosomiasis within the snail host and, at the same time, broaden our knowledge of the evolution of innate immunity. Recently, considerable progress has been made in exploring the mechanisms of snail internal defense at the molecular level (Mitta et al. 2005; Vergote et al. 2005; Goodall et al. 2006; Humphries and Yoshino 2006; Raghavan and Knight 2006; Lockyer et al. 2007), but still much remains unknown.
This paper focuses on our recent studies of B. glabrata genes encoding peptidoglycan (PGN) recognition proteins (PGRPs; including long and short forms) and gram-negative bacteria binding protein (GNBP). GNBP, also known as β-1,3-glucan binding protein (βGBP) and β-1,3-glucan recognition protein (βGRP), actually comprises a family of proteins that possess the bacterial glucanase-like domain that is able to sense and bind the β-glucan of the microbial cell wall. Since purification of a βGBP (Ochiai and Ashida 1988) and a PGRP protein (Yoshida et al. 1996) from the silkworm Bombyx mori was first reported, extensive studies on the structure and function of the two protein families have been made. One of important discoveries has been to reveal the role of PGRPs and GNBPs in the Toll and Imd immune pathways. Studies focused on Drosophila demonstrated that PGRPs and GNBPs play an essential role as pattern recognition receptors (PRRs) in upstream control of the Toll/Imd pathways (Gobert et al. 2003; Hoffmann 2003; Lemaitre and Hoffmann 2007). Generally, PGRP short forms (e.g., PGRP-SA and -SD) in conjunction with GNBPs (e.g., PGRPs 1 and 3) are involved in interaction with pathogen-associated molecular patterns, such as lysine (Lys)-type PGN on the cell surface of gram-positive bacteria, and activate the Toll pathway (Gobert et al. 2003; Lemaitre and Hoffmann 2007). Long forms (e.g., PGRP-LC and -LE), after interaction with diaminopimelic acid (DAP)-type PGN derived from gram-negative bacteria, regulate the Imd pathway (Zaidman-Rémy et al. 2006; Lemaitre and Hoffmann 2007). More specifically, the full-length PGRP-LE acts as an intracellular receptor for monomeric PGN, whereas a version of PGRP-LE containing only the PGRP domain functions extracellularly to enhance PGRP-LC-mediated PGN recognition on the bacterial cell surface (Kaneko et al. 2006). Consequently, different types of antimicrobial peptides are produced to combat invading microbes.
In addition to activation of the pathways, recent studies indicate that at least some PGRPs (e.g., PGRP-LB, SC1B) have amidase activity and can hydrolyze PGN, the elicitor of the signaling pathways. Subsequently, the pathway associated with the degraded PGN is down-regulated (Mellroth et al. 2003; Zaidman-Rémy et al. 2006; Royet and Dziarski 2007).
It is also likely that PGRPs and GNBPs and associated pathways play a role in defense against pathogens other than bacteria or fungi. GNBP is significantly up-regulated in mosquitoes upon infection with malaria parasites (Dimopoulos et al. 1997) and is involved in melanotic encapsulation (Wang et al. 2005). More recent studies suggested that Anopheles Rel, a NF-kB transcription factor, plays an important role in limiting the development of Plasmodium in mosquitoes (Meister et al. 2005; Frolet et al. 2006). However, the definitive roles of PGRPs and GNBPs in other invertebrates have yet to be clarified.
Given the ample precedents set from other studies with respect to the importance of PGRPs and GNBPs in innate immunity, we felt it was important to explore the possible presence and roles of homologous molecules in B. glabrata, the snail host of S. mansoni. We also tested their role(s) in defense against pathogens, including S. mansoni, one of the causative agents of human schistosomiasis.
Materials and methods
Organisms used in the study
The M line and BS-90 strains of the snail B. glabrata and digenetic trematodes Echinostoma paraensei and S. mansoni were maintained in the laboratory as described by Loker and Hertel (1987). Both trematodes use the snail B. glabrata as an intermediate host for their life cycles. M line and BS-90 snails are susceptible and resistant to S. mansoni, respectively. Three microbes, the gram-positive bacterium Staphylococcus aureus (wild type), the gram-negative bacterium Escherichia coli (DH5-alpha strain), and the yeast Saccharomyces cerevisiae (S288C strain) were from the teaching collection of the Department of Biology, University of New Mexico, USA.
Extraction of genomic DNA and RNA
The M line snail used for genomic DNA extraction was a putatively homozygous snail produced from a parent M line snail after 20 generations of self-fertilization. The genomic DNA from this snail was extracted using the CTAB method (Winnepenninckx et al. 1993).
To test the expression of BgPGRP and BgGNBP genes in response to infection with E. paraensei or S. mansoni, RNA was extracted from the whole bodies of 12–15 juveniles of M line or BS-90 snails (6–9 mm in shell diameter) at 2 days post-exposure (dpe) to either trematode or from unexposed control snails using the NucleoSpin RNA II kit (Clontech). In the case of E. paraensei infection, only those snails that had been confirmed by microscopy to be infected were used for RNA extraction. For S. mansoni infection, all snails that were exposed to miracidia were used because it is difficult to identify the infection status at early stages of schistosome infection. The number of miracidia used to infect snails is 10–20 for S. mansoni and 20–30 for E. paraensei.
To test the role of microbial challenge on the expression of BgPGRPs and BgGNBP, 20 M line or BS-90 snails of comparable size (6–9 mm) were injected with 5 μl suspension of S. aureus, E. coli, S. cerevisiae, or phosphate-buffered saline (as control) according to our procedure described previously (Jiang et al. 2006). The approximate number of S. aureus, E. coli, and S. cerevisiae injected per snail was estimated to be 3–4×106, 2–9×106, and 2–5×104, respectively. RNA was extracted from snails at 6-h post-injection.
The concentration and purity of genomic DNA and RNA were determined spectrophotometrically.
Generation of full-length complementary DNAs
For construction of the cDNA library which was used for cloning full-length cDNAs, an RNA sample was derived from ten M line snails at 4 dpe to E. paraensei. GeneRacer (Invitrogen) was used to perform 5′ and 3′ RACE (rapid amplification of cDNA ends). Additionally, some sequences were confirmed from a Bluescript cDNA library which had been generated from M line snails that had been infected for 4 days with E. paraensei (Léonard et al. 2001) using gene specific primers and the vector primer M13 reverse or T7.
For sequencing, amplicons were cloned into the pCR 2.1-TOPO vector (Invitrogen). The profile for the sequencing reactions (BigDye, ABI) was: 96°C for 17 s, 50°C for 12 s, 60°C for 4 min, for a total of 29 cycles. The extension products resulting from cycle sequencing were analyzed on an ABI 377 sequencer (ABI).
Reverse-transcription polymerase chain reaction
The cDNAs were generated using random hexamers from total RNA samples [ThermoScript reverse-transcription polymerase chain reaction (RT-PCR) kit; Invitrogen]. PCR was performed on the cDNA using the profile of 94°C for 2 min, 94°C for 40 s, 50°C for 40 s, 72°C for 1 min, for 32 cycles in total.
Southern and Northern blot analyses
Three probes were PCR-amplified from the corresponding full-length cDNAs. The probe sizes for BgPGRP-LA, BgPGRP-SA, and BgGNBP were 435, 486, and 497 bp, respectively. Primer sequences and location of the probes are listed in Table 1.
Table 1.
Primers used for the study
| Primer | Sequence (5′-3′) | Approximate location and usage |
|---|---|---|
| PGRPf10 | AGACTAGACCCTAGTTTGCTCCC | Upstream of BgPGRP-LA; probe for BgPGRP-LA |
| PGRPallR1 | TCCCAGCCACGGCCCTCA | |
| PGRPf9 | ACCCTAGTTTGCTCCCTAATCTATAG | Upstream of BgPGRP-LA; RT-PCR reaction |
| PGRPr11 | CACTTAATCAACATCTTGACCGTGT | |
| pET23PGsF1 | GTGTCCTACGATTGTGACTAGAC | Almost entire coding region; probe for BgPGRP-SA |
| pET23PGsR1 | GTGTGGCCAGCCCTTGATAAG | |
| QgluF1 | TGACATCATGGAGTCCAGAGGTA | Downstream of BgGNBP; probe for BgGNBP |
| BgGluR1 | GTTCTGAACTCTACATAATCCAC |
For a given gene, the same probe was used for both Southern and Northern blotting. Southern and Northern blot analyses were conducted using modified versions of the SouthernMax™ and NorthernMax™ methods (Ambion), with details provided in our previous report (Zhang and Loker 2004). In the case of Northern blotting, an equal amount of RNA (25 μg) from different snail sources described above was loaded into each lane.
After separation by electrophoresis, DNA or RNA were transferred to a nylon membrane and hybridized to a [α-32P]dATP-labeled cDNA probe at 42°C for overnight. The membrane was washed using low stringency and high stringency wash buffers based on the manufacturer's instructions (Ambion). For re-probing, the membrane was washed twice with the probe degradation buffer provided by the manufacture to remove previous radioactive signals (Strip-EZ™, Ambion).
Sequence analyses
Nucleotide (nt) sequences were checked for accuracy using Chromas v 1.42 (http://www.technelysium.com.au/chromas). Sequence alignments and generation of contiguous sequences were performed using Sequencer (version 4.2; Gene Codes Corporation). BLAST searches were performed using the NCBI Blast program (http://www.ncbi.nlm.nih.gov/blast).
To know how PGRPs and GNBP characterized from B. glabrata relate to other homologues, we collected amino acid (aa) sequences of PGRPs, GNBPs, and bacterial glucanases available in GenBank. As PGRPs have been found to be present in both vertebrates and invertebrates, we obtained four human PGRPs to represent the vertebrate homologues for the analyses. For phylogenetic analyses, aa sequences of the conserved domains were used. The conserved domains, the PGRP domain for PGRPs, and the glycosyl hydrolases family 16 (glycol_hydro_16; or glucanase-like) domain for GNBPs were determined using the SMART program (http://smart.embl-heidelberg.de; Schultz et al. 1998) in which domains are determined based on the Pfam database of protein families (http://www.sanger.ac.uk/Software/Pfam/).
Prediction of the presence of a glycosylphosphatidylinositol (GPI)-anchored protein was performed using GPI modification site predication (http://mendel.imp.ac.at/gpi/gpi_server.html; Eisenhaber et al. 1999). The secondary structure prediction was performed using programs available at http://www.compbio.dundee.ac.uk/~www-jpred/ and http://www.predictprotein.org.
Alignment of homologous sequences for the phylogenetic analyses was performed in ClustalX 1.83 (Thompson et al. 1997). The alignment was entered into PAUP* 4.0b10 (Swofford 1998) for phylogenetic analyses. Phylogenetic relationships based on aa sequences were determined using maximum parsimony (MP) and minimum evolution under mean characteristic difference-distance. The bootstrap support values after 1,000 replicates were obtained based on the MP approach.
Results
The long form of PGRP gene (BgPGRP-LA) gene produces three isoforms, two via alternative splicing
Our initial effort to characterize the PGRP gene was based on an EST sequence in GenBank [GenBank accession number (GAN): CK988696; Mitta et al. 2005]. PCR amplification was performed using primers designed from the EST. PCR products were subsequently cloned. After sequencing of the clones, we have identified four cDNAs that contain the conserved PGRP domain. Further alignment comparison revealed that one cDNA (described in the following section) was quite different from the other three in terms of nt identity and structure of the gene product. The three cDNAs described in this section are very closely related; the homologous regions among these three cDNAs were almost identical. To confirm the sequences, we designed primers located in the 5′ and 3′ untranslated regions (UTRs) of the contiguous sequence. Using these primers, we performed PCR with cDNA as template, cloned the PCR products, and sequenced them. The resulting alignment suggested that the three cDNAs are transcribed from one gene and likely resulted from the process of alternative splicing (Figs. 1a and 2a).
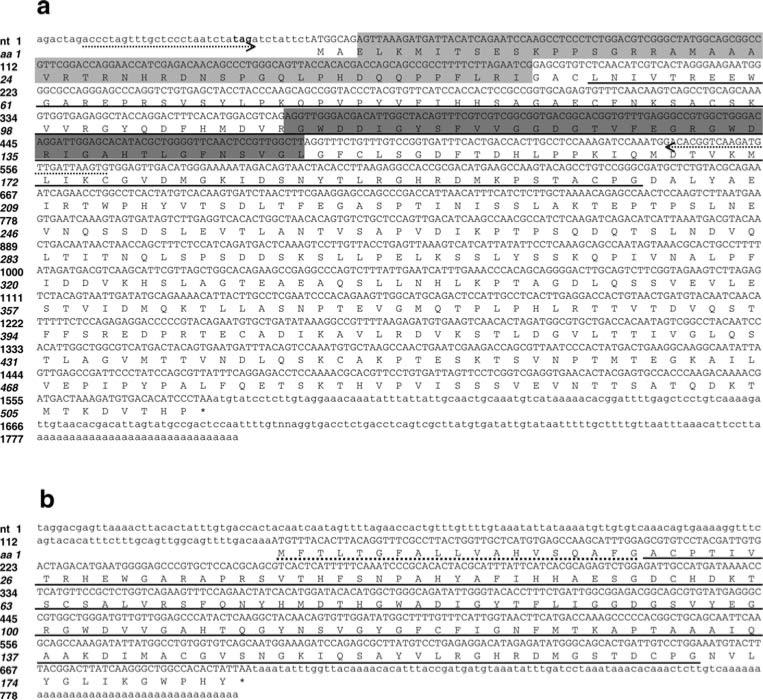
Fig. 1.
Nucleotide and deduced amino acid sequences of a BgPGRP-LA (GAN: EF452347) and b BgPGRP-SA (GAN: EF452346). As compared to BgPGRP-LA, the light shaded region is spliced out of BgPGRP-LA1 (GAN: EF452348). For BgPGRP-LA2 (GAN: EF452349), the sequences in both shaded boxes are spliced out. The primers used for RT-PCR to detect the three isoforms of BgPGRP-LA are indicated by dotted arrow lines. Three nucleotides (tag) which are bolded at the 5′UTR is a stop codon at the open reading frame. The signal peptide (SP) sequence and the putative PGRP domain are underlined by dotted and solid lines, respectively. Numbers in the left margin designate nucleotides (Roman) and amino acids (italicized)
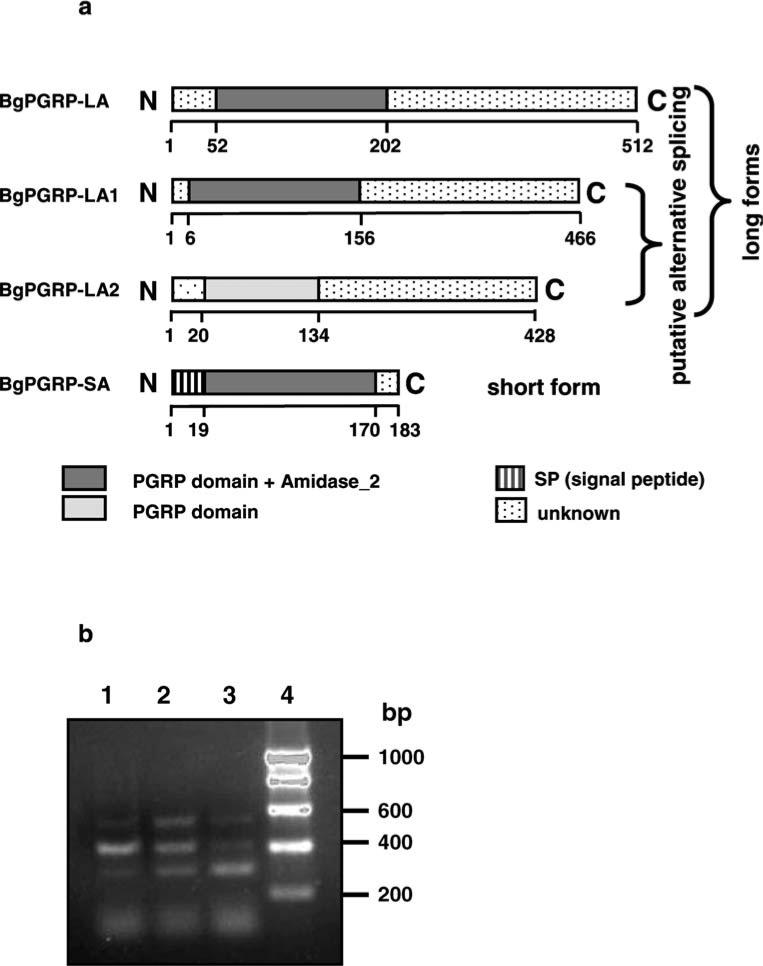
Fig. 2.
a Schematic representation of BgPGRP-LA, -LA1, -LA2, and -SA (not drawn to scale). The SP and the putative PGRP and amidase_2 domains were predicted by the SMART program. b The three alternatively spliced forms of BgPGRP-LA revealed by RT-PCR using cDNA as template. The location of the RT-PCR primers is indicated in Fig. 1a. RNA was extracted from M line snails at 2 dpe to S. mansoni (lane 1), 2 dpe to E. paraensei (lane 2) or from unexposed control snails (lane 3). Lane 4 is DNA size markers in base pairs (bp). Band intensity was found to be variable among replicates so does not simply reflect the magnitude of the response to each parasite
To further confirm the above information, we designed primers spanning the truncated regions to perform RT-PCR on the cDNA (see Fig. 1a for the location of primers). Again, we observed three PCR bands, and their size was consistent from what was expected given the cDNA results (Figs. 1a and 2b). This further verified that three transcripts of PGPR indeed are produced. We have not observed additional transcripts using three different methods [i.e., screening clones, RT-PCR, and Northern blots (see below)]; thus, it is likely that the long form is directly transcribed from a PGRP gene, and the two short ones are generated from the same gene via alternative splicing.
The three forms have an N-terminal PGRP domain and a long C-terminal domain with no homology to any known protein or domain in Pfam, the database of protein families. Based on the structure of PGRPs reported from various species, the three PGRPs identified from B. glabrata could be classified as “long form” PGRPs because long form PGRPs possess a PGRP domain and a long region with no homology to any known domain (Royet and Dziarski 2007). We therefore designated the three PGRPs, from longest to shortest, as BgPGRP-LA, BgPGRP-LA1, and BgPGRP-LA2 (Fig. 2a). They encode proteins that contain 512, 466, and 428 aa, respectively; they all lack a signal peptide, suggestive of an intracellular presence. Alternative splicing occurs within the PGRP region (Figs. 1a and 2a).
For the PGRP domain, the statistical E value from the SMART program for our sequences relative to others for BgPGRP-LA, -LA1 and -LA2 was 4.90e-64, 4.98e-64, and 2.10e-07, respectively. The location of the putative PGRP domain in the three proteins is provided in Fig. 2a. Moreover, NCBI protein blast also strongly suggested that they possess the conserved PGRP domain; the E values for the above three snail PGRPs (BgPGRP-LA, LA1 and -SA) are 2e-48, 1e-48, and 2e-23, respectively.
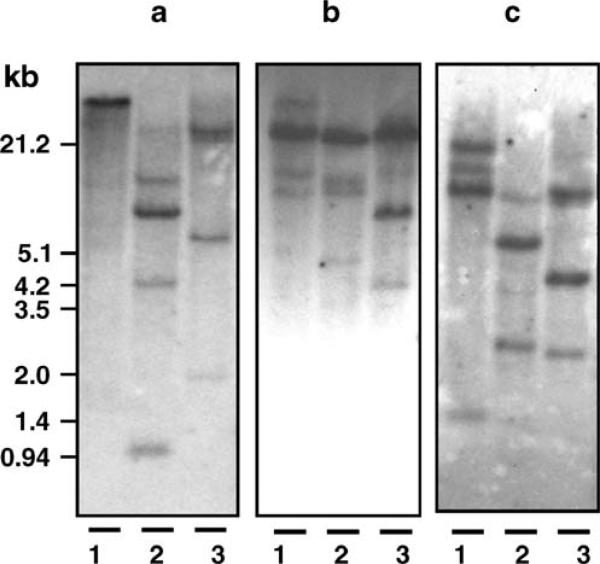
Southern analysis using cDNA located at the N-terminal region of BgPGRP-LA (435 bp) as a probe reveals two to four bands based on the use of three restriction enzymes (Fig. 3a). Because the source of DNA used in the Southern blotting was from a snail derived from 20 generations of self-fertilization, it is likely that most of its genes were homozygous. With that assumption (also applied to the Southern analyses described below), it is estimated that two to four BgPGRP-LA loci are present in the snail genome.
Fig. 3.
Southern blot analyses of a BgPGRP-LA, b BgPGRP-SA, and c BgGNBP. Three restriction enzymes, EcoRI (lane 1), HindIII (lane 2) and PstI (lane 3) were used to digest genomic DNA. Size markers in kilobases (kb) are indicated on the left
The short form PGRP gene (BgPGRP-SA) encodes a secreted protein
During our screening of clones, we also found a short form of PGRP, which encodes a putative 183 aa protein (Fig. 1b). It is different from the above BgPGRP-LAs in that the putative protein consists solely of a PGRP domain (from 22 to 164 aa) predicted by both SMART (3.89e-66) and NCBI blast program (2e-36), and a 19 aa signal peptide, but does not possess the long C-terminal region found in the BgPGRP-LAs. We therefore designated it as BgPGRP-SA (Figs. 1b and 2a). The aa identity of the PRGP domain between BgPGRP-SA and BgPGRP-LA was ~55%.
Southern analysis using the entire coding region of BgPGRP-SA (486 bp) as a probe revealed three to four bands based on use of three restriction enzymes (Fig. 3b). It was estimated that three to four BgPGRP-SA loci are present in the snail genome. The nt variation between BgPGRP-SA and -LA genes is extensive; the nt sequences of the PGRP domain between the two genes could not be aligned by either the NCBI Blast 2 sequences program or Sequencer. It is therefore unlikely that the probe for PGRP-SA would cross-hybridize to the BgPGRP-LA gene and vice versa.
The BgGNBP gene produces a secreted protein
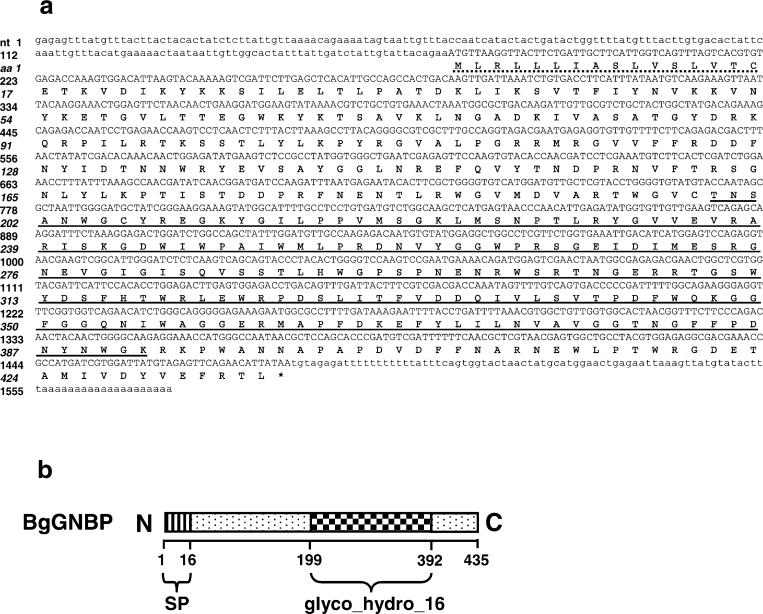
Using the same approaches as described above, we have cloned a GNBP gene from B. glabrata using, as a starting point, an EST sequence (GAN: CK988667; Mitta et al. 2005). We have designated it as BgGNBP. The BgGNBP gene encodes a putative protein comprised of 435 aa residues, with a 16 aa signal peptide (Fig. 4a,b). A 183 aa region at the C terminus of BgGNBP shows a high degree of identity to glycosyl hydrolases of the family 16 domain (glyco_hydro_16 or glucanase-like domain; from 199 to 392 aa) with an E value of 1.1e-08. This conserved domain is the landmark for GNBPs (Hoffmann 2003). The NCBI protein blast search also strongly suggested that BgGNBP is a β-1,3-glucan recognition protein with a high support (2e-91) and possesses a conserved β-glucanase-like domain (4e-12). A computational analysis suggested that the BgGNBP is unlikely to be a GPT-anchored protein because no potential GPI-modification site was found.
Fig. 4.
a Nucleotide and deduced amino acid sequences of BgGNBP (GAN: EF452345). The SP sequence is indicated by a dotted line, and the glucanase-like domain (Pfam: glyc_hydro_16) is underlined. b Schematic diagram of BgGNBP (not drawn to scale)
Southern analysis using cDNA located at the C-terminal region of BgGNBP (497 bp) as a probe indicated that three to four bands are present in the genome, suggesting that about three to four loci are present in the snail genome (Fig. 3c).
Canonical residues essential for enzymatic activity are well conserved in three of the four BgPGRPs and the BgGNBP
The computational analysis of BgPGRPs using the SMART and NCBI programs suggested that BgPGRP-LA, -LA1, and -SA possess both PGN recognition function and amidase activity. Based on the same approaches, BgPGRP-LA2 should lack such enzymatic activity, but have PGN recognition capability (Fig. 2a).
We have further analyzed the B. glabrata sequences for canonical residues known to be critical for amidase activity in Drosophila (Mellroth et al. 2003) and bacteriophage T7 isozyme, a zinc-dependent amidase (N-acetylmuramoyl-l-alanine amidase; Cheng et al. 1994). Figure 5a shows the alignment of the four BgPGRPs, DmPGRPs, and T7 isozyme, which also shows the basic structure of PGRPs (Kim et al. 2003; Reiser et al. 2004; Lim et al. 2006). Whereas five residues [i.e., His (H), Val (Y), His (H), Lys (K), and Cys (C)] are required for the catalytic activity of T7 isozyme, it has been shown that in the Drosophila catalytic PGRPs (e.g., DmPGRP-SC1B), four of these zinc-coordinating residues are retained, but one (italicized) is changed, as follows: H, Y, H, T, C. The fourth amino acid residue (K) in T7 isozyme is replaced by the residue T in Drosophila (Mellroth et al. 2003; Royet and Dziarski 2007). The four residues (H, Y, H, and C) required for amidase activity of the catalytic DmPGRPs have been preserved in BgPGRP-LA, -LA1, and -SA, suggesting that the three BgPGRPs would have a PGN-hydrolyzing amidase activity. However, BgPGRP-LA2 has lost the second conserved residue (Y) due to alternative splicing, suggesting that it would lack catalytic activity (Fig. 5a). These findings are consistent with the prediction made by the SMART and NCBI blast programs.
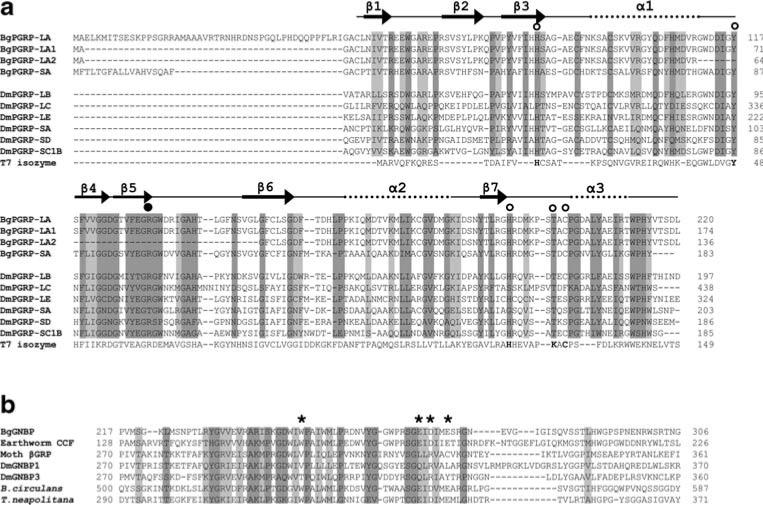
Fig. 5.
a Structure-based sequence alignment of PGRPs guided by the crystal structures of DmPGRP-LB, -SA, and -LE (Kim et al. 2003; Reiser et al. 2004; Lim et al. 2006). The open circles show the five conserved amino acids, and the filled circle indicates the Arg residue critical for the discrimination of DAP-type PGN. The dark and light shaded boxes show regions of sequence identity and of conserved replacements (BLOSUM62 matrix) of greater than 80%, respectively, in the sequences aligned. The five conserved residues critical for enzymatic activity in T7 isozyme are in bold. The numbers on the right side indicate the position from the start codon in each aa sequence. Dm Drosophila melanogaster. The GenBank accession numbers: DmPGRP-LB (Q9VGN3); DmPGRP-LC (Q9GNK5); DmPGRP-LE (Q9VXN9); DmPGRP-SA (Q9VYX7); DmPGRP-SD (Q9VS97); DmPGRP-SC1B (Q95SQ9); bacteriophage T7 isozyme (P00806). b Alignment of the glucanase-like region of the representative GNBPs. The dark and light shaded boxes show regions of sequence identity and of conserved replacements (BLOSUM62 matrix) of greater than 85% (6/7), respectively. Asterisks indicate the residues required for glucanase activity, as assessed from bacterial studies. The numbers on the both sides indicate the position of amino acids related to the start codon of the aa sequence. Bacillius circulans β-1,3-glucanase A1 (P23903), Thermotoga neapolitana β-glucanase (Z47974), Moth (M. sexta) βGRP (AAF44011), DmGNBP1 (AAF33849), DmGNBP3 (AAF33851)
Furthermore, we examined our sequences to determine the presence or absence of an Arg (R) in the BgPGRPs, as this residue plays a significant role in the affinity of PGRPs to DAP-type PGN (Lim et al. 2006; Chang et al. 2006). Our sequence alignment showed that three of the snail PGRPs (BgGRP-LA, -LA1, and -SA) possess the R at the corresponding position, whereas BgPGRP-LA2 does not.
As the crystal structures for some DmPGRPs are known (Kim et al. 2003; Reiser et al. 2004; Lim et al. 2006), we have aligned our BgPGRPs to those DmPGRPs (Fig. 5a). We have also indicated the known positions of the seven β strands and three α helices for the Drosophila sequences on our alignment. We note for BgPGRP-LA, -LA1, and -SA that many of the residues in six of the seven β strands and in all three of the α helices are relatively conserved with those of Drosophila. Interestingly, the alignment shows that the snail sequences located in the region comparable to the β2 region are at variance with the Drosophila sequences; identity and similarity across all residues is less than 80% (Fig. 5a). The secondary structure predictions obtained using the two independent computational programs described in “Materials and methods” are consistent with our above observations in predicting for the snail sequences that six of the seven β strands and the α three helices are indeed located in the appropriate positions and that the β2 strand region is missing. Because of alternative splicing, BgPGRP-LA2 also lacks the regions homologous to the β4, β5, and β6 strands of DmPGRPs (Fig. 5a). These lines of evidence suggest that BgPGRP-LA2 is quite different from the other three BgPGRPs in terms of structure and function. Future crystal analysis is needed to confirm above information.
With respect to BgGNBP, we have compared it to the glucanase-like domains of GNBPs that are currently known (also see below “Evolutionary and functional implications for PGRPs and GNBPs as revealed by phylogenetic analysis and presence/absence of the conserved residues”). The glucanase-like domain of all GNBP proteins is very similar to the catalytic domain of bacterial β-1,3-glucanase. Previous studies revealed that four conserved residues [Trp (W), Glu (E), Asp (D), and E] located in this domain are required for enzymatic activity in bacteria (Yahata et al. 1990). Our sequence alignment revealed that the BgGNBP possesses all four residues (Fig. 5b).
Down-regulation of BgPGRP-SA and BgGNBP in M line and BS-90 snails after injection with gram-positive or gram-negative bacteria or yeast
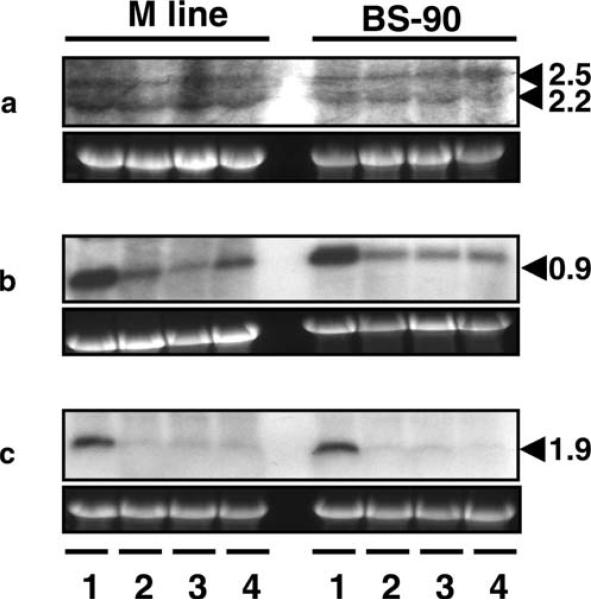
To gain insight into the role in internal defense of the three genes (BgPGRP-LA, BgPGRP-SA, and BgGNBP) identified in this study, we tested whether injection of microbes triggers changes in gene expression. We injected M line and BS-90 snails with the gram-positive bacterium S. aureus, the gram-negative bacterium E. coli, or the yeast S. cerevisiae. It was shown by Northern blot analysis that none of the three microbes altered the gene expression pattern of BgPGRP-LAs in either snail strain (Fig. 6a). Two weak transcripts of BgPGRP-LAs were observed on the blot. Based on the size of the mRNAs, it is likely that the two mRNAs detected were BgPGRP-LA and -LA1. BgPGRP-LA2 expression was too low to be detected. The sizes of the bands recovered on Northern blots were bigger than those inferred from examination of cDNAs, suggesting that the natural 5′UTR of the BgPGRP-LAs may be longer than those on the cDNA we obtained. This may be due to any of the following factors: truncation of the 5′UTR or PolyA on our cDNAs, or, to some extent, the accuracy of the RNA marker.
Fig. 6.
Northern blots showing expression of a BgPGRP-LAs, b BgPGRP-SA, and c BgGNBP in M line or BS-90 snails (6–9 mm). RNA was extracted from snails that were injected with PBS (lane 1), S. aureus (lane 2), E. coli (lane 3), or S. cerevisiae (lane 4; for details, see “Materials and methods”). Lower panels show rRNA stained with GelRed used as loading controls. Size markers in kilobases (kb) are indicated on the right (same as Fig. 7)
However, expression of BgPGRP-SA and BgGNBP, both of which encode extracellular proteins, was down-regulated in both M line and BS-90 snails upon microbe injection (Fig. 6b,c). In addition, Northern blots suggested that all three genes are constitutively expressed, although the expression of the BgPGRP-LAs was consistently the lowest in all of the conditions assayed. According to our observations with Northern blotting and RT-PCR, BgPGRP-SA and BgGNBP are constitutively expressed at a relatively high level, and the relative expression level of BgPGRP-SA was higher than that of BgGNBP in sham-injected M line and BS-90 snails.
Responses of BgPGRPs and BgGNBP in M line and BS-90 snails after exposure to E. paraensei or S. mansoni
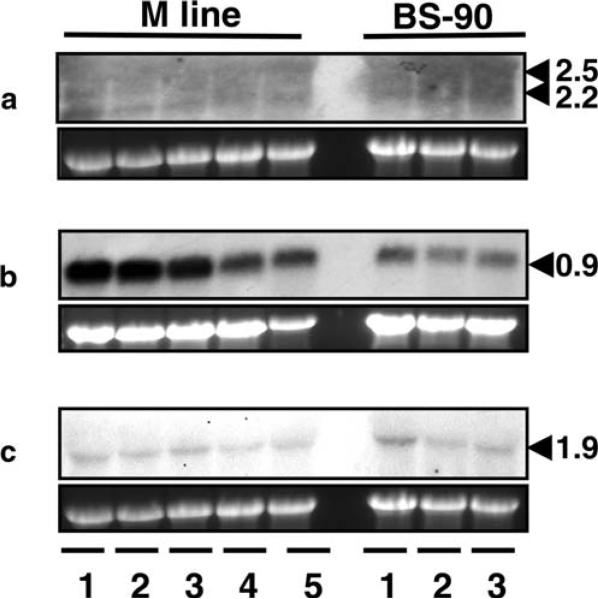
We further examined whether expression of the three genes could be activated by exposure to the trematodes E. paraensei or S. mansoni: both species use B. glabrata as an intermediate host. M line or BS-90 snails were exposed to E. paraensei or S. mansoni and RNA was obtained from the snails at 2 dpe. Northern blot analysis revealed no changes in expression of BgPGRP-LA genes in either strain of snails regardless of the parasite used for infection (Fig. 7a). However, the expression level of BgPGRP-SA in both snail strains at 2 dpe to E. paraensei was slightly reduced compared to the unexposed snails. Interestingly, BgPGRP-SA expression returned to the basal level or was slightly up-regulated by 12 and 17 dpe to E. paraensei (Fig. 7b). BgGNBP expression did not change in the two snail strains exposed to either parasite (Fig. 7c).
Fig. 7.
Northern blots showing the expression of a BgPGRP-LAs, b BgPGRP-SA, and c BgGNBP in M line and BS-90 snails (6–9 mm) exposed to E. paraensei or S. mansoni. For M line snails, lanes 1 and 2 show mRNA from snails at 12 and 17 dpe to E. paraensei. Lanes 3 to 5 show the expression level of three genes in snails that were not exposed (lane 3) or were exposed for 2 days to E. paraensei (lane 4) or to S. mansoni (lane 5). For BS-90 snails, lanes 1 to 3 show mRNA abundance of the three genes from snails that were not exposed (lane 1) or that were exposed for 2 days to E. paraensei (lane 2) or to S. mansoni (lane 3). Please note that the BgGNBP band is faint because the membrane had been stripped three times. Other Northern blots revealed that if the membrane was first probed by BgGNBP, it showed high expression level in all RNA samples, and differential expression was not observed (data not shown). In the case of BgPGRP-SA, the bands were intense, even though the membrane may have been previously stripped (the one shown here was stripped three times). For BgPGRP-LAs, the bands were always faint even on new membranes. Among the three genes, expression from lowest to highest was BgPGRP-LAs, BgGNBP, and BgPGRP-SA
Evolutionary and functional implications for PGRPs and GNBPs as revealed by phylogenetic analysis and presence/absence of the conserved residues
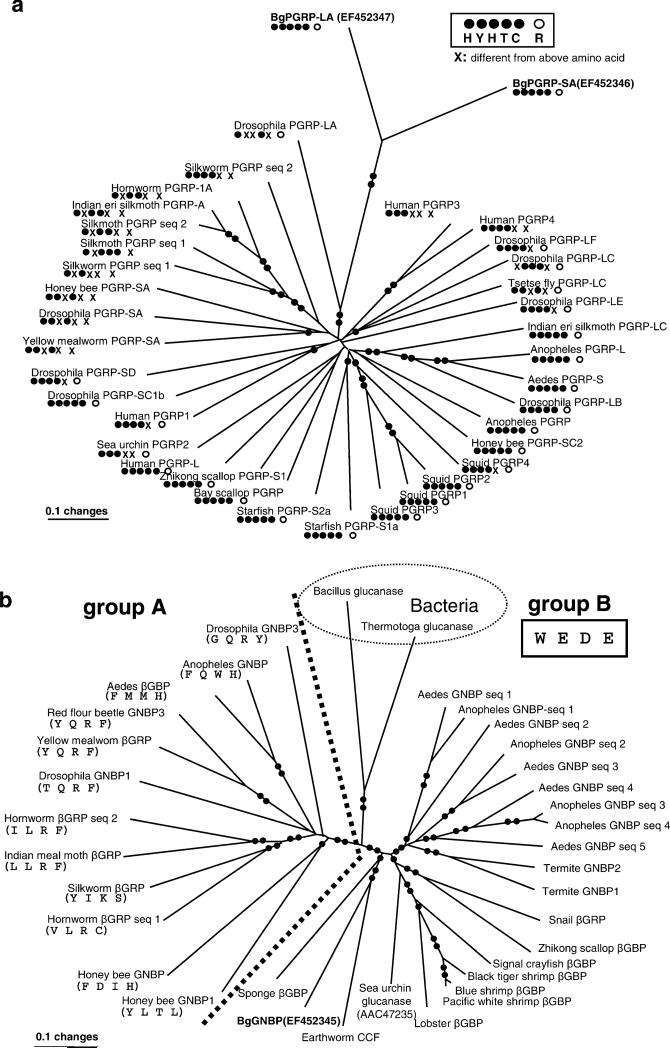
All PGRPs, both long and short forms, contain a conserved PGRP domain. As the conserved PGRP domain might appear in either N- or C-terminal regions, it is difficult to make a proper alignment based on the entire sequence. So, we compared just the conserved PGRP domains (~160 aa after alignment), which were identified and delineated by the SMART program. The phylogenetic analysis revealed that BgPGRP-LA and -SA clustered with a high bootstrap value (100%) despite their obvious differences (only ~55% aa identity). They were most closely related to DmPGRP-LA (GAN: NP_996029; a total of 299 aa), a Drosophila membrane protein whose function has not, thus far, been reported. BgPGRPs did not group with PGRPs identified from other molluscan species (i.e., squid and scallop; Fig. 8a).
Fig. 8.
A gene tree using minimum evolution (ME) showing relationships among a PGRPs or b GNBPs based on amino acid sequences. For each sequence, a protein name and a common name of species are provided. Bootstrap values less than 50% are not shown. Single- or double-filled circles indicated on the branches indicate bootstrap support of 50–80% and 81–100%, respectively. For a depicting relationships among PGRP sequences, the presence/absence of each of the five conserved residues associated with amidase activity is provided under the protein's name, as is the presence/absence of the Arg residue responsible for discrimination of the DAP-type PGN (see Fig. 5a, b for the positions of the residues). In b showing relationships among GNBPs, the delineation between groups A and B is shown with a dotted line (100% bootstrap value). In group A, the presence/absence of the four conserved residues associated with glucanase activity is provided under each protein's name. In group B, the four residues are the same (W, E, D, E) for all sequences. The GenBank accession numbers for all sequences in a are as follows (starting from Human PGRP3, clockwise direction). Human PGRP3 (AAI28115), human PGRP4 (AAI07158), Drosophila melanogaster PGRP-LF (NP_648299), Drosophila PGRP-LC (NP_996030), tsetse fly Glossina morsitans PGRP-LC (ABC25065), Drosophila PGRP-LE (NP_573078), Indian eri silkmoth Samia cynthia PFRP-LC (BAF03521), mosquito Anopheles gambiae PGRP-L (XP_321943), mosquito Aedes aegypti PGRP-S (ABF18154). Drosophila PGRP-LB (NP_731576), Anopheles PGRP (Q7PP76), honey bee Apis mellifera PGRP-SC2 (XP_395941), squid Euprymna scolopes PGRP4 (AAY27976), squid PGRP2 (AAY27974), squid PGRP1 (AAY27973), squid PGRP3 (AAY27975), starfish Asterias rubens PGRP-S1a (DQ222477), starfish PGRP-S2a (DQ222478), Bay scallop Argopecten irradians PGRP (AY437875), Zhikong scallop Chlamys farreri PGRP-S1 (AAY53765), human PGRP-L (NP_443122), sea urchin Strongylocentrotus purpuratus PGRP-2 (XP_796422), human PGRP1 (NP_005082), Drosophila PGRP-SC1B(AAG23736), Drosophila PGRP-SD (NP_648145), yellow mealworm Tenebrio molitor PGRP-SA (BAE78510 ), Drosophila PGRP-SA (NP_572727), honey bee PGRP-SA (XP_001123180), silkworm Bombyx mori PGRP seq 1 (BAA77209), silkmoth Antheraea mylitta PGRP seq 1 (ABG72708), silkmoth PGRP seq 2 (ABG72709), Indian eri silkmoth PGRP-A (BAF03522), hornworm Manduca sexta PGRP-1A (AAO21509), silkworm PGRP seq 2 (AAL32058), Drosophila PGRP-LA (NP_996029). GenBank accession numbers in b are as follows (starting from mosquito Aedes GNBP seq 1, clockwise direction). Aedes aegypti GNBP seq 1 (EAT44801), Anopheles gambiae GNBP seq 1 (Q7QA32), Aedes GNBP seq 2 (EAT41280), Anopheles GNBP seq 2 (Q7PQA4), Aedes GNBP seq 3 (EAT44802), Aedes GNBP seq 4 (EAT 38985), Anopheles GNBP seq 3 (Q7QCT6), Anopheles GNBP seq 4 (Q7QCT7), Aedes GNBP seq 5 (EAT38986), Termite Nasutitermes fumigatus GNBP2 (AAZ08496), Termite GNBP1 (AAZ08483), Snail B. glabrata βGBP (EF121824), Zhikong scallop βGBP (AAP82240), signal crayfish Pacifastacus leniusculus βGBP (CAB65353), black tiger shrimp Penaeus mondon βGBP (AAM21213), blue shrimp Litopenaeus stylirostris βGBP (AAM73871), Pacific white shrimp Litoppenaeus vannamei βGBP (AAW51361), lobster Homarus gammarus βGBP (CAE47485), sea urchin glucanase (AAC47253), earthworm Eisenia fetida CCF (AAC35887), sponge Suberites domuncula βGBP (CAE54585), honey bee GNBP 1 (XP_001121634), honey bee GNBP (XP_395368), hornworm βGRP seq 1 (AAF44011), silkworm βGBP (BAA92243), Indian meal moth Plodia interpunctella βGRP (AAM95970), hornworm βGBP seq 2 (AN10151), Drosophila GNBP1 (AAF33849), yellow mealworm βGRP (BAC99308), red flour beetle Tribolium castaneum GNBP (XP_972063), Aedes βGBP (EAT40654), Anopheles GNBP (Q7QIB2), Drosophila GNBP3 (AF33851), Bacillus circulans glucanase (Z47974), Thermotoga neapolitana glucanses (P23903)
Additionally, we have checked the five residues deemed to be critical for amidase activity in the 37 PGRPs used for the phylogenetic analysis (see Fig. 5a for the position of the residues). The first residue (H) is most conserved, with only 1 of 37 sequences deviating at this residue. The second residue (Y) was also relatively invariant (6 of 37 varied at this position), as was the third residue (H), with 5 of 37 variant residues. The fourth residue (T) is in 35 of 37 sequences and is never a K as it is in the T7 bacteriophage sequence. The last residue associated with amidase activity is a C, and this was less conserved, being present in 19 of 37 sequences, in most cases (17 of 37) being replaced by a ser (S). The amino acid substitutions we noted with respect to the phylogenetic position of the organism from which the sequence was obtained did not reveal a clear pattern in the PGRPs surveyed (Fig. 8a).
The overall topology of the GNBP tree based on the conserved glucanase-like domain (~300 aa after alignment) of 36 GNBP sequences revealed two distinct groups with 100% bootstrap support, designated as groups A and B. Group A comprises homologues derived from arthropods only. Group B contains GNBP homologues from deuterostomes, protostomes, sponges, and bacteria. BgGNBP was most similar to the coelomic cytolytic factor (CCF) of the earthworm Eisenia fetida, followed by the sponge homologue. CCF is a glucan- and lipopolysaccharide (LPS)-binding protein that triggers the prophenoloxidase-activating pathway (Beschin et al. 1998). The amino acid identities of the polysaccharide-binding motif and the glucanase-like motif, which together form the first carbohydrate recognition domain in CCF, are 50 and 70% identical to the corresponding motifs of BgGNBP. An additional domain located in the C-terminal region of CCF, which can recognize β-1,4-N-acetylucosamine-linked saccharides and muramic acid, has 41% identity to the corresponding region of BgGNBP sequence (Bilej et al. 2001). The two additional GNBP homologues from molluscs that are available in GenBank, one from B. glabrata (GAN: EF121824) and the other from scallop (GAN: AAP82240), grouped together with 88% support, but this group did not cluster with BgGNBP. Our computational analysis revealed that unlike BgGNBP which is postulated to be secreted, the latter two molluscan homologues are intracellular proteins, suggesting that the function of BgGNBP may be different from the two intracellular homologs (Fig. 8b).
Notably, we found that the glucanase-like domain from members of group B possesses all four conserved residues (also see Fig. 5b for the position of the residues) shown to be essential for bacterial glucanase activity. Two glucanase domains from bacteria clustered with the group B glucanase-like domains with 92% support. A homolog from the sponge Suberites domuncula, a member of the most basal animal phylum, also belongs to group B, which is comprised of 24 of the 36 known GNBPs. In each of the 12 members of group A, all four conserved residues in the glucanase-like domain have been changed (Fig. 8b), thus, making it easy to classify individual GNBP sequences into group A or B. The biological significance of this intriguing pattern is unknown.
Discussion
Innate immune defenses against pathogens are initiated by PRRs that bind conserved moieties present in a wide spectrum of microorganisms but absent in the host. One example of such a pathogen structure is PGN, the major constituent of the cell wall of both gram-positive and gram-negative bacteria, which is bound by PGRPs. Although there is no evidence in Drosophila suggesting GNBPs have bacteriocidal enzymatic activity, they are able to recognize and bind to LPS and β-1,3-glucan, the components of the walls of gram-negative bacteria and fungi, respectively (Kim et al. 2000), and this may lead subsequently to activation of immune signaling pathway. PGRPs and GNBP along with other proteins (for example LPS-binding protein) have been confirmed to be PRRs in invertebrates (Kanost et al. 2004; Lemaitre and Hoffmann 2007). In B. glabrata, no PRRs in these categories have been characterized in terms of structure or function. In this study, we report the characterization of four BgPGRPs (BgPGRP-LA, -LA1, -LA2, and -SA) and one BgGNBP from this medically important species and explore their responses to microbes and metazoan parasites. To our knowledge, an N-terminal location of a PGRP domain, as in the BgPGRP-LAs, is here reported for the first time.
PGRPs are grouped into long and short forms. Both forms are present in mammals and insects (Kurata 2004). The three long forms of BgPGRPs do not have a signal peptides, thus, are presumed to be intracellular in their action. We cannot rule out the possibility that they may be secreted despite their absence of signal peptides. For example, Drosophila PGRP-LE has no signal peptide yet in addition to a cytoplasmic form; for reasons not yet clear, it has also been found to be present in a secreted form (Werner et al. 2000; Lim et al. 2006; Royet and Dziarski 2007). Interestingly, a recent study demonstrated that DmPGRP-LE can function by two distinct mechanisms. The extracellular DmPGRP-LE (a truncated form containing only the PGRP domain) binds to the monomeric DAP-type PGN tracheal cytotoxin (TCT) and carries it to the cell surface where the complex interacts with DmPGRP-LC to induce signal transduction. However, the intracellular full-length version of DmPGRP-LE functions independently of DmPGRP-LC. It acts as an intracellular TCT receptor and activates the Imd pathway (Kaneko et al. 2006). Like DmPGRP-LE, BgPGRP-LA encodes three isoforms including two truncated forms, none of which have a signal peptide. Based on this information, it is likely that the location and function of the individual BgPGRP-LA may be different. Future studies using specific antibodies raised against recombinant forms of BgPGRP-LAs will be helpful in determining their extra- or intracellular localization and their possible functions in snails.
In our study, based on the presence of canonical residues and our computational analyses, the two long forms of PGRP (BgPGRP-LA and -LA1) and the short form PGRP-SA (BgPGRP-SA) are predicted to have both PGN recognition capability and amidase activity, like bacterial amidases that can degrade PGN. Some PGRP family members in Drosophila such as PGRP-SC1B and -LB have conserved T7 lysozyme-like catalytic activity and are able to cleave PGN (Mellroth et al. 2003; Zaidman-Rémy et al. 2006); other PGRPs, however, have lost enzymatic activity, yet still serve as microbe sensors via their ability to recognize PGN (Royet and Dziarski 2007).
Recent studies with Drosophila suggest that recognition of DAP-type PGN depends on the presence of an Arg residue in the PGN-binding groove (Chang et al. 2006; Lim et al. 2006). Our sequence analysis shows that the Arg residue is conserved in BgPGRP-LA, -LA1, and -SA, raising the possibility that BgPGRPs can also recognize DAP-type PGN. However, it should be kept in mind that other studies have shown that some PGRPs, such as DmPGRP-SD, that retain the conserved Arg residue (see Fig. 5a) nonetheless respond instead to gram-positive bacteria and activate the Toll pathway (Bischoff et al. 2004), whereas other PGRPs have different amino acids in the corresponding position, and their specificity cannot be predicted at present (Royet and Dziarski 2007).
The BgPGRPs we found are quite divergent from those reported from other species including their possession of a unique N-terminal PGRP domain and absence of a putative β2 strand. However, the SMART and NCBI blast programs identified a PGRP domain in our sequences with strong support and canonical residues consistent with the enzymatic activity were subsequently identified to be present. Additionally, the reality of our sequences is supported by the report from another laboratory of identical aa sequence from two different B. glabrata sources, BS-90 snails (GAN: EF079963) and B. glabrata embryonic cells (Bge cells; GAN: EF079962). The deduce 466 aa sequence was identical to our BgPGRP-LA1, an alternatively spliced form of BgPGRP-LA, from the M line strain of B. glabrata. This also suggests the considerable conservation of this BgPGRP protein in the B. glabrata. Moreover, a stop codon has been found in 5′UTR, but none has been found in the open reading frame. Together, it seems unlikely that our BgPGRPs are derived from pseudogenes. The divergence between the snail PGRPs and PGRPs of other animals may imply the alternative function of the snail PGRPs.
So far, we have identified only one transcript of BgPGRP-SA, although multiple loci in the genome were suggested by Southern blot analysis. Northern blots indicated a high level of expression for BgPGRP-SA, and given this form is probably secreted (all short forms known from other animals are secreted), snail hemolymph likely contains BgPGRP-SA protein. A constitutive presence of this protein in the circulating blood would facilitate a ready response to invading microbes.
In Drosophila, at least 13 PGRPs with 17 isoforms have been found (Royet and Dziarski 2007). In our Southern blots, only one of the visible bands was intense, the others being weakly detected. This phenomenon was also observed in the Southern blot probed by BgGNBP. Our explanation is that some BgPGRPs present in the genome could not be uncovered by our PCR-based cloning approaches because the nt identity among these genes (e.g., BgPGRP-LA or BgPGRP-SA) was too low for efficient hybridization and PCR amplification. Similar considerations likely apply for BgGNBP. For example, two known B. glabrata GNBP nucleotide sequences with only three nt differences between them (GANs: EF121824 and EF121825) are quite different from the BgGNBP reported here. The deduced amino acid identity between BgGNBP and those homologues is 42%. Therefore, it can be predicted that for both BgPGRPs and BgGNBP, there are more loci present in the genome, but that the level of sequence identity among the members of each family is low.
Our expression studies revealed that the BgPGRP-LAs were expressed at very low levels in both infected and non-infected snails. BgPGRP-SA was down-regulated after infection with gram-positive and gram-negative bacteria and yeast in both M line and BS-90 snails, but did not change expression levels in BS-90 snails after infection with either E. paraensei or S. mansoni. However, in M line snails, the expression of BgPGRP-SA was initially slightly down-regulated, returning to normal levels (with slight up-regulation) at later times during the course of trematode infection (12 and 17 dpe). This response pattern is intriguing. One hypothesis is that trematodes in their early stages of development may be harmed by high levels of BgPGRP-SA, and they may consequently manipulate the host to produce lower levels of BgPGRP-SA.
The responsiveness of PGRPs after exposure to various microorganisms also differs considerably between various host species. In Drosophila, expression of most PGRP short forms was up-regulated in response to bacterial infection, whereas PGRP long forms, except for PGRP-LB, encode constitutive proteins (Werner et al. 2000). It has recently been shown that the different members of the PGRP family recognize different types of PGN on bacterial cell walls (Lemaitre and Hoffmann 2007). In addition to humoral immune responses such as activation of Toll/Imd pathways, PGRPs also play a role in cellular immune responses. For example, preliminary evidence suggested that DmPGRP-LC is involved in phagocytosis of bacteria in Drosophila (Ramet et al. 2002). Silkworm PGRP short form and Drosophila PGRP-LE initiated activation of the prophenoloxidase cascade, which generates antimicrobial melanin and reactive oxygen species (Yoshida et al. 1996; Takehana et al. 2002). Not surprisingly, the data suggest that different members of the PGRP family function in different ways within a species and that PGRP functions will vary from one species to another. Similar considerations may apply to BgPGRPs.
With respect to GNBPs, numerous studies indicate that they play a role in defense against pathogens in invertebrates. A significant down-regulation of GNBPs (GNBPs 1, 2 and 3) in Drosophila was observed at the early phase (3–6 h) of E. coli infection (Kim et al. 2000). In the case of the mosquito Armigeres subalbatus, down-regulation was observed at 3- and 6-h post-infection, but up-regulation appeared at 24- to 48-h post-infection (Wang et al. 2005). A GNBP in the mosquito A. gambiae (GAN: AJ001042) was up-regulated after exposure to E. coli, Micrococcus luteus, and the malaria parasite Plasmodium berghei, but not after exposure to S. cerevisiae (Dimopoulos et al. 1997). In addition, a βGRP from the moth Manduca sexta could agglutinate microorganisms and activate the phenoloxidase cascade (Ma and Kanost 2000). In shrimp, it has been shown that GNBP was not induced by injection of β-1,3-glucan, curdlan, and heat-killed Vibrio harveyi in Penaeus monodon (Sritunyalucksana et al. 2002), but up-regulated in P. stylirostris after infection with the white spot virus (Roux et al. 2002). Clearly, there are pronounced differences among host species in their GNBP responses to pathogens.
GNBPs are secreted proteins with a C-terminal β-glucan binding domain similar to β-1,3- and β-1,4-bacterial glucanases (Yahata et al. 1990) and an N-terminal domain reported to bind to β-1,3-glucan (Ochiai and Ashida 1999; Hoffmann 2003). Our analyses revealed that all conserved residues required for bacterial glucanase activity are present in BgGNBP. The two additional molluscan homologues, one from B. glabrata and the other from scallops, also possess the four conserved residues, but are predicted to be intracellular proteins by our computational analysis. This is uncommon for GNBP proteins. For example, all Drosophila GNBPs possess signal peptides and predicted to be secreted proteins (or membrane-bound proteins because of the GPI-anchor). Functional differences between the putative intracellular and extracellular GNBPs are unknown. A computational analysis suggested that BgGNBP has no GPI-anchored sites, implying that it is unlikely to be membrane-bound. However, caution is required, as the same computational program performed for DmGNBP1 did not reveal any obvious GPI-anchored sites, yet biochemical evidence revealed that it exists in both soluble and membrane-bound GPI-anchored forms in Drosophila cells (Kim et al. 2000). Therefore, we cannot rule out the possibility of a GPI-anchored form of BgGNBP being present in B. glabrata cells.
For our phylogenetic analyses, both PGRPs and GNBPs are gene families, so it must be kept in mind that our analysis will include both homologs and paralogs that make it difficult to relate the evolutionary histories of the genes to the evolutionary histories of the organisms bearing the genes. In our PGRP tree, the two examples from B. glabrata (BgPGRP-LA and -SA) identified in this study were most similar to a gene from Drosophila rather than to the few known PGRPs from other molluscan species. Moreover, the two B. glabrata homologues, although clearly PGRPs-based on our sequence analysis, are divergent from those of other animals.
In the case of GNBPs, the two homologues from B. glabrata included in the analysis are distant from one another. The BgGNBP recovered in this study was most similar to an earthworm (phylum Annelida) sequence. Our analysis suggests that the known GNBPs can be divided into two groups, one having all the critical residues for glucanase activity conserved (group B), and the other, comprised thus far only of insect sequences, having replaced or lost all those residues (group A). All three Drosophila GNBPs have lost the conserved residues, so it is understandable that they do not have catalytic activity. It is intriguing that group B GNBPs come from a broad range of animal species, from sponges to deuterosomes and protostomes. However, the situation may be more complex than implied by this dichotomy, as a few studies of group B GNBPs have shown that enzymatic activity has been lost, as for example with earthworm CCF (Beschin et al. 1998), or βGBP of the crayfish Pacifastacus leniusculus (Lee et al. 2000). A gene cloned from the eggs of sea urchins was named as a glucanase gene, the only so-named gene from a metazoan, but there has been no experimental evidence thus far demonstrating the enzymatic prosperities of the gene product (Bachman and McClay 1996).
Although two types of GNBPs with or without conserved residues have been discussed in the literature, our analysis shows for the first time that these split into two identifiable groups in our phylogenetic analysis. It seems likely that the members of the two groups will have functional differences, but this will require further study, particularly to assess the glucanase activity of GNBPs of group B.
In conclusion, members of Toll/Imd pathway are relatively conserved across animal phyla and regulate the expression of genes involved in immune responses (Hoffmann 2003; Lemaitre and Hoffmann 2007). Over the past decade, NF-kB signal transduction associated pathways have been found to be key components of both adaptive and innate immune responses. PGRPs and GNBPs are key recognition and regulatory components of both Toll/Imd pathways. As noted in “Introduction”, molluscan species that have been studied are believed to possess these pathways suggesting that B. glabrata would as well. The characterization of BgPGRPs and BgGNBP are suggestive of this possibility, but further study is needed to confirm their functional roles and to identify other components of the pathways. As suggested by the studies described above, PGRPs and GNBPs are likely to be functionally heterogeneous. Further studies are needed to define further the function of the various isoforms of BgPGRPs and BgGNBPs we now know to be produced by B. glabrata.
Acknowledgments
We would like to thank Dr. Sara Brant for assistance with the phylogenetic analyses, Dr. Barbara Stout for critically reading of the manuscript, and Dr. Coen Adema for helpful discussions, and two anonymous reviewers for valuable comments. This work was supported by NIH RO1AI067686 (S-MZ) and NIH Grant Number RR-1P20RR18754 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources.
Footnotes
Nucleotide sequence data reported are available in the GenBank database under accession numbers EF452345–EF452349.
References
- Bachman ES, McClay DR. Molecular cloning of the first metazoan beta-1,3-glucanase from eggs of the sea urchin Strongylocentrotus purpuratus. Proc Natl Acad Sci USA. 1996;93:6808–6813. doi: 10.1073/pnas.93.13.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beschin A, Bilej M, Hanssens F, Raymakers J, Van Dyck E, Revets H, Brys L, Gomez J, De Baetselier P, Timmermans M. Identification and cloning of a glucan- and liopoplysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J Biol Chem. 1998;273:24948–24954. doi: 10.1074/jbc.273.38.24948. [DOI] [PubMed] [Google Scholar]
- Bilej M, De Baetselier P, Van Dijck E, Stijlemans B, Colige A, Beschin A. Distinct carbohydrate recognition domains of an invertebrate defense molecule recognize Gram-negative and Gram-positive bacteria. J Biol Chem. 2001;276:45840–45847. doi: 10.1074/jbc.M107220200. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- Cheng XD, Zhang X, Plugrath JW, Studier FW. The structure of bacteriophage-T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA-polymerase. Proc Natl Acad Sci USA. 1994;91:4034–4038. doi: 10.1073/pnas.91.9.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Briant L, Montagnani C, Hez S, Devaux C, Roch P. Oyster IKK-like protein shares structural and functional properties with its mammalian homologues. FEBS Lett. 1999;453:293–298. doi: 10.1016/s0014-5793(99)00737-1. [DOI] [PubMed] [Google Scholar]
- Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappa B-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25:677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Goodall CP, Bender RC, Brooks JK, Bayne CJ. Biomphalaria glabrata cytosolic copper/zinc superoxide dismutase (SOD1) gene: Association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni. Mol Biochem Parasitol. 2006;147:207–210. doi: 10.1016/j.molbiopara.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ. Identifying components of the NF-kappa B pathway in the beneficial Euprymna scolopes Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryseels B, Polmman K, Clernx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Humphries JE, Yoshino TP. Schistosoma mansoni excretory-secretory products stimulate a P38 signalling pathway in Biomphalaria glabrata embryonic cells. Intl J Parasitol. 2006;36:37–46. doi: 10.1016/j.ijpara.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wu X. Characterization of a Rel/NF-kB homologue in a gastropod abalone, Haliotis diversicolor supertexta. Dev Comp Immunol. 2006;31:121–132. doi: 10.1016/j.dci.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Loker ES, Zhang S-M. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 2006;30:855–866. doi: 10.1016/j.dci.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silvermman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Kim YY, Ryu JH, Han SJ, Choi KH, Nam KB, Jang JH, Lemaitre B, Brey PT, Lee WJ. Gram-negative bacteria binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- Kim MS, Byun M, Oh B-H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- Kurata S. Recognition of infection non-self and activation of immune responses by peptidoglycan recognition protein (PGRP) family members in Drosophila. Dev Comp Immunol. 2004;28:89–95. doi: 10.1016/s0145-305x(03)00121-6. [DOI] [PubMed] [Google Scholar]
- Lee SY, Wang R, Söderhäll K. A lipopolysaccharide-and β-1,3-glucan-binding protein from hemocytes of the freshwater crayfish Pacifastacus leniusculus. J Biol Chem. 2000;275:1337–1343. doi: 10.1074/jbc.275.2.1337. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann JA. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:679–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Léonard PM, Adema CM, Zhang S-M, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS. Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol. 2007;151:18–27. doi: 10.1016/j.molbiopara.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker ES, Hertel LA. Alternation in Biomphalaria glabrata plasma induced by infection with the digenetic trematode Echinostoma paraensei. J Parasitol. 1987;75:505–513. [PubMed] [Google Scholar]
- Loker ES, Adema CM, Zhang S-M, Kepler TB. Invertebrate immune systems—not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Kanost MR. A β-1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. 2000;275:7505–7514. doi: 10.1074/jbc.275.11.7505. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, PrestonHurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Meister S, Kanzok SM, Zheng XI, Luna C, Li T-R, Hoa NT, Clayton JR, White KP, Kafatos FC, Christophides GK, Zheng L. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, Guillou Y, Bouchut A, Coustau C. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29:393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Montagnani C, Kappler C, Reichhart JM, Escoubas JM. Cg-Rel, the first Rel/NF-kappa B homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas. FEBS Lett. 2004;56:75–82. doi: 10.1016/S0014-5793(04)00124-3. [DOI] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1988;263:12056–12062. [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A pattern recognition protein for peptidoglycan: cloning the cDNA and the gene of the silkworm, Bombyx mori. J Biol Chem. 1999;274:11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- Raghavan N, Knight M. The snail (Biomphalaria glabrata) genome project. Trends Parasitol. 2006;22:148–151. doi: 10.1016/j.pt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Reiser J-B, Teyton L, Wilson IA. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 Å resolution. J Mol Biol. 2004;340:909–917. doi: 10.1016/j.jmb.2004.04.077. [DOI] [PubMed] [Google Scholar]
- Roux M, Pain A, Klimpel KR, Dhar AK. The lipopolysaccharide and β-1,3-glucan binding protein gene is upregulated in white spot virus-infected shrimp (Penaeus stylirostris). J Virol. 2002;76:7140–7149. doi: 10.1128/JVI.76.14.7140-7149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Bian G, Raikhel SA. A Toll receptor and a cytokine, Toll5A and Spz1C, are involved in Toll antifungal immune signaling in the mosquito Aedes aegypti. J Biol Chem. 2006;281:39388–39395. doi: 10.1074/jbc.M608912200. [DOI] [PubMed] [Google Scholar]
- Song LS, Xu W, Li CH, Li HL, Wu LT, Xiang JH, Guo XM. Development of expressed sequence tags from the bay scallop, Argopecten irradians irradians. Mar Biotechnol. 2006;8:161–169. doi: 10.1007/s10126-005-0126-4. [DOI] [PubMed] [Google Scholar]
- Sritunyalucksana K, Lee SY, Söderhäll K. A β-1,3-glucan binding protein from the black tiger shrimp, Penaeus monodon. Dev Comp Immunol. 2002;26:237–245. doi: 10.1016/s0145-305x(01)00074-x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Version 4.0b. Sinauer Associates; Sunderland, MA: 1998. PAUP*. Phylogenetics analysis using parsimony (* and other methods). [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S. Overexpression of a pattern-recognition receptor, peptidoglycan recognition protein-LE, activates Imd/Relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy A, Guo XM, Ford SE. Discovery of genes expressed in response to Perkinsus marinus challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) oysters. Gene. 2004;338:121–131. doi: 10.1016/j.gene.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Terwilliger DP, Buckley KM, Mehta D, Moorjani PG, Smith LC. Unexpected diversity displayed in cDNA expressed in the immune cells of the purple sea urchin, Strongylocentrotus purpurats. Physiol Genomics. 2006;26:134–144. doi: 10.1152/physiolgenomics.00011.2006. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergote D, Bouchut A, Sautiere PE, Roger E, Galinier R, Rognon A, Coustau C, Salzet M, Mitta G. Characterisation of proteins differentially present in the plasma of Biomphalaria glabrata susceptible or resistant to Echinostoma caproni. Int J Parasitol. 2005;35:215–224. doi: 10.1016/j.ijpara.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Walker AJ. Do trematode parasites disrupt defence-cell signaling in their snail hosts? Trends Parasitol. 2006;22:154–159. doi: 10.1016/j.pt.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Wang XG, Fuchs JF, Infanger LC, Rocheleau TA, Hillyer JF, Chen CC, Christensen BM. Mosquito innate immunity: involvement of beta 1,3-glucan recognition protein in melanotic encapsulation immune responses in Armigeres subalbatus. Mol Biochem Parasitol. 2005;139:65–73. doi: 10.1016/j.molbiopara.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnepenninckx B, Backeljau T, DeWachter R. Extraction of high-molecular-weight DNA from mollusks. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- Yahata N, Watanabe T, Nakamura Y, Yamamoto Y, Kamimiya S, Tanaka H. Structure of the gene encoding beta-1,3,-glucanase A1 of Bacillus circulans WL-12. Gene. 1990;86:113–117. doi: 10.1016/0378-1119(90)90122-8. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M. Purification of peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- Zaidman-Rémy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang S-M, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the snail Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-M, Adema CM, Kepler TB, Loker ES. Diversification of Ig genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]