Abstract
Using a novel microfluidic chamber that allows the isolation of axons without contamination by nonaxonal material, we have for the first time purified mRNA from naive, matured CNS axons, and identified the presence of >300 mRNA transcripts. We demonstrate that the transcripts are axonal in nature, and that many of the transcripts present in uninjured CNS axons overlap with those previously identified in PNS injury-conditioned DRG axons. The axonal transcripts detected in matured cortical axons are enriched for protein translational machinery, transport, cytoskeletal components, and mitochondrial maintenance. We next investigated how the axonal mRNA pool changes after axotomy, revealing that numerous gene transcripts related to intracellular transport, mitochondria and the cytoskeleton show decreased localization 2 d after injury. In contrast, gene transcripts related to axonal targeting and synaptic function show increased localization in regenerating cortical axons, suggesting that there is an increased capacity for axonal outgrowth and targeting, and increased support for synapse formation and presynaptic function in regenerating CNS axons after injury. Our data demonstrate that CNS axons contain many mRNA species of diverse functions, and suggest that, like invertebrate and PNS axons, CNS axons synthesize proteins locally, maintaining a degree of autonomy from the cell body.
Introduction
Important functional roles for local protein synthesis in mammalian neurons have been demonstrated, including synaptic plasticity occurring in hippocampal dendrites (Sutton and Schuman, 2006), growth cone turning in developing retinal and sensory mammalian axons (Campbell and Holt, 2001; Wu et al., 2005), and growth cone regeneration in adult sensory axons (Willis and Twiss, 2006). Whereas local protein synthesis in dendrites, developing axons, and adult sensory axons is established, it is unclear whether cortical axons have a similar capacity. In situ hybridization studies have shown that mRNA is present in developing cortical axons and then decreases with maturity, eventually becoming undetectable (Bassell et al., 1994; Kleiman et al., 1994). However, due to the limited resolution of previous in situ hybridization methods, it is unclear whether mRNA transcripts are truly absent from mature cortical axons.
Importantly, adult mammalian axons contain translational machinery (Koenig et al., 2000; Sotelo-Silveira et al., 2008). In response to injury, adult dorsal-root ganglion (DRG) axons synthesize proteins locally (Zheng et al., 2001; Hanz et al., 2003; Verma et al., 2005), and this local translation is required for growth cone regeneration (Verma et al., 2005). The local synthesis of proteins occurs rapidly following axotomy, with maximal 3H-leucine incorporation occurring at 1 h following injury (Verma et al., 2005), suggesting translation of preexisting axonal mRNAs. In support of this, importin β, vimentin and RanBP1 transcripts are present in naive axons and are translated locally following injury in DRGs (Hanz et al., 2003; Perlson et al., 2005; Yudin et al., 2008). Other locally synthesized proteins have been identified in injury-conditioned DRGs using a sciatic nerve crush model (Willis et al., 2005). Although a number of studies have investigated axonal protein synthesis in response to injury, little is known about protein synthesis in the mature uninjured axon. An important first step to elucidating functions of axonal protein synthesis will be to identify transcripts that are present in naive (uninjured) axons, including axons of the CNS.
The identification of transcripts in mature cortical axons has thus far been precluded by several technical factors. In situ hybridization and radiolabeling have traditionally been limited in sensitivity, and microarrays or reverse transcriptase PCR (rt-PCR) have been limited by the technical difficulty of isolating mature CNS axons without contamination by postsynaptic or glial material (Piper and Holt, 2004). We have overcome these technical impediments by using a microfluidic chamber that allows harvesting of cortical axons free of dendritic, somal and glial material (Taylor et al., 2005). Here, we apply this technique to prepare mRNA from cortical axons and use microarrays to identify and catalog the transcripts. As a first step, we identify transcripts present in naive matured cortical axons. Further, we investigate how the mRNA composition is altered in regenerating axons, using the microfluidic chamber as an in vitro model for axonal injury and regeneration (Taylor et al., 2005). Last, we validate the microarray findings using a new highly sensitive fluorescence in situ hybridization (FISH) method to visualize mRNA within the cortical axons.
Materials and Methods
Cell culture in microfluidic chambers.
For microarray experiments, cortical and hippocampal dissociated neurons were prepared from embryonic Sprague Dawley rats [embryonic day 18 (E18)] and plated into microfluidic chambers, as described previously (Rhee et al., 2005; Taylor et al., 2003, 2005). For FISH, both postnatal day 0 (P0) hippocampal neurons and E18 cortical and hippocampal neurons from Sprague Dawley rats were used. The P0 hippocampal neurons were isolated as previously published (Sutton et al., 2007).
The microfluidic chamber consists of a poly(dimethylsiloxane) (PDMS) piece with a bas relief pattern of microfluidic channels atop a polylysine-coated cover glass. The microfluidic features consist of two 1.5-mm-wide parallel channels (100 μm high), each channel having access ports (or wells) at either end. A PDMS barrier separating the two parallel channels contains 157 small microgrooves (7.5 μm wide, 3 μm high) which connect the two channels or compartments. Dissociated neurons are added into one of the compartments (“cell body compartment”); the suspension of dissociated neurons then flows through the compartment and neurons attach to the glass after 5–10 min. After 3–4 d, axons grow through the microgrooves and into the second channel or 'axonal compartment'. The small dimensions of the microgrooves allow axons and dendrites to grow through the microgrooves, but prevent the passage of larger cell bodies. Neurobasal medium (Invitrogen) supplemented with B27 and Glutamax was used as growth medium for the microfluidic chambers. Neither serum nor growth factors were added to either compartment at any time.
For the axotomized cultures, axons were removed via aspiration after 11 d in culture, as described previously (Taylor et al., 2005; Park et al., 2006). The medium in the axonal compartment was carefully withdrawn, and then added back following the aspiration of the axons from this compartment.
Immunocytochemistry.
Cultures were fixed and processed as described previously (Taylor et al., 2005). Primary antibody dilutions were as follows: monoclonal anti-MAP2 1:1000 (Sigma), polyclonal anti-tau 1:500 (gift from L. Binder, Northwestern University Medical School, Chicago, IL), polyclonal anti-β-tubulin 1:1000 (Abcam), monoclonal anti-β-tubulin isotype III 1:1000 (Sigma), rabbit anti-β-catenin antibody 1:500, goat anti-neurexin III (C-17) antibody 1:500 (Santa Cruz Biotechnology), and rabbit anti-eEF1A1 antibody 1:400 (Abcam). Briefly, cultures were fixed in 4% paraformaldehyde for 20 min, washed twice in PBS, and then permeabilized in PBS with 0.2% Triton X-100 for 5 min. Cultures were blocked in PBS with 2–10% BSA for 15–30 min at room temperature. Cultures were incubated in primary antibody diluted in PBS with 1% BSA, at 4°C overnight or for 1 h at room temperature. Cultures were rinsed 3 times (5 min each), and incubated for 1 h at room temperature with secondary antibody (Alexa Fluor 488, 568, or 647, 1:500; Invitrogen) diluted in PBS.
Fluorescent in situ hybridization.
The PDMS pieces were removed after 13–14 d in culture and fixed immediately using 4% formaldehyde/4% sucrose in PBS for 20 min, followed by ethanol dehydration. After rehydration, cells were permeabilized using a detergent solution (Panomics) for 5 min. To visualize axons, immunocytochemistry was performed with either monoclonal or polyclonal anti-β-tubulin antibodies. The blocking solution and antibody solution contained 1% Ultrapure BSA (Ambion). Chambers were incubated with primary antibody for 30 min at 37°C, or 1 h at room temperature, followed by secondary antibody incubation (Alexa Fluor 647, 1:500, 1 h at room temperature; Invitrogen). The sense control of the neuronal cell bodies was performed without immunocytochemistry. For FISH processing, cultures were subjected to protein digestion using proteinase K (Panomics) (1:5000) for 10 min at room temperature, followed by in situ hybridization using probes designed by Panomics (supplemental Methods, available at www.jneurosci.org), following manufacturer's instructions. Briefly, probes were diluted 1:100 in hybridization buffer supplied by Panomics, incubated at 40°C (3–5 h), washed, hybridized with preamplification oligos (1:100) at 40°C (25 min), washed, hybridized with amplification oligos (1:100) at 40°C (15 min), washed, and finally hybridized with the label oligos (1:100) at 40°C (15 min). Each probe set consists of 10 or more nucleotide sequence pairs (supplemental Methods, available at www.jneurosci.org). The oligonucleotide probe pairs are designed to hybridize to adjacent segments on the target RNA allowing the hybridization of a preamplification probe (Panomics) which spans the hybridized probe pair. This signal is further amplified by amplification probes (Panomics), then label probes (Panomics); the result is up to a 500-fold amplification for low abundance mRNAs.
Confocal imaging.
Confocal images were captured on either a Bio-Rad Radiance 2100 or an Olympus IX-70 laser scanning confocal microscope using λ-strobing or sequential imaging to avoid nonspecific cross-excitation or -detection. For imaging the FISH samples, the confocal aperture was opened to setting 3 on the Olympus IX-70 to increase signal detection of the small puncta and the argon laser intensity was set at 60%, higher than the usual settings.
RNA isolation.
The isolation method for axonal RNA is described previously (Taylor et al., 2005; Park et al., 2006) and is shown in supplemental Figure S1, available at www.jneurosci.org as supplemental material. To isolate RNA from the axonal compartment, the majority of the media on the axonal side was removed. The medium was then aspirated from the cell body compartment until it was completely dry. With continual aspiration applied to the cell body compartment, 100–150 μl of RNAqueous-Micro lysis solution (Ambion) was added to one of the axonal wells. Continual aspiration to the cell body side during RNA extraction prevents cellular material from the cell body compartment from entering into the axonal compartment. After 5–10 s, the lysis solution was collected with a pipette from the connecting well on the axonal side. The withdrawn volume of lysis solution was approximately a quarter to a third of the initial lysis solution volume; this amount was added to a microcentrifuge tube containing 50 μl of fresh lysis solution and mixed well by pipetting. The excess lysis solution serves to prevent further degradation of the RNA. Subsequent RNA purification was performed according to manufacturer's instructions for the Ambion RNAqueous-Micro kit. RNA was eluted in a final volume of 10 μl. The same procedures were used to isolate neuronal RNA from the cell body compartment except without continual aspiration of the adjacent compartment.
RNA amplification and microarrays.
Isolated RNA was pooled from three microfluidic chambers for each GeneChip sample. A total of 12 GeneChips were used (axonal, n = 7; regenerating axons, n = 3; neuronal, n = 3). Although yields for RNA from the axonal and neuronal samples were not equivalent, samples were processed equivalently to ensure accurate comparisons. For GeneChip samples, isolated RNA was subjected to two rounds of amplification using an Ambion MessageAmp II aRNA kit. First, cDNA was made from pooled RNA samples using an oligo(dT) primer with a T7 promoter, followed by antisense RNA generation by in vitro transcription for 14 h with T7 RNA polymerase. The antisense RNA underwent a second round of amplification which included biotin-labeling for the GeneChip hybridization to produce labeled antisense amplified RNA (aRNA). The quality of the aRNA for all samples was assessed using an Agilent Bioanalyzer after the second round of amplification.
Fragmentation and hybridization of the aRNA was performed by the UCI DNA & Protein MicroArray Facility, University of California, Irvine. 20 μg of aRNA was fragmented to an average strand length of 100 bases (range 35–200 bases), and hybridized to Rat230.2 GeneChip following manufacturer protocols (Affymetrix GeneChip Expression Analysis Technical Manual). Quality control parameters were all within expected ranges.
Microarray analysis.
Expression values were derived from the CEL image files by applying the robust multiarray average (RMA) algorithm (Irizarry et al., 2003a,b) using GeneSpring software (Agilent Technologies). Expression values across samples were normalized to axonal control samples run in each processing batch, followed by filtering to remove unreliable probe sets and those with unknown function. To identify the types of mRNA transcripts present in axons, probe sets were filtered on “Presence Call” (GeneSpring software, Agilent) to include only those probe sets called “present” in at least 70% of the axonal samples, reducing the probe set list from 31,099 to 5000. Probe sets were further filtered to include only those probe sets with known function (2484 probe sets remaining). Using RMA-calculated expression values, the probe set list was sorted and ranked according to descending expression level. Actb was used as a reference ranking, as we had previously detected this transcript in matured axons using quantitative rt-PCR (qPCR). We included probe sets with 30% lower ranking than Actb resulting in a final list of 308 probe sets for which transcripts were considered “reliably localized” in axons. To identify potential functional roles of axonally localized transcripts, the probe set list was submitted to DAVID (http://david.abcc.ncifcrf.gov/) and categorized using Gene Ontology (GO) annotations to identify enriched functional categories of genes (using functional annotation clustering). The p value for each identified GO terms was calculated using the Fisher Exact test to measure the level of enrichment within GO terms.
To investigate changes in transcript availability in regenerating axons, probe sets were again filtered on Presence Call, retaining only those probe sets called present in at least 70% of the regenerating axonal samples, followed by removal of probe sets that did not show >20% change relative to naive axons, and statistical thresholding [FDR (false discovery rate) < 0.05] with multiple testing correction using the Benjamini and Hochberg method. The DAVID web site was used to categorize all changed probe sets using the functional annotation clustering feature.
Heat maps were generated based on sample clustering using Pearson Correlation as a similarity measure and an average linkage clustering algorithm (separation ratio 1, minimum distance 0.001).
cDNA synthesis and PCR.
PCR validation was undertaken for H1histone (H1f0), β actin (Actb), synaptophysin (Syp), map kinase 8 interacting protein (Mapk8ip) and exocyst 3 (Exoc3). Primer sequences are included in supplemental Methods, available at www.jneurosci.org. H1f0 and Actb underwent end-point rt-PCRs as detailed previously (42 cycles) (Taylor et al., 2005). qPCR was used to assess levels of Actb, Syp, Mapk8ip, and Exoc3. For Mapk8ip and Exoc3 validation, isolated RNA underwent 1 round of amplification using the Ambion MessageAmp II aRNA kit (Austin, TX). cDNA was made from pooled RNA samples using an oligo(dT) primer with a T7 promoter, followed by antisense RNA generation by in vitro transcription for 14 h with T7 RNA polymerase. Reverse transcription was then performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Actb and Syp underwent reverse transcription using the Transcriptor First Strand cDNA Synthesis Kit (Roche) using random hexamer primers and 10 μl of total RNA (unamplified) for each cDNA synthesis reaction [25°C (10 min); 55°C (30 min); 85°C (5 min)]. A reaction without reverse transcriptase (−rt) was included as a negative control in every run. For absolute quantification of Actb and Syp, cDNA concentration was quantified using a Nanodrop Spectrophotometer, and the Nanodrop reading from the –rt control was subtracted from the positive reactions to obtain the cDNA concentration.
For qPCR, primers and prevalidated probes were designed using Roche's Universal Probe Library web site (supplemental Methods, available at www.jneurosci.org). Csf-1 was selected as an axonal housekeeping gene based on our observation that this gene is maintained at similar levels in naive and regenerating axonal samples, according to our microarray data. For qPCR, each 20 μl of PCR contained 5.4 μl of water, 0.2 μl probe (10 μm), 0.4 μl of 20 μm primer mix, and 4 μl of (5×) TaqMan Master Mix and 1.5 μg of cDNA for Syp and Actb targets (for absolute quantification), or 10 μl of aRNA for the Mapk8ip or Exoc3 targets (for relative quantification). qPCRs were preincubated at 95°C (10 min), followed by 50 amplification cycles of 95°C (10 s), 60°C (30 s), and 72°C (1 s) using a Roche LightCycler 2.0 instrument. 50 amplification cycles were used to ensure detection of low abundance targets. The reactions were cooled for 30 s at 40°C following the amplification cycles.
Copy numbers of Actb and Syp were calculated using absolute quantification with external standards. Standard curves were generated using purified rt-PCR targets, using RNA isolated from standard cortical and hippocampal cultures and the primers listed in the supplemental Methods, available at www.jneurosci.org. Five tenfold dilutions were used in triplicate, and the second derivative maximum method was used to calculate the standard curves.
Results
Actin decreases, but synaptophysin increases, in matured cortical axons
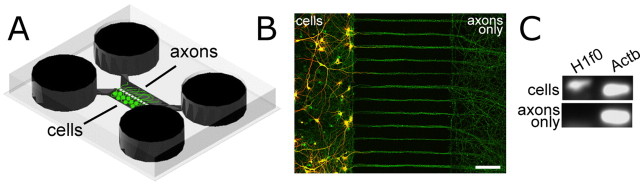
The multicompartment microfluidic chamber has previously been used to isolate rat cortical and hippocampal axons from their cell bodies and dendrites (Taylor et al., 2005). Neurons, plated into one compartment of the microfluidic chamber, extend processes through >150 small microgrooves to an adjacent axonal compartment. Due to the cross-sectional area and length of the microgrooves, only axons are able to extend into the axonal compartment (Fig. 1 A,B). Importantly, the growth and isolation of axons does not require serum or other growth factors. In previous work, we also developed procedures to isolate axonal mRNA (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) (Taylor et al., 2005). Using this microfluidic chamber, we detected both β-actin (Actb) and synaptophysin (Syp) mRNA in pure preparations of immature cortical axons (6 d in vitro) using end-point rt-PCR (42 cycles) (Taylor et al., 2005).
Figure 1.
The compartmentalized microfluidic chamber is used to isolate pure axonal mRNA from mammalian cortical neurons. A, A schematic of the microfluidic chamber (∼22 mm square). B, Fluorescent micrograph of neurons cultured within the microfluidic chamber showing the ability to isolate axons free of somata and dendrites (MAP2 = red; tau = green). Due to the length of the microgrooves, only axons extend into the axonal compartment. Axonal mRNA was harvested from within this axonal compartment. Scale bar, 100 μm. C, rt-PCR analysis of mRNA collected from neurons and axons within the microfluidic chamber showing that H1 histone mRNA is present in cells, but not the harvested axons. β-Actin mRNA is present in both cells and axons.
To determine whether axonal mRNA was present in more mature cortical axons, we isolated axonal mRNA from neurons 13 d in culture, showing that Actb mRNA was present in the harvested axons without any detectable somatic mRNA (Fig. 1 C). Next, we assessed how the transcript levels changed in axons as the neurons mature to form functional synaptic connections. qPCR was used to assess Actb and Syp transcript levels at 7 and 13 d in culture. Actb was detectable at both culture ages, but levels decreased significantly from 7 to 13 d (mean copies per sample in 7 d axons: 25 ± 6, n = 15 measurements; mean copies per sample in 13 d axons: 10 ± 1, n = 5 measurements; two-tailed t test p value = 0.0092), consistent with previous findings in cortical and hippocampal axons showing declining levels of available mRNA with maturity (Bassell et al., 1994; Kleiman et al., 1994). In contrast, mRNA levels of Syp were undetectable at 7 d, with levels increasing at 13 d (mean copies per sample in 13 d axons: 124 ± 78, n = 5 measurements), indicating the availability of this transcript in matured cortical axons. Although we previously detected Syp in immature (6 d) axons using end-point rt-PCR, qPCR is less sensitive at extremely low copy number (e.g., <10 transcripts) suggesting that axons at 7 d have copies of Syp transcripts, but at extremely low levels. Together, these results demonstrate that mRNA transcripts are at detectable levels in cortical axons at 13 d in vitro, and suggest that the composition of the mRNA pool at this culture age is different from the mRNA pool present in developing axons.
The axonal mRNA pool is distinct from the neuronal pool
We next sought to identify and categorize other mRNA transcripts localized to axons using microarray expression profiling. A pure axonal mRNA preparation was obtained from 13-d-old neurons within the microfluidic chambers and amplified using two in vitro transcription amplification steps. In parallel, for quality control purposes, we also isolated neuronal mRNA from the cell body compartment of the microfluidic chambers (consisting of a mixed mRNA pool - primarily from cell bodies with a minor proportion of mRNA from dendrites and axons).
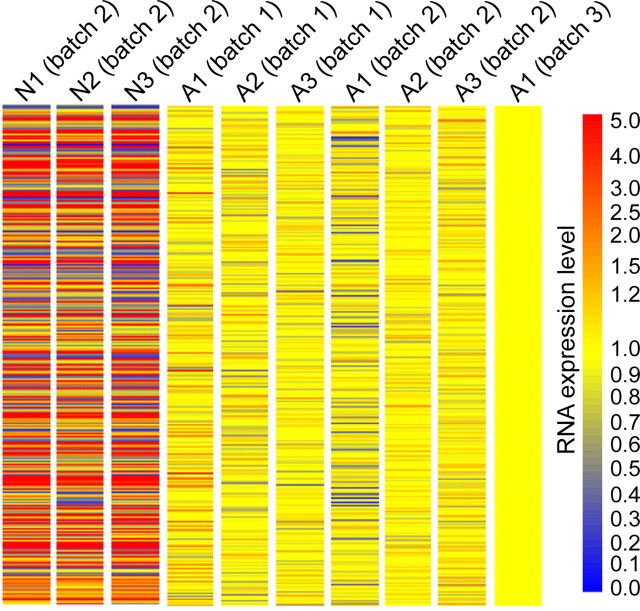
After processing the microarrays (7 axonal; 3 neuronal), we identified mRNA transcripts present in axons by filtering to include probe sets present in at least 70% of the axonal microarrays with recognized function (2484 probe sets remaining). Using this list of probe sets, we created a heat map of RMA-calculated expression for all the neuronal and axonal microarrays normalized to the axonal microarrays (Fig. 2). The results clearly show a distinct difference between the axonal and neuronal populations, with there being both increased and decreased levels of transcripts in the neuronal microarrays relative to the axonal ones. For comparison, the number of probes sets present in the neuronal samples (i.e., present in at least 70% of the neuronal microarrays with recognized function) is 6702. Of these 6702 probe sets, 2051 (or 31%) were also identified in the axonal pool.
Figure 2.
Microarray analysis shows that neuronal and axonal mRNAs isolated from within the microfluidic chamber have distinct profiles. A heat map of RMA-calculated expression values for 2484 transcripts which were present in 70% of axonal microarrays and had recognizable functions. Each column corresponds to a microarray sample (N, neuronal sample; A, axonal sample). Expression values are normalized to sample A1 (batch 3) as this microarray sample was run simultaneously with regenerating axons in a subsequent experiment and was used to evaluate changes in mRNA composition between naive and injured conditions (shown later in Fig. 4). Neuronal and axonal mRNA was harvested from the microfluidic chamber after 13 d in culture.
Using RMA-calculated expression values, the probe set list identified from the axonal samples was sorted and ranked according to descending expression level. Actb was used as a reference ranking, as we previously detected this transcript in axons using qPCR, resulting in a final list of 308 probe sets for which transcripts were considered reliably localized to axons (supplemental Table S1, available at www.jneurosci.org as supplemental material). Importantly, many transcripts that are exclusively dendritic and/or somatic were not detected or were below our criteria (supplemental Table S2, available at www.jneurosci.org as supplemental material), confirming that the transcripts identified in our microarray analysis are from axons.
Naive axons are enriched for translation, mitochondria, intracellular transport, and cytoskeletal elements
To identify potential functional roles of these axonally localized transcripts, the final probe set list was submitted to DAVID (http://david.abcc.ncifcrf.gov/) and categorized using GO annotations using the functional annotation clustering feature to identify enriched categories (p < 0.001). Using these criteria we identified four functional categories which had >20 transcripts identified as reliably localized in axons. The identified GO categories (Fig. 3 A) reveal that translation, mitochondrion, intracellular transport and cytoskeleton transcripts are highly enriched in naive axons. It is important to note that in addition to these main categories, many other small clusters of transcripts likely play important roles in axons which are not shown in Figure 3. For example, 157 transcripts of the 308 mRNAs reliably localized to axons were not included in these four main functional categories. Supplemental Table S3, available at www.jneurosci.org as supplemental material, includes a complete list of enriched categories for transcripts present in axons (p < 0.05).
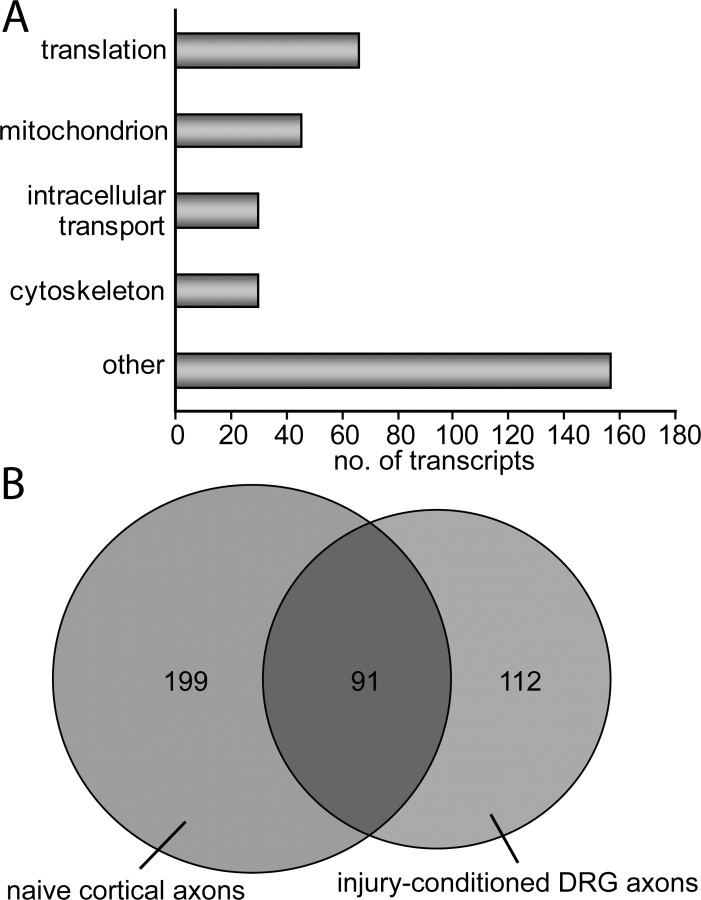
Figure 3.
GO analysis shows that within the mRNA population reliably localized to naive cortical axons multiple functions are enriched and that many of these transcripts overlap with adult DRG axons. A, Enriched GO terms include translation (p = 1.7 × 10−30), mitochondrion (p = 6.7 × 10−10), intracellular transport (p = 7.5 × 10−5), and cytoskeleton (p = 6.0 × 10−4). Notably, there are many transcripts not included in these enriched functional categories. B, Venn diagram showing overlapping transcripts in the naive matured cortical axons and adult injury-conditioned DRG axons identified in (Willis et al., 2007).
Notably, the most significantly enriched functional category for transcripts present in cortical axons is translation (p = 1.7 × 10−30). The majority of the transcripts identified in this category are ribosomal protein mRNAs, which have been similarly identified in axons of Aplysia sensory neurons (Moccia et al., 2003) and mammalian DRGs (van Niekerk et al., 2007; Willis et al., 2007) where local protein synthesis in axons has been firmly established. Translation mRNAs have also been identified in hippocampal processes (i.e., both dendrites and axons) isolated after 14 d in culture (Poon et al., 2006). Another example of an identified translation transcript is eukaryotic elongation factor 1a1 (Eef1a1) which is a member of the EF1 family of proteins required for tethering of aminoacyl tRNA to the ribosome during peptide synthesis. Eef1a1 mRNA has been shown to be transported to Aplysia axon terminals following 5-HT applications to cell bodies and synapses and is required for maintenance of long term facilitation (Giustetto et al., 2003). Because the presence of translational machinery is a prerequisite for supporting local protein translation, the presence of these translation transcripts supports a role for local translation in mammalian CNS axons.
Transcripts related to mitochondrial function were also enriched (p = 6.7 ×10−10). Mitochondria are present throughout the entire length of axons and have been observed to localize near active zones. Although mitochondrial DNA encodes some of its own proteins, the mitochondrial transcripts that we identified in the axonal compartment are encoded by nuclear genes. These transcripts include mitochondrial H+ transporting ATP synthase subunits, cytochrome c oxidase subunits, and NADH dehydrogenase components. The presence of nuclear-derived mitochondrial transcripts in the axon suggests that they are locally translated and may participate in maintaining local mitochondrial function and energy production.
In addition to translation and mitochondrial transcript enrichment, transcripts related to intracellular transport were also enriched (p = 7.5 × 10−5) in matured CNS axons. Included in these transport genes are two cytoplasmic light chain dynein transcripts (1 and 2a) and two kinesin transcripts, kinesin family member 5B (kif5B) and kinesin light chain 1 (klc1), all being part of motor complexes involved in transport of cargo and mitochondria within axons. These results suggest that axons may be able to locally synthesize components to transport cargo and organelles.
The axonal compartment of the cortical neurons was also found to be enriched in cytoskeletal gene transcripts (p = 6.0 × 10−4). Many of the cytoskeletal transcripts that we identified in matured CNS axons include gene transcripts that have previously been identified in non-CNS axons from various species (Chun et al., 1996; Weiner et al., 1996; Moccia et al., 2003; Willis et al., 2005). Previously unidentified transcripts were also discovered. For example, we identified β-catenin (Ctnnb1) mRNA in matured cortical axons, which is required for synaptic vesicle clustering at presynaptic terminals (Bamji et al., 2003) and links the synapse to the actin cytoskeleton (Kwiatkowski et al., 2007).
Together, these data reveal that gene transcripts of diverse functional categories are localized to matured cortical mammalian axons. The presence of numerous biosynthesis genes required for RNA translation demonstrates that axons have some of the genetic machinery at their disposal for local protein translation. In addition, the presence of nuclear mitochondrial genes suggests that axons draw upon nuclear-encoded mitochondrial genes to meet local energy needs, such as translation and general axonal function. Finally, axons rely on maintenance of their cytoskeleton for intracellular transport. The presence of both intracellular transport and cytoskeletal transcripts suggests that local protein synthesis plays a role in maintaining these critical functions within axons.
Many mRNA transcripts are similar to transcripts found in regenerating adult PNS axons
We next investigated whether there is overlap between the cortical axonal mRNAs identified in our microarray analysis and those previously identified in vertebrates in other neuronal culture types, culture ages, or species (supplemental Table S2, available at www.jneurosci.org as supplemental material). Our data validates and extends previous findings of axonal mRNAs and axonally synthesized proteins. For example, cytoskeletal mRNAs such as Actb and Tubb are known to be present and translated in rat and chick DRG axons (Lee and Hollenbeck, 2003; Willis et al., 2005), but have not previously been observed in adult CNS axons. In addition, our microarray analysis identified many transcripts in naive cortical axons which are similarly present in adult injury-conditioned rat DRG axons (Willis et al., 2007). We identified 91 of the same (or closely related) transcripts in both our cortical axon prep and the adult injury-conditioned DRG axons (Fig. 3 B) (supplemental Table S2, available at www.jneurosci.org as supplemental material). Notably, these 91 common transcripts were identified despite differences in preparation, microarray type and analysis methods (Willis et al., 2007).
These findings reveal that there are compositional similarities between the naive matured cortical axons and regenerating adult PNS axons, suggesting that the presence of certain gene transcripts may be a core feature common to both types of axons. These data also suggest a common functional role of axonal protein synthesis.
Composition of local mRNA transcripts significantly changes after axotomy
In PNS axons, effective growth cone regeneration after injury requires local protein synthesis in the axon (Verma et al., 2005; Blizzard et al., 2007). Given the similarities in the mRNA population between PNS axons and cortical axons suggested by our data, we sought to elucidate how local axonal capacity for protein translation is altered after injury in cortical neurons, by comparing axonal mRNA composition between naive and injured axons using the microfluidic chambers. To keep the same harvesting age for the regenerating axons, we axotomized the cortical axons at day 11 then isolated the regenerating axons 2 d later (i.e., 13 d in vitro) when there was sufficient axonal regrowth. Axons from naive cultures were isolated and processed simultaneously with regenerating axons to control for batch processing variability.
We identified all transcripts that showed significant changes (>20%) in the level of localization after injury. Of the 866 resulting axonal transcripts, 55.4% showed increased localization after injury and 44.6% showed decreased localization (supplemental Table S4, available at www.jneurosci.org as supplemental material), demonstrating that approximately equivalent numbers of transcripts showed increased and decreased localization following injury. A heat map (Fig. 4 A) shows the expression values of these 866 transcripts for all the naive and regenerating axonal samples (normalized to expression levels in naive axons) and shows clear distinction between these samples. There are several explanations as to why more transcripts were identified as significantly changed between naive and injured axons (n = 866) compared with the reliably localized transcripts (n = 308). First, the number of reliably localized transcripts was limited by the relatively conservative criteria of using Actb expression as a reference value. Second, most of the transcripts that showed increased levels in injured axons were not identified in naive axons (only eight of the transcripts showing increased levels following injury were identified as reliably localized transcripts in naive axons), suggesting that these transcripts were initially at low levels in naive axons and then increased following injury.
Figure 4.
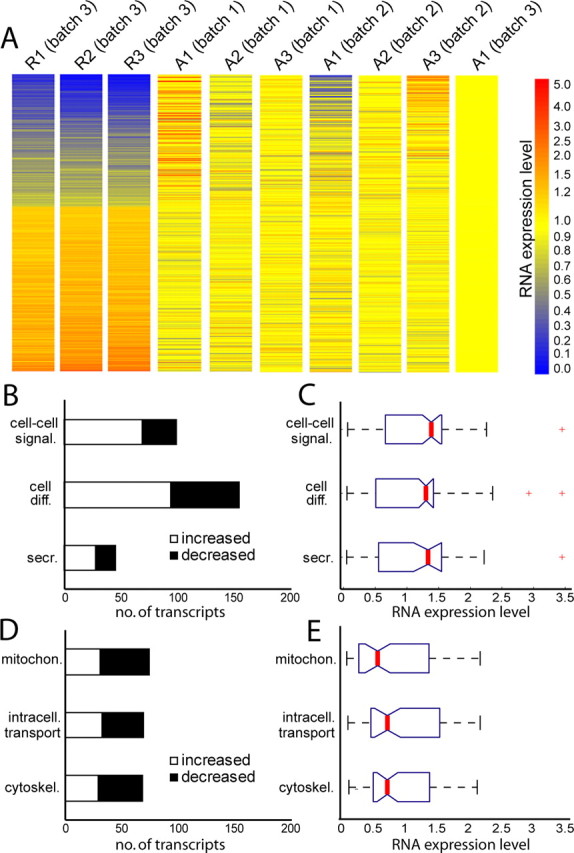
The mRNA pool in regenerating matured cortical axons is distinct from naive matured cortical axons and supports an altered functional role for axonal protein synthesis 2 d following axotomy. A, A heat map showing the RMA-calculated expression level of 866 transcripts identified as >20% changed between naive and regenerating axons. Expression levels are normalized to axonal microarray sample, A1 (batch 3), as this sample was run simultaneously with the regenerating axonal samples (R1–R3). All samples used RNA isolated within the microfluidic chamber at 13 d in vitro. For the regenerating axons, axons were removed at 11 d in vitro, then harvested at 13 d. B, Enriched functional categories showing overall increased localization following injury include cell–cell signaling (p = 3.8 × 10−17), cell differentiation (p = 5.0 × 10−12), and secretion (p = 2.6 × 10−5). Criteria for selecting changed transcripts: (>20% change, FDR <0.05). C, Boxplot showing expression values following injury (normalized to naive axons) for all probe sets identified in categories listed in (B). Median expression levels (red lines) are all >1. D, Enriched functional categories showing overall decreased localization following injury include mitochondrion (p = 2.1 × 10−5), intracellular transport (p = 1.5 × 10−6), and cytoskeleton (p = 2.5 × 10−4). E, Boxplot showing expression values following injury for all probe sets identified in categories listed in D. Median expression levels are all <1. For boxplots, red lines indicate medians; tops and bottoms of each “box” are 25th and 75th percentile of the samples; “whiskers” show extent of data; outliers are denoted in red and consist of data >1.5 times interquartile ranges.
The DAVID web site was again used to categorize significantly changed probe sets using the functional annotation clustering feature (p < 0.01) for enriched, non-overlapping GO terms (Fig. 4 B,C). Notably, although translation transcripts were enriched in the naive axons, this category was not significantly changed following injury (p = 0.30), suggesting that the capacity for local translation is not appreciably altered in axons with injury. In contrast, there was a trend showing that axonal targeting and synaptic function transcripts have increased localization to early regenerating axons, whereas mitochondrial, cytoskeleton, and intracellular transport transcripts have, on average, decreased localization. A complete list of GO terms is included in supplemental Table S5, available at www.jneurosci.org as supplemental material (p < 0.05).
Following injury, transcripts with increased localization are enriched for axonal targeting and synaptic function
Enriched categories of genes with increased localization in regenerating axons were predominantly related to cell differentiation (p = 5.0 × 10−12), cell–cell signaling (p = 3.8 × 10−17), and secretion (p = 2.6 × 10−5). These GO categories have a higher proportion of transcripts with increased localization in regenerating axons (Fig. 4 B), as well as higher median expression values following injury (Fig. 4 C). Interestingly, a common functional theme of many of these genes with increased localization is to support axonal targeting, synapse formation and synaptic function.
In the GO category of cell differentiation, our microarray analysis identified numerous transcripts that are known to localize to axon terminals and are required for proper nervous system development. For example, we identified two ephrin ligand transcripts, ephrin B1 (Efnb1) and ephrin A3 (Efna3), that were increased in regenerating cortical axons. Ephrin signaling is of particular interest as it has been identified as an important axon guidance mechanism. During development, axons grow toward higher concentrations of ephrin ligands (such as Efnb1 and Efna3), guided by ephrin receptors localized in axon growth cones. In addition to the role of ephrin signaling in axon guidance during development, recent data demonstrate that ephrin ligands, including Efnb1, are involved in synapse maturation and function after the axon has contacted the target cell (Lim et al., 2008). In parallel with upregulation of ephrin ligand transcripts, our analysis revealed upregulation of two probe sets for Poliovirus receptor-related 1 (Pvrl1) in the regenerating cortical axons. Pvrl1 (also called nectin-1) belongs to the nectin family of cell adhesion molecules, is asymmetrically localized to axons, and like ephrin, is involved in guidance of the axon trajectory, as well as axon-dendritic adhesion (Okabe et al., 2004; Beaudoin, 2006). This data are consistent with the in vivo condition, where cell differentiation genes are upregulated in cortical axons within days following injury (von Gertten et al., 2005).
Like cell differentiation genes, categories of cell–cell signaling and secretion were enriched after injury, in particular relating to synaptogenesis and neurotransmitter release. Some examples of mRNAs with increased localization following injury that play important roles in presynaptic function are synapsin III (Syn3), involved in regulation of neurotransmitter release and synaptogenesis, and neurexin 3 (Nrxn3), which localizes to presynaptic terminals and plays a role in synapse function. In addition, we found that ‘major histocompatibility complex class I (MHC-1) in rat’ (Rt1) had increased localization in regenerating axons. Interestingly, MHC-1 mRNA and protein are expressed by CNS neurons in vivo, where they are involved in nonimmune functions such as neuronal development and activity-dependent plasticity (Boulanger and Shatz, 2004). Further, in PNS neurons following axotomy, MHC-1 is rapidly upregulated locally around the lesion (Maehlen et al., 1988; Olsson et al., 1989), aiding regeneration following injury (Oliveira et al., 2004). Finally, several of the secretion transcripts that we identified and show increased localization, are linked to neurotransmitter release and presynaptic function. These transcripts include a frequenin homolog (Freq) (involved in exocytosis) (Choe and Ehrlich, 2006), Rab14 (may play a role in vesicle trafficking and neurotransmitter release) (Junutula et al., 2004), Rims1 (regulates exocytosis at the synapse) (Kaeser et al., 2008), and exocyst complex component 3 (Exoc3), which is involved in later stages of vesicle trafficking (Vik-Mo et al., 2003).
Together, these data suggest that there is increased capacity for axonal outgrowth and targeting in regrowing axons soon after injury, and increased support for both synapse formation and presynaptic function.
Mitochondrial transcripts are primarily downregulated in regenerating axons
Our earlier results revealed that mitochondrial transcripts were highly enriched in naive axons. Interestingly, in the pool of transcripts showing altered localization in regenerating axons, mitochondrial-related transcripts are significantly enriched (p = 2.1 × 10−5) early after injury, with the majority of mitochondrial transcripts showing reduced localization (Fig. 4 D). The median reduction in expression of these transcripts in the regenerating axons is 0.55 of the naive levels. The change in balance of mitochondrial transcripts suggests that there may be altered mitochondrial function in regenerating axons.
Numerous ATP synthase, H+ transporting, mitochondrial genes are downregulated in axons following injury (Atp5f1, Atp5g1, Atp5jd, Atp5b, Atp5o). Similarly, regenerating axons show decreased availability of multiple cytochrome c oxidase subunit transcripts (Cox7b, Cox5a, Cox6c, Cox8a), as well as somatic cytochrome c (Cycs), and ubiquinol cytochrome c reductase core proteins 1 and 2 (Uqcrc1, Uqcrc2) transcripts. Interestingly, whereas a majority of the mitochondrial transcripts were downregulated after injury, in particular genes related to electron transport function, several transcripts showed increased localization, such as mitofusin 1 and 2 (Mfn1, Mfn2), which are required for mitochondrial fusion and proper mitochondrial morphology.
Altogether, the alterations in transcript levels for mitochondrial genes suggest that there is a modification of mitochondrial functioning in axons following injury. However, this does not necessarily implicate shutdown or reduction of mitochondrial energy production. Indeed it may be that regenerating axons rely less on oxidative phosphorylation/electron transport as a source of ATP and may regenerate in a more glycolytic mode, analogous to the low oxygen tension that is associated with prenatal development (Rust, 1994). In addition to an energy-generation role, mitochondria are present in the presynaptic terminal where they play an important role in making calcium available for neurotransmitter release. These data suggest that there might also be a change in locally available calcium for neurotransmitter release during axonal regeneration.
Cytoskeletal and intracellular transport mRNAs change in axons following injury
Our data further revealed that transcripts showing altered localization in the regenerating axon were enriched for cytoskeletal (p = 2.5 × 10−4) and intracellular transport functions (p = 1.5 × 10−6). In both of these categories, there were more changed transcripts with decreased expression levels than increased following injury (Fig. 4 D). Additionally, the median level of change for the identified transcripts was 0.7 of the expression level in naive axons (Fig. 4 E).
Altered cytoskeletal elements included decreased localization of actin and microtubule transcripts in regenerating axons and increased localization of intermediate filament protein transcripts. Decreased actin-related transcripts included Actb, actin related protein 2/3 complex subunit 1b (Arpc1b), and Ctnnb1. Paralleling the change in actin-related transcripts, several microtubule-related transcripts showing decreased localization in regenerating axons included β-tubulins, microtubule-associated proteins, and microtubule-associated motors such as dynein light chains (Dlc2 and Dncli1) and kinesin light chain (Klc1). Interestingly, transcripts for several intermediate filament proteins are increased in regenerating axons, including multiple keratin intermediate filaments. Notably, following injury, protein expression of other intermediate filaments (i.e., vimentin and peripherin) increases in sciatic nerve axoplasm partially via local translation in the axon (Perlson et al., 2005). The case of vimentin highlights the concept that locally translated proteins may have unique functions in addition to their predicted protein function, since it was shown that following local synthesis vimentin is proteolyzed and its proteolytic fragment provides a scaffold for the importin retrograde signaling complex.
Like those for cytoskeletal elements, genes for intracellular transport function were changed in regenerating axons. For example, several members of the Ras oncogene family (Rab2, Rab31, Rab11A, Rab6A, Rab14) which are involved in protein transport and trafficking between the Golgi and ER, were changed (up and down). Some of the intracellular transport mRNAs which show altered localization in axons following injury are involved in vesicle trafficking within axons, consistent with a role in presynaptic function. Notably, the scaffolding protein Mapk8ip, a mitogen-activated protein kinase (MAPK) that binds cargo to kinesin, was decreased in regenerating cortical axons. In contrast to Mapk8ip, chondroitin sulfate proteoglycan 2 (Vcan) was increased in regenerating cortical axons, consistent with the in vivo literature demonstrating that Vcan is increased around the lesion site following a knife lesion to the cerebral cortex (Asher et al., 2002).
Together, these data suggest that there is a change in the cytoskeletal dynamics and composition in regenerating axons early after injury, as well as altered intracellular transport. These functions would be important for support of growth cones and axon path-finding, as well as for the function of presynaptic terminals during and after synaptogenesis, in the regenerating axons.
qPCR validation of upregulated and downregulated mRNAs in injured axons
qPCR was used to validate microarray data demonstrating that the mRNA pool in matured cortical axons undergoes both upregulated and downregulated change of transcript levels after injury, using independent cohorts of both regenerating and naive axons. Two genes were selected for qPCR validation: Exoc3, one of a number of secretory function genes, involved in targeting exocytotic vesicles that showed increased localization after injury, and Mapk8ip, a intracellular transport gene with decreased localization in regenerating axons that binds cargo to kinesin (Hirokawa and Takemura, 2005). Consistent with the microarray data demonstrating a 2.2-fold increase in axonal Exoc3 transcripts following axotomy, qPCR revealed an increase in Exoc3 mRNA localization in injured axons (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Similarly, consistent with the microarray data showing a 0.6-fold reduction in localized Mapk8ip transcript following injury, qPCR data show that Mapk8ip mRNA is decreased in regenerating axons (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Together with the microarray data, these results demonstrate that matured cortical axons contain a relatively large and diverse pool of readily available mRNA, and that this pool of axonal mRNAs in matured cortical axons is fundamentally altered following injury.
Validation of axonally localized transcripts using FISH
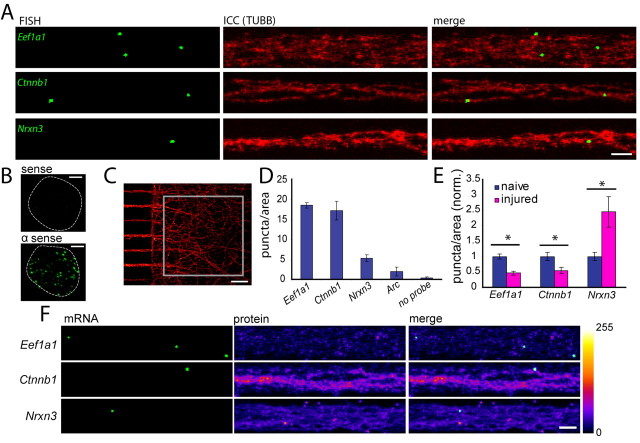
To further validate the microarray results, the localization of transcripts to matured uninjured axons was verified using a new highly specific FISH method. Three candidate genes were selected which have functions in translation, cytoskeleton, and cell–cell signaling (Eef1a1, Ctnnb1, and Nrxn3, respectively). Eef1a1 is required for translation and has been suggested to be locally synthesized in dendrites following LTP stimulation (Tsokas et al., 2005). Ctnnb1 is required for presynapse formation, linking the cell surface protein N-cadherin to the actin cytoskeleton (Kwiatkowski et al., 2007). Nrxn3, a presynaptic cell surface protein that has many isoforms, including a secreted isoform, is involved in synaptic function (Südhof, 2008). Our microarray analysis showed that Eef1a1 and Ctnnb1 mRNAs are reliably localized in naive axons and that their transcript levels are reduced in injured axons (23% and 5.5%, respectively, of naive levels, average of two probe sets). Conversely, Nrxn3 mRNA was not identified as a reliably localized transcript in naive axons, but the transcript level was increased considerably in the injured axons (253%) according to our microarray results. FISH was undertaken to assess detection and expression levels of these target mRNAs in naive and regenerating cortical axons, as predicted by the microarray data.
FISH for the target mRNAs was first combined with immunocytochemistry for tubulin to visualize the target mRNA with respect to the axon cytoskeleton (Fig. 5 A). A dense punctate pattern of staining was visible with FISH for all target transcripts, suggesting a high specificity of hybridization as well as extremely low background. The absence of signal detected with a sense β-catenin probe set further demonstrates the specificity of the hybridization (Fig. 5 B). The puncta were fairly evenly spread out in both the naive and injured axons (data not shown). Fluorescently labeled puncta localized adjacent to the tubulin staining (Fig. 5 A). The levels of puncta were quantified for multiple target mRNAs to compare the relative expression levels predicted by the microarray data (Fig. 5 D). Consistent with the microarray data, FISH analysis confirmed that Eef1a1 and Ctnnb1 were reliably localized in naive axons, whereas Nrxn3, which did not meet our criteria as a reliably localized transcript in naive axons, had few puncta, as predicted. Similarly, Arc, which is a dendritically localized mRNA and was not identified as reliably localized in axons, showed low to negligible levels in axons.
Figure 5.
Fluorescence in situ hybridizations for eukaryotic elongation factor 1α1, β-catenin, and neurexin III shows the localization of their transcripts in matured axons, their change following injury, and validates the findings of the microarray studies. A, Fluorescent micrographs of FISH results showing mRNA puncta (green) present in cortical axons. The axons are also immunolabeled with tubulin (TUBB) (red). ICC, Immunocytochemistry. Scale bar, 5 μm. B, Fluorescent micrographs of cell bodies following FISH for both sense and antisense probes for Ctnnb1. White dashed lines delineate the boundary of the cell body. Scale bar, 5 μm. C, Boxed region is an example of the axonal region used for quantification of transcript levels in (D) and (E). Scale bar, 50 μm. D, Number of puncta per area of naive axons, showing a similar mRNA expression level trend as the microarray analysis. E, Puncta levels changed significantly (*) following injury (normalized to average puncta level in naive axons) (Eef1a1 p = 0.00018; Ctnnb1 p = 0.010; Nrxn3 p = 0.019; two-tailed Student's t test). The percentage of area of β-tubulin protein was not significantly changed between naive and injured axons (data not shown). F, Localization of mRNA and corresponding protein. The protein signal is pseudocolored using the intensity lookup table, “Fire.” Scale bar, 5 μm. Error bars indicate SEM.
Next, microarray data revealing changes in transcript levels between naive and injured axons were validated by FISH. Again, the FISH results parallel the microarray data, showing significantly decreased levels of Eef1a1 (46%) and Ctnnb1 (54%) transcripts in axons following injury, and increased levels of Nrxn3 mRNA in injured axons (245%), compared with naive levels (Fig. 5 E).
Finally, to simultaneously visualize the localization of both the target mRNA and protein, FISH for the target mRNAs was combined with immunocytochemistry, using specific antibodies for EEF1A1, CTNNB1, and NRXN3 (Fig. 5 F). Ctnnb1 mRNA shows some colocalization with its protein and there appears to be a lesser degree of colocalization for Eef1a1. Overall, there does not appear to be a large increase in protein localized to the target mRNA in either the naive or injured axons (data not shown). Since it is generally thought that during mRNA transport transcripts are translationally silenced and then activated when they reach their target (Besse and Ephrussi, 2008), it is possible that at the time of fixation many of the transcripts have not reached their target and therefore are not actively translated. Nonetheless, it appears that there are numerous transcripts available in case of subsequent protein demand.
Combined, the FISH results show that Eef1a1, Ctnnb1, and Nrxn3 transcripts are localized to axons, validating the microarray analysis. The FISH transcript levels reflect the same trend as indicated by the microarray expression levels. These results also validate our microarray analysis of changing levels of transcript following injury.
Discussion
We have purified mRNA from naive, matured cortical axons for the first time and identified the presence of >300 mRNA transcripts. We demonstrate that the transcripts are axonal in nature and that many of the transcripts present in naive CNS axons overlap with those previously identified in PNS injury-conditioned DRG axons (Willis et al., 2005, 2007). Whereas the similarity between the naive CNS axons and regenerating PNS axons (91 common transcripts) is quite striking, when we compared the most abundant transcripts in regenerating CNS axons with those identified in regenerating DRG axons (Willis et al., 2007), there were only six common transcripts. There may be several technical reasons as to why there are differences between regenerating cortical and DRG axons, including, but not limited to, (1) differences in time points following injury when we isolated the axonal RNA (2 d following injury for cortical axons compared with 7 d following injury for DRG axons), (2) differences in injury methods (injury-conditioning for the DRGs consisted of a sciatic nerve crush in the intact animal), and (3) differences in microarray technology or analysis. Another explanation is that the differences between regenerating cortical and DRG axons may be due to a fundamental difference between the two types of axons and their regenerative capacity.
In the naive axon, transcripts related to translation were highly enriched within the axonal mRNA pool, supporting the idea that the machinery necessary for protein synthesis exists within CNS axons. Translational mRNAs have similarly been identified in other axons, such as PNS injury-conditioned DRGs, where local protein synthesis is necessary for axonal regeneration after injury (Verma et al., 2005). Unlike injury-conditioned DRG axons, our results suggest that the capacity for local translation is not appreciably altered in regenerating CNS axons. Although it should be noted that our results were obtained using neurons cultured in vitro and these results need to be confirmed in vivo.
In addition to translation, transcripts related to intracellular transport and the cytoskeleton were enriched in naive cortical axons. Some of the transport transcripts included those that are known to be axonally localized and involved in transport of axonal cargo and mitochondria, including transcripts of microtubule-associated motors (e.g., dynein and kinesin). Many of the cytoskeletal transcripts that we identified in cortical axons have been previously reported in non-CNS axons. After injury, the mRNA pool shifts, including a decreased in the localization of actin and microtubule transcripts, accompanied by an increase in the localization of transcripts for intermediate filament proteins. These data suggest that there is a change in the cytoskeletal dynamics and composition in regenerating axons. It will be critical to determine whether the cytoskeletal and intracellular transport genes are ultimately increased in axons to normal levels (or beyond) at later time points during regeneration, to support axonal growth. The local translation of these transcripts would allow axons to rapidly generate proteins where they may be needed for normal function and during regeneration without the delay that would be incurred by transport from the cell body.
Several genes are present in matured cortical axons that mediate cell–cell signaling and secretion, as well as genes involved in axonal targeting and synaptic function. Interestingly many of the transcripts that show increased localization after injury regulate vesicle trafficking, exocytosis and neurotransmitter release, including synapsin 3, neurexin 3, nectin-1, and multiple ephrin ligands. Together, these data suggest that there is increased capacity for axonal outgrowth and targeting in regrowing axons early after injury, and increased support for both synapse formation and presynaptic function.
We also identified mitochondrial transcripts as highly enriched within the mRNA pool identified in naive axons. Mitochondria have the ability to synthesize only 13 of their own proteins, and import hundreds of proteins encoded by nuclear genes (Scarpulla, 2008). It is commonly held that mitochondrial biogenesis occurs when the mitochondria are in the neighborhood of the nucleus. However, nuclear-encoded mitochondrial genes have been reported in giant squid axons as well as presynaptic terminals and axons of primary sympathetic neurons, predicting that these transcripts are locally translated in regions of high energy demand (Gioio et al., 2001; Hillefors et al., 2007; Aschrafi et al., 2008). Given the length of many axons, it seems plausible that mitochondrial proteins encoded by nuclear genes may need to be made locally within the axon. Interestingly, numerous mitochondrial transcripts show decreased axonal localization after injury, in particular ATPases involved in the electron transport chain, whereas mitochondrial transcripts related to mitochondrial fusion and proper mitochondrial morphology show increased localization. The decreased localization of electron transport chain transcripts suggests that neurons may regrow in a more glycolytic mode, which may be analogous to the low oxygen tension that is associated with prenatal development (Rust, 1994). Our data demonstrating the presence of nuclear encoded mitochondrial genes in cortical axons is a novel finding, and suggests that mitochondria maintain a level of autonomy within the axon that allows mitochondria to be maintained and regulated under local control mechanisms.
In summary, our data demonstrate that CNS axons contain many mRNAs of diverse functions, suggesting that, like invertebrate and PNS axons, CNS axons may synthesize proteins locally. It will be essential to demonstrate that the mRNAs that we identified in cortical axons are translated, and to elucidate the mechanisms regulating their translation. This study provides the first evidence that matured cortical axons contain their own mRNA that can serve to support translation, transport, the cytoskeleton, mitochondrial maintenance, and synaptic function within the axonal compartment.
Footnotes
This work was supported by a Christopher and Dana Reeve Foundation Grant and the National Institute of Aging (AG00538). A.M.T. was partially supported by a National Institutes of Health Ruth L. Kirschstein National Research Service Award Predoctoral Fellowship Award. A.M.T. thanks Erin Schuman [Caltech/Howard Hughes Medical Institute (HHMI)] for use of her laboratory for the FISH experiments and Lin Chen (Caltech/HHMI) for preparation of the E18 and P0 cultures for the FISH experiments. University of California, Irvine owns the patent for the multicompartment microfluidic chambers.
References
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22:2225–2236. doi: 10.1523/JNEUROSCI.22-06-02225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, Reichardt LF. Role of beta-catenin in synaptic vesicle localization and presynaptic assembly. Neuron. 2003;40:719–731. doi: 10.1016/s0896-6273(03)00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Beaudoin GM., 3rd Con-nectin axons and dendrites. J Cell Biol. 2006;174:7–9. doi: 10.1083/jcb.200606024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Blizzard CA, Haas MA, Vickers JC, Dickson TC. Cellular dynamics underlying regeneration of damaged axons differs from initial axon development. Eur J Neurosci. 2007;26:1100–1108. doi: 10.1111/j.1460-9568.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci. 2004;5:521–531. doi: 10.1038/nrn1428. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Choe CU, Ehrlich BE. The inositol 1,4,5-trisphosphate receptor (IP3R) and its regulators: sometimes good and sometimes bad teamwork. Sci STKE. 2006;2006:re15. doi: 10.1126/stke.3632006re15. [DOI] [PubMed] [Google Scholar]
- Chun JT, Gioio AE, Crispino M, Giuditta A, Kaplan BB. Differential compartmentalization of mRNAs in squid giant axon. J Neurochem. 1996;67:1806–1812. doi: 10.1046/j.1471-4159.1996.67051806.x. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1{alpha} mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci U S A. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Junutula JR, De Maziére AM, Peden AA, Ervin KE, Advani RJ, van Dijk SM, Klumperman J, Scheller RH. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, Kwon HB, Chiu CQ, Deng L, Castillo PE, Südhof TC. RIM1alpha and RIM1beta are synthesized from distinct promoters of the RIM1 gene to mediate differential but overlapping synaptic functions. J Neurosci. 2008;28:13435–13447. doi: 10.1523/JNEUROSCI.3235-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman R, Banker G, Steward O. Development of subcellular mRNA compartmentation in hippocampal neurons in culture. J Neurosci. 1994;14:1130–1140. doi: 10.1523/JNEUROSCI.14-03-01130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig E, Martin R, Titmus M, Sotelo-Silveira JR. Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J Neurosci. 2000;20:8390–8400. doi: 10.1523/JNEUROSCI.20-22-08390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Weis WI, Nelson WJ. Catenins: playing both sides of the synapse. Curr Opin Cell Biol. 2007;19:551–556. doi: 10.1016/j.ceb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- Lim BK, Matsuda N, Poo MM. Ephrin-B reverse signaling promotes structural and functional synaptic maturation in vivo. Nat Neurosci. 2008;11:160–169. doi: 10.1038/nn2033. [DOI] [PubMed] [Google Scholar]
- Maehlen J, Schröder HD, Klareskog L, Olsson T, Kristensson K. Axotomy induces MHC class I antigen expression on rat nerve cells. Neurosci Lett. 1988;92:8–13. doi: 10.1016/0304-3940(88)90733-1. [DOI] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E E Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe N, Shimizu K, Ozaki-Kuroda K, Nakanishi H, Morimoto K, Takeuchi M, Katsumaru H, Murakami F, Takai Y. Contacts between the commissural axons and the floor plate cells are mediated by nectins. Dev Biol. 2004;273:244–256. doi: 10.1016/j.ydbio.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Oliveira AL, Thams S, Lidman O, Piehl F, Hökfelt T, Kärre K, Lindå H, Cullheim S. From The Cover: A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proc Natl Acad Sci U S A. 2004;101:17843–17848. doi: 10.1073/pnas.0408154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson T, Kristensson K, Ljungdahl A, Maehlen J, Holmdahl R, Klareskog L. Gamma-interferon-like immunoreactivity in axotomized rat motor neurons. J Neurosci. 1989;9:3870–3875. doi: 10.1523/JNEUROSCI.09-11-03870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Microfluidic culture platform for neuroscience research. Nat Protocols. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Piper M, Holt C. RNA translation in axons. Annu Rev Cell Dev Biol. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of Process-Localized mRNAs from Cultured Rodent Hippocampal Neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SW, Taylor AM, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Patterned cell culture inside microfluidic devices. Lab Chip. 2005;5:102–107. doi: 10.1039/b403091e. [DOI] [PubMed] [Google Scholar]
- Rust RS. Energy metabolism of developing brain. Curr Opin Neurol. 1994;7:160–165. doi: 10.1097/00019052-199404000-00013. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira J, Crispino M, Puppo A, Sotelo JR, Koenig E. Myelinated axons contain beta-actin mRNA and ZBP-1 in periaxoplasmic ribosomal plaques and depend on cyclic AMP and F-actin integrity for in vitro translation. J Neurochem. 2008;104:545–557. doi: 10.1111/j.1471-4159.2007.04999.x. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Microfluidic multicompartment device for neuroscience research. Langmuir. 2003;19:1551–1556. doi: 10.1021/la026417v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik-Mo EO, Oltedal L, Hoivik EA, Kleivdal H, Eidet J, Davanger S. Sec6 is localized to the plasma membrane of mature synaptic terminals and is transported with secretogranin II-containing vesicles. Neuroscience. 2003;119:73–85. doi: 10.1016/s0306-4522(03)00065-4. [DOI] [PubMed] [Google Scholar]
- von Gertten C, Flores Morales A, Holmin S, Mathiesen T, Nordqvist AC. Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci. 2005;6:69. doi: 10.1186/1471-2202-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Zorn AM, Krieg PA, Bittner GD. Medium weight neurofilament mRNA in goldfish Mauthner axoplasm. Neurosci Lett. 1996;213:83–86. doi: 10.1016/0304-3940(96)12860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Twiss JL. The evolving roles of axonally synthesized proteins in regeneration. Curr Opin Neurobiol. 2006;16:111–118. doi: 10.1016/j.conb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]