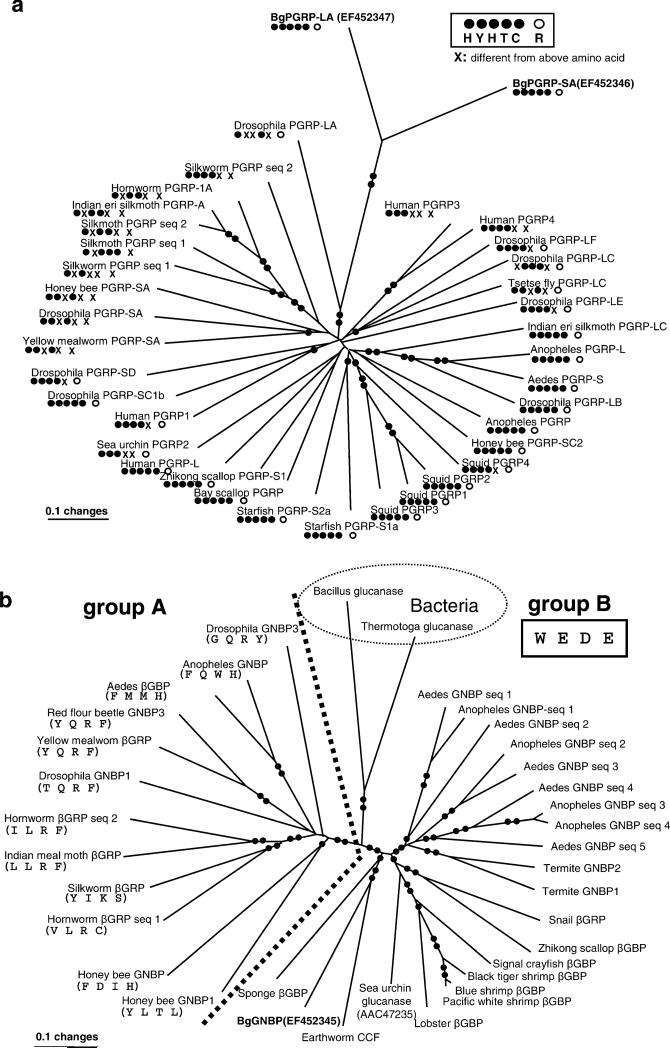
Fig. 8.
A gene tree using minimum evolution (ME) showing relationships among a PGRPs or b GNBPs based on amino acid sequences. For each sequence, a protein name and a common name of species are provided. Bootstrap values less than 50% are not shown. Single- or double-filled circles indicated on the branches indicate bootstrap support of 50–80% and 81–100%, respectively. For a depicting relationships among PGRP sequences, the presence/absence of each of the five conserved residues associated with amidase activity is provided under the protein's name, as is the presence/absence of the Arg residue responsible for discrimination of the DAP-type PGN (see Fig. 5a, b for the positions of the residues). In b showing relationships among GNBPs, the delineation between groups A and B is shown with a dotted line (100% bootstrap value). In group A, the presence/absence of the four conserved residues associated with glucanase activity is provided under each protein's name. In group B, the four residues are the same (W, E, D, E) for all sequences. The GenBank accession numbers for all sequences in a are as follows (starting from Human PGRP3, clockwise direction). Human PGRP3 (AAI28115), human PGRP4 (AAI07158), Drosophila melanogaster PGRP-LF (NP_648299), Drosophila PGRP-LC (NP_996030), tsetse fly Glossina morsitans PGRP-LC (ABC25065), Drosophila PGRP-LE (NP_573078), Indian eri silkmoth Samia cynthia PFRP-LC (BAF03521), mosquito Anopheles gambiae PGRP-L (XP_321943), mosquito Aedes aegypti PGRP-S (ABF18154). Drosophila PGRP-LB (NP_731576), Anopheles PGRP (Q7PP76), honey bee Apis mellifera PGRP-SC2 (XP_395941), squid Euprymna scolopes PGRP4 (AAY27976), squid PGRP2 (AAY27974), squid PGRP1 (AAY27973), squid PGRP3 (AAY27975), starfish Asterias rubens PGRP-S1a (DQ222477), starfish PGRP-S2a (DQ222478), Bay scallop Argopecten irradians PGRP (AY437875), Zhikong scallop Chlamys farreri PGRP-S1 (AAY53765), human PGRP-L (NP_443122), sea urchin Strongylocentrotus purpuratus PGRP-2 (XP_796422), human PGRP1 (NP_005082), Drosophila PGRP-SC1B(AAG23736), Drosophila PGRP-SD (NP_648145), yellow mealworm Tenebrio molitor PGRP-SA (BAE78510 ), Drosophila PGRP-SA (NP_572727), honey bee PGRP-SA (XP_001123180), silkworm Bombyx mori PGRP seq 1 (BAA77209), silkmoth Antheraea mylitta PGRP seq 1 (ABG72708), silkmoth PGRP seq 2 (ABG72709), Indian eri silkmoth PGRP-A (BAF03522), hornworm Manduca sexta PGRP-1A (AAO21509), silkworm PGRP seq 2 (AAL32058), Drosophila PGRP-LA (NP_996029). GenBank accession numbers in b are as follows (starting from mosquito Aedes GNBP seq 1, clockwise direction). Aedes aegypti GNBP seq 1 (EAT44801), Anopheles gambiae GNBP seq 1 (Q7QA32), Aedes GNBP seq 2 (EAT41280), Anopheles GNBP seq 2 (Q7PQA4), Aedes GNBP seq 3 (EAT44802), Aedes GNBP seq 4 (EAT 38985), Anopheles GNBP seq 3 (Q7QCT6), Anopheles GNBP seq 4 (Q7QCT7), Aedes GNBP seq 5 (EAT38986), Termite Nasutitermes fumigatus GNBP2 (AAZ08496), Termite GNBP1 (AAZ08483), Snail B. glabrata βGBP (EF121824), Zhikong scallop βGBP (AAP82240), signal crayfish Pacifastacus leniusculus βGBP (CAB65353), black tiger shrimp Penaeus mondon βGBP (AAM21213), blue shrimp Litopenaeus stylirostris βGBP (AAM73871), Pacific white shrimp Litoppenaeus vannamei βGBP (AAW51361), lobster Homarus gammarus βGBP (CAE47485), sea urchin glucanase (AAC47253), earthworm Eisenia fetida CCF (AAC35887), sponge Suberites domuncula βGBP (CAE54585), honey bee GNBP 1 (XP_001121634), honey bee GNBP (XP_395368), hornworm βGRP seq 1 (AAF44011), silkworm βGBP (BAA92243), Indian meal moth Plodia interpunctella βGRP (AAM95970), hornworm βGBP seq 2 (AN10151), Drosophila GNBP1 (AAF33849), yellow mealworm βGRP (BAC99308), red flour beetle Tribolium castaneum GNBP (XP_972063), Aedes βGBP (EAT40654), Anopheles GNBP (Q7QIB2), Drosophila GNBP3 (AF33851), Bacillus circulans glucanase (Z47974), Thermotoga neapolitana glucanses (P23903)