Abstract
Metallopeptides containing both the complex Cu2+-glycyl-glycyl-histidine (Cu-GGH) and the sequence WRWYCR were shown to possess antimicrobial activity against a variety of pathogenic bacteria, as well as bind to and cleave a variety of nucleic acids, suggesting potential mechanisms for antimicrobial activity that involve binding and/or irreversible cleavage of bacterial nucleic acids.
Current major classes of antimicrobial agents include β-lactams, aminoglycosides, macrolides, tetracyclines, fluoroquinolones, and various peptides,1-3 with the evolution of drug-resistance in pathogenic bacteria as a rising concern.4-7 The most widely used modes of action include inhibition of protein biosynthesis and cell wall formation through binding of ribosomal RNA and proteins involved in cell wall synthesis, respectively.8 Since nucleic acids (both DNA and RNA) mediate many critical cellular processes that include the storage, transmission, and translation of genetic information, binding of specific nucleic acid targets has been a common mechanism of action employed by many antibiotics. However, to date, strikingly few of the vast number of potential nucleic acid targets in bacteria have been exploited. Due to the rise of bacterial drug-resistance there is a clear general need for new antibiotics that function on distinct targets and through novel modes of action.
Recent work demonstrated antimicrobial activity for peptide WRWYCR.2 The activity of this peptide was ascribed to binding and trapping of bacterial Holliday junctions (HJ DNA) that arise during homologous recombination, a fundamental cellular process.1, 2, 9 Herein we investigate the antimicrobial activity, nucleic acid binding, and nucleolytic properties of peptide WRWYCR-NH2, as well as derivative metallopeptides containing both the catalytically active N-terminal complex Cu2+-GlyGlyHis (Cu-GGH)10-20 and the WRWYCR targeting peptide, and test for antimicrobial activity against various pathogens (Table 1).
Table 1. Antimicrobial activity and nucleic acid binding of metallopeptides containing the catalytic Cu-GGH complex and/or the nucleic acid-binding sequence WRWYCR.
| Complex | Minimal inhibitory concentration (μM) for bacteriocidal activity | Apparent KD value (μM) for nucleic acid binding | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.coli | S. nterica | MRSA | P. aeruginosa | K. pneumoniae | E. faecium | A. baumanii | Fl-16S A-site RNA | Fl-RRE IB RNA | Fl-HCV IIB RNA | Fl-HCV IV RNA | pUC19 plasmid DNA (bp) | |
| 1 | 17 | 8 | 66 | 33 | 66 | 8 | 132 | 15 ± 1 | 11.6 ± 0.6 | 10.3 ± 0.4 | 12.6 ± 0.6 | 4.7 ± 0.4 |
| Cu-2 | 3 | 6 | 3 | 25 | 25 | 25 | 25 | 11 ± 2 | 9 ± 1 | 10 ± 2 | 10 ± 2 | 4.1 ± 0.5 |
| Cu-3 | 3 | 1 | 11 | 11 | 11 | 3 | 11 | 13 ± 3 | 7.0 ± 0.6 | 8.8 ± 0.8 | 12 ± 1 | 2.5 ± 0.8 |
| Free Cu(II) | >2016 | >2016 | >2016 | >2016 | >2016 | >2016 | >2016 | --a | --a | --a | --a | n.d. |
| Cu-GGH | >476 | >476 | >476 | >476 | >476 | >476 | >476 | 230 ± 40 | 320 ± 30 | 400 ± 100 | 260 ± 80 | n.d. |
| ampicillin | 23 | 92 | 183 | >367 | >367 | >367 | >367 | n.d. | n.d. | n.d. | n.d. | n.d. |
| kanamycin | 8 | 1 | 264 | 33 | 66 | >264 | >264 | n.d. | n.d. | n.d. | n.d. | n.d. |
Although binding of free Cu2+ to Fl-RNA constructs was apparent (micromolar range) from fluorescence-monitored titration progress curves, the data were too complex for fitting to a simple binding equation. n.d. = not determined. Specific strains for the bacteria used are summarized in Supp. Info. No toxicity was observed against human cells.
The metallopeptides had favorable bacteriocidal activity, with minimal inhibitory concentrations in the low micromolar range (Table 1) and KD values for DNA- and RNA-binding in the same range. This suggests that antimicrobial activity may have arisen from relatively nonselective binding of a variety of nucleic acids, rather than specific binding to HJ DNA. While the reported solution binding affinity between the dimer form (disulphide form) of peptide WRWYCR and HJ DNA is ∼3 orders of magnitude stronger (nanomolar KD)2 than the observed binding affinities between the monomer form of WRWYCR and the DNA and RNA constructs used in our experiments (micromolar KD values),1, 9 the relatively low in vivo concentration of cellular HJ DNA, relative to all other nucleic acid targets, as well as a decreased propensity for disulphide bonding in the cytosol (reducing), makes binding of HJ DNA a less probable mechanism of antimicrobial activity. Rather, it appears that both monomeric WRWYCR and derivative metallopeptides bind nonselectively, but with moderately high affinity to a wide variety of DNA and/or RNA targets, or even nucleoside triphosphates, as observed for other cationic/hydrophobic peptides.21 These widespread binding events may cause inhibition of critical cellular processes, with commensurate decreases in cell viability.
An important observation in our experiments was that incorporation of an N-terminal Cu-GGH ATCUN (amino terminal copper/nickel) complex resulted in enhanced activity against certain pathogenic strains, beyond that expected based on the antimicrobial activity of either free Cu2+ or Cu-GGH, suggesting a unique mechanism of activity. Furthermore, in vitro experiments demonstrated that the Cu-GGH-containing metallopeptides Cu-GGHGWRWYCR-NH2 (Cu-2) and Cu-GGHWRWYCRGGK-NH2 (Cu-3) possessed catalytic nuclease activity toward a variety of nucleic acid targets (both plasmid DNA and several structured RNA targets), while the peptide WRWYCR-NH2 (1) did not. These results highlight nuclease activity as a probable mechanism of enhanced antimicrobial activity for the metallopeptides.10, 11, 14, 15, 17, 22-24
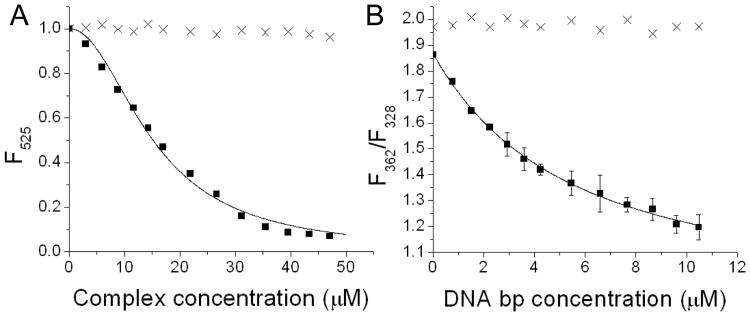
The DNA-binding affinities of peptide WRWYCR-NH2, as well as the metallopeptides Cu-GGHGWRWYCR-NH2 and Cu-GGHWRWYCRGGK-NH2, were determined by performing titrations of each monomer with supercoiled pUC19 plasmid DNA and monitoring the intrinsic tryptophan fluorescence from each species. Upon DNA-binding, an intrinsic blueshift occurred in the fluorescence emission of each peptide that was related to a decrease in the local dielectric constant.19, 25, 26 The blue-shift in tryptophan fluorescence emission was plotted as a function of DNA concentration, and the resulting plots were fit to a binding equation in order to determine the apparent dissociation constant (KD) for each interaction (Figure 1). Control experiments, in which buffer was added instead of DNA, did not give a blue-shift in tryptophan fluorescence; a control experiment with free tryptophan (instead of the full peptide), revealed a similar blue-shift in fluorescence upon DNA-binding, although with a higher apparent KD, as expected (supporting information).
Figure 1.
(A) Titration of Fl-16S A-site RNA with either 1 (■) or the same volumes of buffer lacking peptide (×), monitored by a decrease in fluorescence intensity at 525 nm. (B) Titration of 1 with either supercoiled plasmid DNA (■) or the same volumes lacking DNA (×), monitored by a blueshift in the fluorescence emission of 1. All apparent KD values are shown in Table 1.
RNA-binding affinities were determined in a similar manner. Four different RNAs, each containing a 5′-fluorescein label, were separately titrated with each species, and a decrease in the intrinsic fluorescein emission was plotted as a function of peptide concentration, allowing apparent KD values to be determined (Figure 1). The fluorescein-labelled RNAs were: a stem loop RNA derived from E. coli ribosomal 16S A-site RNA (Fl-16S A-site RNA), HIV RRE RNA stem loop IIB (Fl-RRE IIB RNA), and stem loops IIB and IV from HCV IRES RNA (Fl-HCV IIB RNA and Fl-HCV IV RNA, respectively). Control experiments were performed using free fluorescein (lacking attached RNA) and/or Cu-GGH (lacking the nucleic acid-targeting sequence), and strong binding was not observed (supporting information). Binding of RNA by complexes containing WRWYCR typically appeared to occur with an apparent stoichiometry between 1 and 2, as seen by the sigmoidal shape of most RNA-binding curves for complexes containing WRWYCR (supporting information).
All apparent DNA- and RNA-binding affinities (apparent KD values) are summarized in Table 1 alongside minimal inhibitory concentrations for antimicrobial activity (MIC values), for the same complexes. A monomeric state was ensured for each species by the presence of the reducing agent TCEP (1 mM) in all binding experiments. The apparent KD values for nucleic acid-binding by species containing WRWYCR were strikingly similar to the MIC values for antimicrobial activity, although the attachment of Cu-GGH to peptides containing WRWYCR was found to lower MIC values (increase antimicrobial activity) while apparent KD values were relatively unchanged. Significantly, no toxicity was observed against control human cells.
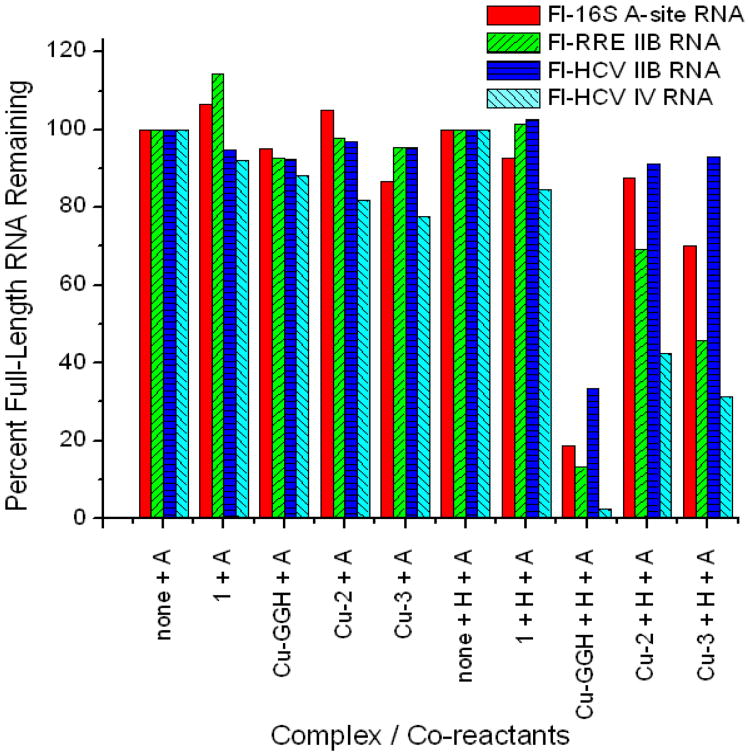
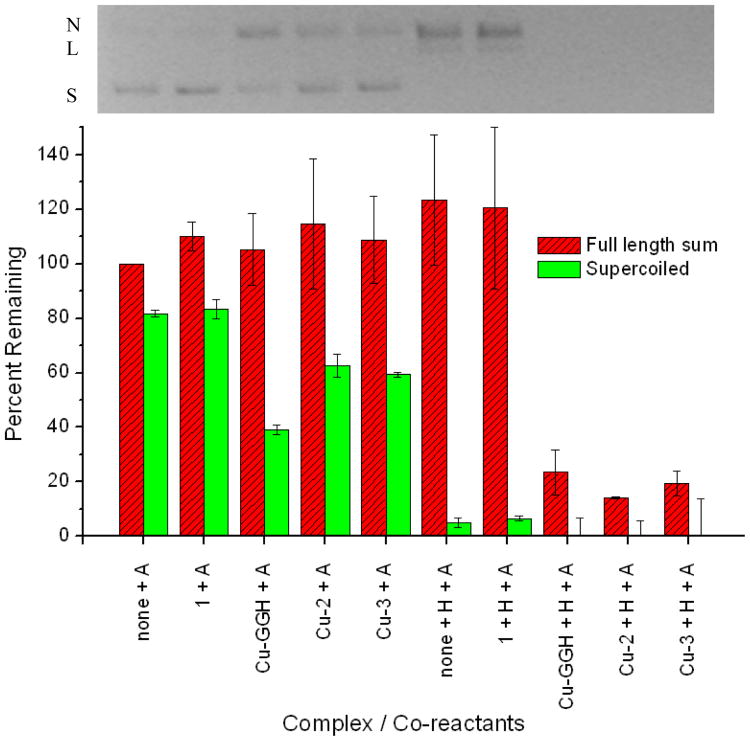
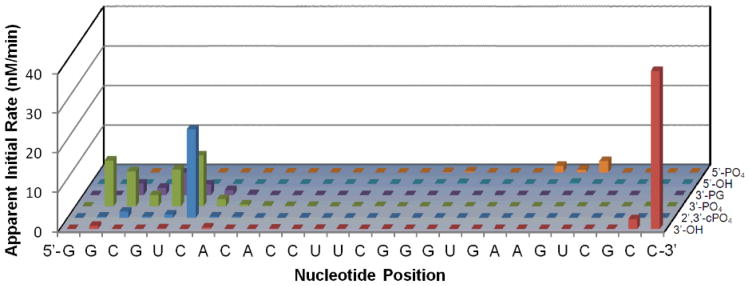
In addition to binding nucleic acids with relatively high affinity, Cu-2 and Cu-3 were observed to possess significant levels of ribonuclease and nuclease activity (Figures 2 to 4), due to incorporation of the catalytic Cu-GGH complex and consistent with our previous studies of ribonuclease mimics, where cleavage appeared to be mediated by a metal-associated reactive oxygen species.10, 11, 14, 15, 17, 22 Cleavage of various stem loop RNAs by Cu-2 and Cu-3 was observed in the presence of the mild biological reducing agent ascorbate, and this cleavage was augmented by addition of the oxidant H2O2 (Figure 2)—the biological redox co-reactants (including ambient O2) provide the driving forces for the catalytic activity of Cu-GGH. Mass spectrometric analysis of RNA cleavage products defined the apparent initial cleavage rates and also identified oxidative cleavage pathways (primarily hydrogen abstraction)24 and distributions of reaction products (Figure 3 and supporting information). Interestingly, fluorophore-labeled Fl-HCV IV RNA appeared to be most susceptible to cleavage, among the stem loop RNAs tested, possibly due to a more favorable alignment of its backbone with the catalytic Cu-GGH center within the RNA-metallopeptide complex, although the exact reason remains unclear. Nicking of plasmid DNA by Cu-2 and Cu-3 also occurred readily in the presence of ascorbate, and was again enhanced significantly by the presence of H2O2, which led to a dramatic increase in linearization and overall cutting of DNA (Figure 4). Peptide 1, which lacks the Cu-GGH complex but binds all tested nucleic acids with a similar affinity as Cu-2 and Cu-3, did not promote cleavage of either RNA or DNA (Figures 2 and 4, respectively).
Figure 2.
Cleavage of stem-loop RNA constructs by each metallopeptide, in the presence of co-reactants: 1 mM ascorbate (A), or 1 mM H2O2 + 1 mM ascorbate (H + A). The percentage of full-length RNA remaining after each 5.5 h incubation was quantified by PAGE analysis. Initial concentrations were 1 μM RNA, 1 μM complex, 1 mM each co-reactant.
Figure 4.
Nicking and total cleavage of plasmid DNA by each metallopeptide, in the presence of co-reactants. The gel above corresponds to the data in the bar plot below (average of duplicate trials). After 4 h incubation, cleavage resulted in nicking of plasmid DNA, followed by linearization, and finally to a distribution of smaller fragments (not shown above). Initial concentrations were 10 μM base pairs DNA (defined as 100%), 0.1 μM complex, and 1 mM each co-reactant. ‘Full length sum’ is the sum of the percentages of supercoiled (S), nicked (N), and linear (L) forms of DNA.
Figure 3.
Cleavage of Fl-16S A-site RNA by Cu-2 and co-reactants, analyzed by MALDI-TOF MS, showing the apparent initial rates of formation of cleavage fragments containing the listed nascent overhangs at each position within the sequence (front to back: 3′-hydroxyl, 2′,3′-cyclic phosphate, 3′-phosphate, 3′-phosphoglycolate, 5′-hydroxyl, 5′-phosphate). Mass spectra are shown in the supporting information.
The observation that species containing WRWYCR bind non-selectively, and with moderately high affinity, to a variety of nucleic acids is most likely a result of the combination of hydrophobic and positively-charged amino acids within the sequence.21 These residues provide a combination of potential π-stacking and salt-bridge contacts, respectively, ideal for binding of nucleic acids, which contain hydrophobic bases and negatively-charged backbones. Furthermore, it is likely that certain structured nucleic acids, such as HJ DNA and many RNAs, provide unique alignments of bases and phosphate groups that may provide higher-affinity interactions with WRWYCR. Regardless of the selectivity toward different nucleic acids, it is apparent from this work that almost all nucleic acid polymers are prone to at least moderately high-affinity binding by WRWYCR.
Incorporation of the catalytic Cu-GGH complex into peptides containing the sequence WRWYCR endows these nucleic acid-binding peptides with nucleolytic activity. The resulting irreversible cleavage of cellular DNA and RNA is expected to provide a far more potent challenge to cell viability than reversible binding. In fact, enhanced antimicrobial activity was observed for the Cu-GGH-containing Cu-2 and Cu-3, relative to 1, for most of the pathogenic bacteria tested (Table 1). Therefore, it is very likely that these enhancements in antimicrobial activity were mediated by the introduction of nucleolytic activity.
Other possible explanations for observed enhancements in antimicrobial activity for Cu-2 and Cu-3, relative to 1, include oxidative reactivity with protein targets or redox cofactors and/or initiation of a cellular oxidative stress response due to the oxidative chemistry promoted by Cu-GGH.28 Notably, neither Cu-GGH nor free Cu2+ possessed any significant antimicrobial activity within the concentration range used, demonstrating that attachment to WRWYCR is likely required for cellular uptake and/or in vivo activity of the Cu-GGH domain. Regardless, the demonstrable affinity toward, and cleavage of, various RNA and DNA substrates highlights the possibility of binding and cleavage of nucleic acids as mechanisms of antimicrobial activity.
Metal-based catalytic antibiotics, such as those presented here increase the likelihood for antimicrobial activity promoted by multiple-mechanisms, such as binding and cleavage of multiple nucleic acid targets and/or possible initiation of oxidative stress mechanisms.27-29 Antibiotics with multiple mechanisms of activity have the potential for higher efficacy and broader-spectrum activity, since any given strain of bacteria most likely contains at least one viable drug target. Furthermore, bacteria treated with such molecules are less likely to develop resistance, since a larger number of pathways would need to be circumvented to override their effect. The unique strategies presented herein will likely prove useful for development of other novel advanced antibiotics.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [46 and AA016712]. J.C.J. was supported by an NIH Chemistry/Biology Interface training grant (T32 GM08512).
Footnotes
Electronic Supplementary Information (ESI) available.
Notes and references
- 1.Kepple K, Patel N, Salamon P, Segall A. Nucleic Acids Res. 2008;36:5319–5334. doi: 10.1093/nar/gkn512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunderson C, Boldt J, Authement R, Segall A. J Bacteriol. 2009;191:2169–2176. doi: 10.1128/JB.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izadpanah A, Gallo R. J Am Acad Dermatol. 2005;52:381–390. doi: 10.1016/j.jaad.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole K. Cellular and Molecular Life Sciences. 2004;61:2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J, Wright G. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 7.Holmberg SD, Osterholm MT, Senger KA, Cohen ML. N Engl J Med. 1984;311:617–622. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 8.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 9.Kepple K, Boldt J, Segall A. PNAS. 2005;102:6867–6872. doi: 10.1073/pnas.0409496102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford S, Cowan JA. Chem Comm. 2012 doi: 10.1039/c2cc17377h. [DOI] [PubMed] [Google Scholar]
- 11.Cowan JA. Pure Appl Chem. 2008;80:1799–1810. [Google Scholar]
- 12.Gokhale NH, Bradford S, Cowan JA. J Am Chem Soc. 2008;130:2388–2389. doi: 10.1021/ja0778038. [DOI] [PubMed] [Google Scholar]
- 13.Jin Y, Cowan JA. J Am Chem Soc. 2005;127:8408–8415. doi: 10.1021/ja0503985. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Cowan JA. J Am Chem Soc. 2006;128:410–411. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Cowan JA. J Biol Inorg Chem. 2007;12:637–644. doi: 10.1007/s00775-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, Lewis MA, Gokhale NH, Long EC, Cowan JA. Journal of the American Chemical Society. 2007;129:8353–8361. doi: 10.1021/ja0705083. [DOI] [PubMed] [Google Scholar]
- 17.Joyner JC, Cowan JA. J Am Chem Soc. 2011;133:9912–22. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyner JC, Hocharoen L, Cowan JA. J Am Chem Soc. 2011;134:3396–3410. doi: 10.1021/ja208791f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joyner JC, Keuper KD, Cowan JA. Dalton Trans. 2012;41:6567–6578. doi: 10.1039/c2dt00026a. [DOI] [PubMed] [Google Scholar]
- 20.Joyner JC, Reichfield J, Cowan JA. J Am Chem Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilpert K, McLeod B, Yu J, Elliott M, Rautenbach M, Ruden S, Bürck J, Muhle-Goll C, Ulrich A, Keller S, Hancock R. Antimicrob Agents Chemother. 2010;54:4480–4483. doi: 10.1128/AAC.01664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford S, Kawarasaki Y, Cowan JA. J Inorg Biochem. 2009;103:871–875. doi: 10.1016/j.jinorgbio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Hocharoen L, Cowan JA. Chem Eur J. 2009;15:8670–8676. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joyner JC, Keuper K, Cowan JA. Nucl Acids Res. 2012 doi: 10.1093/nar/gks811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eftink MR, Ghiron CA. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 26.Vivian JT, Callis PR. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meggers E. Chem Commun. 2009:1001–1010. doi: 10.1039/b813568a. [DOI] [PubMed] [Google Scholar]
- 28.Betanzos-Lara S, Liu Z, Habtemariam A, Pizarro A, Qamar B, Sadler P. Angew Chem Int Edit. 2012;51:3897–3900. doi: 10.1002/anie.201108175. [DOI] [PubMed] [Google Scholar]
- 29.Sasmal P, Streu C, Meggers E. Chem Comm. 2013 doi: 10.1039/C2CC37832A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.