Abstract
Neurons must cope with extreme membrane trafficking demands to produce axons with organelle compositions that differ dramatically from those of the cell soma and dendrites; however, the mechanism by which they accomplish this is not understood. Here we use electron microscopy and quantitative imaging of tagged organelles to show that Caenorhabditis elegans axons lacking UNC-16 (JIP3/Sunday Driver) accumulate Golgi, endosomes, and lysosomes at levels up to 10-fold higher than wild type, while ER membranes are largely unaffected. Time lapse microscopy of tagged lysosomes in living animals and an analysis of lysosome distributions in various regions of unc-16 mutant axons revealed that UNC-16 inhibits organelles from escaping the axon initial segment (AIS) and moving to the distal synaptic part of the axon. Immunostaining of native UNC-16 in C. elegans neurons revealed a localized concentration of UNC-16 at the initial segment, although UNC-16 is also sparsely distributed in distal regions of axons, including the synaptic region. Organelles that escape the AIS in unc-16 mutants show bidirectional active transport within the axon commissure that occasionally deposits them in the synaptic region, where their mobility decreases and they accumulate. These results argue against the long-standing, untested hypothesis that JIP3/Sunday Driver promotes anterograde organelle transport in axons and instead suggest an organelle gatekeeper model in which UNC-16 (JIP3/Sunday Driver) selectively inhibits the escape of Golgi and endosomal organelles from the AIS. This is the first evidence for an organelle gatekeeper function at the AIS, which could provide a regulatory node for controlling axon organelle composition.
NEURONS have a unique cell biology that presents daunting membrane trafficking challenges. For example, they must selectively transport two classes of regulated secretory vesicles (synaptic vesicles and dense core vesicles) long distances into axons, but only after the vesicles have completed their maturation process in the cell soma, during which they arise from, and interact with, other organelles in the soma. Neurons must also restrict, or even prevent, the flow of some organelles, such as Golgi, lysosomes, and endosomes, into the distal synaptic region of axons, which are relatively devoid of these organelles compared to cell somas. However, under special conditions, such as the need for axon repair or growth, neurons may require these organelles in axons. The potential hazards of excessive organelle transport into axons may include organelle traffic jams within narrow axons, reduced synaptic vesicle production as synaptic vesicle proteins are transported away from the cell soma before they are assembled into mature vesicles, and the disruption of membrane trafficking pathways in the synaptic region of axons caused by the inappropriate presence of cell soma organelles.
A crucial regulatory domain for controlling axon composition is the region at or near the junction of the cell soma and axon, designated the axon initial segment (AIS). The available evidence suggests that this region is not a uniform structure, but rather a cluster of specialized subdomains that perform at least two major functions (Grubb and Burrone 2010). First, in animals with sodium channels, this region serves as the site of action potential initiation. Second, it provides a barrier or filtering function for establishing the molecular composition of axons. The barrier/filtering function is known to affect the movement of molecules along the plasma membrane (Winckler et al. 1999). However, a role for the AIS in regulating organelle flow into axons has not yet been established.
If the AIS has an organelle barrier function, genetic studies in model organisms could identify its molecular components by isolating mutants in which specific organelles accumulate at abnormally high levels in the distal regions of axons. Genetic studies have found that mutations in the Sunday Driver gene SYD (the Drosophila ortholog of JIP3) cause massive axonal accumulations of various unidentified membrane compartments (Bowman et al. 2000). However, that study proposed that the axonal accumulations resulted from stalled organelle transport rather than overactive or unregulated organelle transport. The logic behind the stalled transport model is based on the fact that fly SYD mutants die as larvae and thus must be derived from heterozygous mothers. The model predicts that a small amount of wild-type SYD from maternally contributed mRNA permits cargo to enter the axons of homozygous mutant larvae, but that organelle transport stalls as the maternal supply is depleted, leaving the organelles stranded in the axons (Bowman et al. 2000). However, this interpretation has not been tested, and it is unclear why a gradual stall in transport would cause massive accumulations of organelles in axons instead of normal or even lower-than-normal levels of stalled organelles.
Studies of Caenorhabditis elegans mutants with impaired UNC-16 (the worm Sunday Driver/JIP3 ortholog) seem to contradict the stalled transport model. unc-16 loss-of-function mutations cause synaptic vesicle proteins to move into axons by a transport mechanism that is independent of the KIF1 motor that normally transports mature synaptic vesicles (Byrd et al. 2001), and unc-16 mutations are associated with axonal accumulations of unidentified membranous cisternae (Brown et al. 2009). These phenotypes cannot be a function of maternal UNC-16, because unc-16 null mutants are homozygous viable and thus have no maternal contribution of wild type unc-16 mRNA.
In the current study we used time lapse imaging of tagged organelles in the axons of living animals to show that UNC-16 inhibits organelle transport into axons, rather than promoting organelle transport as predicted by the stalled transport model. We show that the accumulation of organelles in UNC-16 (JIP3) mutant axons results from the impairment of a previously unrecognized organelle gatekeeper function that selectively restricts the flow of Golgi and endosomal organelles, but not endoplasmic reticulum (ER) membranes, beyond the axon initial segment. A JNK-1 MAP kinase signaling pathway contributes to UNC-16’s organelle gatekeeper function, suggesting that wild-type animals regulate the organelle content of their axons in response to signals.
Materials and Methods
Worm culture and strains
Worm culture and manipulation essentially followed previously described methods (Brenner 1974; Stiernagle 2006). Briefly, culture media was modified NGM, containing no added calcium or magnesium, and consisted of the following (per liter): 2 g NaCl, 3.1 g peptone, 3.0 g KH2PO4, 0.5 g K2HPO4, and 20 g Sigma A-7002 agar. After autoclaving and cooling to 55° the following was added (per liter): 1.6 ml 5 mg/ml cholesterol in ethanol, 1.0 ml of 100 mg/ml streptomycin in ddH2O, and 1.25 ml of 10 mg/ml mycostatin suspension in ethanol. Prior studies define the culture plate types “spread plates,” “streak plates,” and “locomotion plates” (Miller et al. 1999; Edwards et al. 2008). Wild-type worms were the N2 strain and the mapping strain CB4856. File S6 lists the complete genotypes of all strains used in this study. We maintained strains containing dhc-1(or195ts) at 14°.
Forward genetic screen
The goa-1(sa734) suppressor screen has been previously described (Edwards et al. 2009). Briefly, we mutagenized L4-stage goa-1 mutants with 46 mM ethyl methanesulfonate (EMS) in M9 (Sulston and Hodgkin 1988). We plated synchronous F2 grandprogeny of the mutagenized animals as young larvae on 24-well culture plates using a repeat pipettor to plate ∼100 animals in each well for a total of 600,000 F2 animals in 10 weekly cycles. We grew the cultures 2 days at room temperature and 4 days at 20° after plating, and screened the F3 generations. We then screened the wells for mutants with improved growth and reduced hyperactive behaviors.
Mapping, identification, and outcrossing of mutations
Before mapping the new suppressor mutations, we separated the mutations from goa-1(sa734) by crossing N2 males to the suppressed strain and isolating putative suppressor single mutants in the F2 generation based on their predicted sluggish phenotype. We kept animals that were both homozygous for the mutation and did not segregate goa-1(sa734) as one time outcrossed strain stocks. We then mapped the ce421 mutation relative to single nucleotide polymorphisms (SNPs) to a 7-Mb region in the center of chromosome III using a previously described method (Schade et al. 2005). The mapping data for 47 mapping lines were as follows (listed in left–right order on the chromosome with the recombination frequency in each interval in parentheses): ceP37 (3/47) ce421 (1/47) ceP168. The SNPs are located as follows (SNP names are followed by genome location, base pair change, and the restriction enzyme that can identify the SNP): ceP37 [III:3,391,534 (G/A Bgl II)] and ceP168 [III:10,361,701 (G/C Mnl I)]. A complementation test with the mutant unc-16(e109), done by crossing e109/+ males to ce421; dpy-11(e221) hermaphrodites, showed noncomplementation for the locomotion and egg laying phenotypes. Similar complementation tests identified ce451 and ce483 as unc-16 alleles. PCR amplification and sequencing of the unc-16 gene coding region in ce421, ce451, and ce483 mutant genomic DNAs revealed a nonsense mutation in each of these mutants. We outcrossed ce421 and ce483 five times each and ce451 once, to N2. dhc-1(or195) (Caenorhabditis Genetics Center), and unc-116(e2310) (also known as e2281; Caenorhabditis Genetics Center) had previously been outcrossed at least four times to wild type and we did not further outcross them.
Plasmids and PCR products
Supporting information, File S1 lists all of the DNA constructs used in this study (plasmids and PCR products) along with construction details. In all constructs involving the cloning of PCR fragments, we sequenced the inserts and used clones containing no mutations in the fragment of interest to establish the plasmid stock. We produced sense–antisense (sas) fusion PCR products for cell-specific RNAi using a previously described method (Esposito et al. 2007). Briefly, we used Herculase II (Stratagene) and Expand 20 kb+ (Roche) and fusion PCR with overlapping primers to fuse the 2.6-kb unc-129 promoter with a 2-kb unc-116 genomic-rich region in forward and reverse orientations to allow sense and antisense RNAs to be formed in motor neurons in which the unc-129 promoter is active.
Transgene production
We prepared plasmids for microinjection using the Qiagen Tip-20 system according to the manufacturer’s instructions, except that we added a 0.1-M potassium acetate/two volume ethanol precipitation step (30 min on ice; 10 min × 15,000 rpm microcentrifuge spin at 3°, wash with 300 μl −20° cooled 70% ethanol, spin for 5 min as above, and air dry the pellet 3–5 min before resuspending in 50 μl ddH2O) after resuspending the isopropanol-precipitated pellet. We prepared high-fidelity PCR products for microinjection by purifying with Wizard PCR Prep (Thermo Scientific) and quantifying with a Nanodrop spectrophotometer (Thermo Scientific). We precipitated fusion PCR products for cell-specific RNAi using the above potassium acetate method and resuspended them in ddH2O. We produced transgenic strains bearing extrachromosomal arrays by the method of Mello et al. (1991). For the colocalization experiments (producing the ceEx329, ceEx346, and ceEx353 arrays), the host was KG2338 unc-16(ce483). For the unc-116(e2310); ceIs56 rescue experiment the host was KG1942. For strains bearing ceEx305 or ceEx309 arrays the host was KG2430. For all other injection experiments, N2 was the host. We used pBluescript carrier DNA to bring the final concentration of DNA in each injection mixture to 175 ng/μl and integrated arrays into the genome as described (Reynolds et al. 2005), using 9100 rad of gamma rays. File S6 lists all of the transgenic arrays in this study, their DNA contents, and the injection concentration of each DNA.
C. elegans strain constructions
We constructed double mutants using the standard method of crossing heterozygous males of mutant A with homozygous hermaphrodites of mutant B and cloning virgin F1 cross progeny. From plates segregating mutant A in their F2 progeny, we cloned mutant A and/or B animals and looked for segregation of the double mutant in the next generation. For doubles with transgenes, we cloned candidate doubles directly from the F2 progeny and chose one line homozygous for the fluorescence marker and behavioral phenotypes in the next generation. To make doubles with the temperature sensitive (ts) sterile mutation dhc-1(or195ts), homozygous or putative homozygous or195 animals were grown at 14° or, if testing for or195 homozygosity, young adults were grown at 25° for 1 day before transferring parental animals back to 14° to restore viability and leaving the eggs that were laid at 25° for 24 hr to test for 100% embryonic lethality. To produce the closely linked unc-116(e2310) unc-16(ce421) double mutant, we cloned 122 Unc-116 mutant animals from the progeny of e2310/ ce421 transheterozygotes and, after incubating these cultures 4 days at room temperature, identified rare cultures segregating double mutants. For double mutants, we verified the homozygosity of both mutations by amplifying and sequencing both loci from genomic DNA.
Statistical analysis
We performed all statistical comparisons using the unpaired t-test, Welch corrected, via Graphpad Instat 3 or Graphpad Prism software.
Live animal assays
The live animal assays have been described in detail in previous studies: locomotion assays (Miller et al. 1999; Reynolds et al. 2005) and population growth rate assays (Williams et al. 2007).
Electron microscopy
Young adult hermaphrodites for each strain were prepared for high-pressure freezing as described (Rostaing et al. 2004). Briefly, 10–15 animals were loaded in a specimen chamber filled with Escherichia coli, immobilized by high-pressure freezing at −180° under high pressure in a Bal-Tec HPM010, and moved to liquid nitrogen. Freeze substitution was performed in a Reichert AFS machine (Leica, Oberkochen, Germany) as described previously for morphological analysis, using tannic acid (0.1%) and 0.5% gluteraldehyde fixative introduced over 96 hr, followed by 2% osmium tetroxide (OsO4) (Weimer and Richmond 2005). Fixed specimens were then embedded in Araldite 502 over a 48-hr period at 65°. Serial sections were cut at a thickness of 40 nm, collected on formvar-covered, carbon-coated copper grids (EMS, FCF2010-Cu), and counterstained in 2.5% aqueous uranyl acetate for 4 min, followed by Reynolds lead citrate for 2 min. Images were obtained on a Jeol JEM-1220 (Tokyo) transmission electron microscope operating at 80 kV. Micrographs were collected using a Gatan digital camera (Pleasanton, CA) at a magnification of ×100. Morphometric analysis of both ventral and dorsal nerve cord serial sections was scored blind. Images were quantified using National Institutes of Health Image software. A synapse was defined as a set of serial sections spanning the presynaptic density plus two flanking sections on each side.
Quantitative fluorescence imaging and image analysis
Growth and mounting of strains:
We produced live adults for imaging fluorescent-tagged proteins by plating 10–14 L2-stage larvae on each of five locomotion plates and growing 5.5–6 days at 20° to produce next generation young adult progeny (growth times and numbers plated were modified as necessary for slow growing mutants). Approximately 50 young adults were selected and transferred to an unseeded plate, from which we loaded them into a 30-μl drop of 30 mg/ml 2, 3-butanedione monoxime (BDM) (Sigma, B0753) (Sieburth et al. 2005) in M9 buffer on a coverslip, which we incubated for 10 min (from the time the animals first touch the drop) on a moistened 1.5-cm Kimwipe square under a Petri plate lid. We then removed ∼27 μl of the solution using a P20 microinjection tip (Eppendorf, 5242 956.003), leaving the worms behind in a small amount of anesthetic. We inverted the coverslip onto a 2% agarose pad (Sulston and Hodgkin 1988), with the agarose dissolved in M9 buffer. After straightening the coverslip slightly and nudging it ∼1 mm to help with dorsal/ventral orientation, we sealed two diagonal corners with a dab of clear nail polish and imaged animals over the next 35–55 min. To grow dhc-1(or195ts)-containing strains for imaging, we produced freshly starved cultures containing a high density of L1 larvae. We then transferred these animals to spread plates and grew them 40–52 hr at 25.5° before selecting young adults for imaging (the exact time of growth was modified based on the growth characteristics of the strain).
Collecting images:
We collected images at 22° using a Nikon Eclipse TE2000-E inverted microscope equipped with a Nikon CFI Apo TIRF 100×/1.49 numerical aperture (N.A.) objective, a Nikon motorized linear-encoded high-resolution z-drive, a SmartShutter ultrafast shutter with a Lambda SC controller and a foot pedal switch (Sutter Instruments), and a motorized filter turret containing GFP, CFP, YFP, and Texas Red image-registered filter cubes (Semrock). Our illumination source was an X-Cite 120Q illuminator (EXFO, Montreal, Canada), and we captured 12-bit images with an ORCA-AG camera (Hamamatsu, Bridgewater, NJ) controlled by Metamorph Premier software (versions 6.3 r1 and 7.7 were used over the course of this study). We only collected images from animals with their ventral or dorsal surfaces facing the objective. We collected dorsal cord images from a region extending posterior from a point ∼10 μm anterior to the vulva. For lines expressing markers from the unc-129 promoter we collected ventral soma images from the DA6 and DB6 neurons (imaged together and quantified as one region). We used Metamorph to acquire a 0.243 μm × 11 slice z-series (automatically closing the shutter between exposures) with 150-msec exposure times. Exposure times were adjusted down for some arrays to prevent chip saturation. We collected all images for an experiment at identical light source powers and repeated collections of the wild-type strain every sixth imaging session in an experiment. Before imaging each strain, we measured the light power of the peak emission wavelength at the objective plane using an XR2100 power meter (Lumen Dynamics) and an XP750 objective plane light sensor (Lumen Dynamics) with the stage position set at a standard distance (z-position) from the objective. We then used the XR2100 light guide insertion port to determine the total light power necessary to achieve the targeted light power at the objective. Total light power was then measured just prior to acquiring each image stack. If necessary, we adjusted the light power to within 1% of the target value by adjusting the extent to which the light guide was pushed into the exit port of the X-Cite 120Q. Typical total light powers were kept relatively low at ∼2.1 W (of ∼4.0 W maximum) to reduce photobleaching.
Processing and quantifying images:
To process the images, we used AutoDeblur Gold CWF (Media Cybernetics) to deconvolve the image stacks using the adaptive point spread function blind method and 10 iterations at the low noise setting. After deconvolving, we produced maximum intensity projections of each image stack and, for display images, set a scaling value that was used for all images in the experiment. To quantify dorsal axon fluorescence intensities per micrometer, we first tightly traced the dorsal cord across the entire field of view using the Trace Region tool in Metamorph (version 6.3, or 7.7 for later experiments) and obtained the total integrated fluorescence in the region. We then copied this region and shifted it onto a region of the animal immediately adjacent to the cord to obtain the background value. We used the Multi-line tool to obtain the length of the region. Data were logged to a spreadsheet, which subtracted the background and computed the total fluorescence per micrometer of cord length. To quantify ventral soma intensities per square micrometer from lines expressing markers from the unc-129 promoter, we traced the DA6 and DAB6 cell somas and the dendrite process between them as a single region, produced a background region as above, and used a spreadsheet to compute the background-corrected intensity per square micrometer. When tracing cell soma areas of CTNS-1-RFP images, we first traced the soma region from a coexpressed marker such as NLP-21-Venus (for ceIs56) or GFP-RAB-5 (for ceIs81) and then copied the region onto the CTNS-1-RFP image (for which it was difficult to see the cell soma boundary). To count lysosomes in cell somas and the dorsal axons for Figure 3 and Figure 5, we used the Metamorph Thresholding feature to define a threshold of intensity for objects to count and applied this threshold level to all images in the experiment. To count lysosomes in commissures we divided each commissure in laterally oriented, unanesthetized animals mounted in polybeads on 10% agarose pad slides (see below) into four equal regions: the first half of the distance between the ventral side of the animal and the center line of the animal; the second half of the distance between the ventral side of the animal and the center line of the animal; the first half of the distance between the center line of the animal and the dorsal side of the animal; and the second half of the distance between the center line of the animal and the dorsal side of the animal. These correspond to regions 2–5 in the Figure 5 schematic. Starting at the posterior-most commissure and proceeding anteriorly, for each commissure we focused on the highest point of the commissure using a GFP filter (the transgene coexpresses soluble GFP and CTNS-1-mCherry), switched to the Texas Red filter, and focused through the entire visible portion of the commissure, noting the number of lysosomal puncta in the four regions using four tally counters. We kept the fluorescence diaphragm on the microscope maximally closed to restrict the excitation light to the commissure of interest. The cell somas, region 1, and the synaptic region (region 6) were generally not visible in laterally oriented animals; lyosome counts of those regions come from ventrally and dorsally oriented animals.
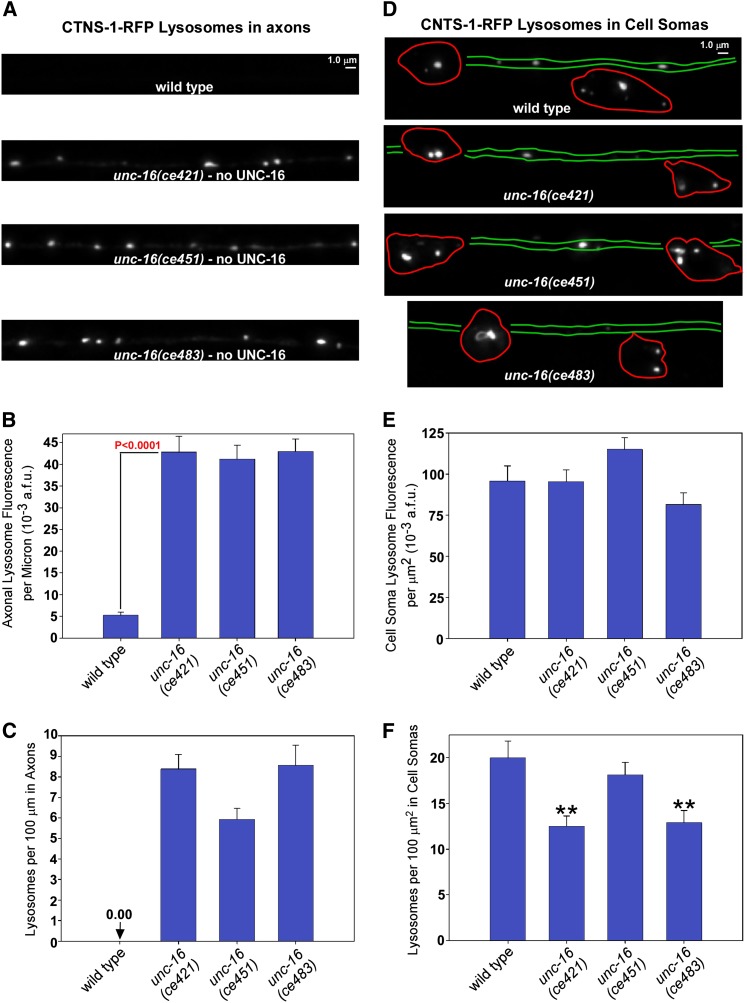
Figure 3.
unc-16 mutants accumulate high levels of lysosomal membranes in their axons with minimal depletion from cell somas. (A and D) Representative images of the DA6/DB6 motor neuron axons and cell somas, respectively, from animals of the indicated genotypes carrying the genomically integrated transgene ceIs56, which expresses CTNS-1-RFP to mark lysosomes. Images were collected at identical light powers and scaled identically. In D, the approximate boundaries of the cell somas and dendrite processes, as determined from analysis of a set of more brightly scaled images, are outlined in red and green, respectively. Since DA6 and DB6 extend dendrites in opposite directions, the region between the somas should contain the dendrites from both cells, but we were unable to resolve them. The spacing between the somas is variable. (B and E) Quantification of CTNS-1-RFP fluorescence in the dorsal axons and ventral somas, respectively, of the indicated genotypes. Graphs show the background-adjusted integrated fluorescence per micrometer of dorsal axon length (B) or background-adjusted integrated cell soma and dendrite fluorescence normalized to area (E). Data are means and standard errors from 14 animals each. The three unc-16 mutants in E are not significantly different from wild type. (C and F) The same data used for graphs B and E were used to count lysosomes defined by a threshold level. Data are means and standard errors from images acquired from 14 animals each. **P < 0.001.
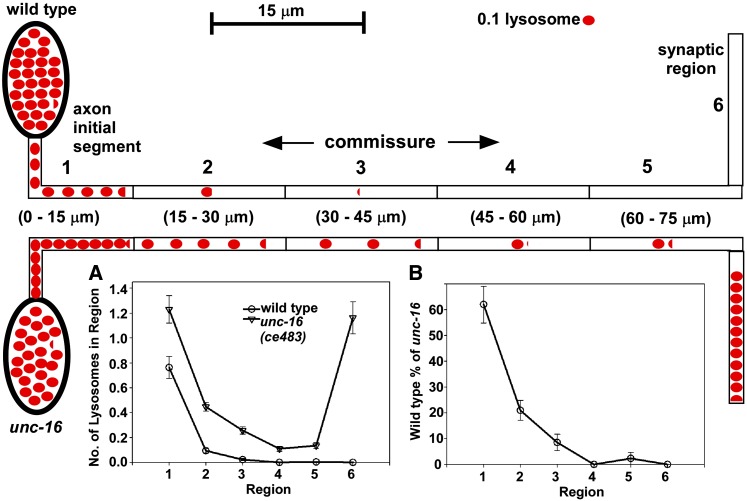
Figure 5.
UNC-16 exerts its gatekeeper function at the axon initial segment, not at the soma–axon junction. (A) Graph comparing the distribution of lysosomes in various regions of wild type and unc-16(ce483) mutants containing the ceIs134 integrated transgene, which expresses CTNS-1-RFP in cholinergic motor neurons. The upper schematic defines the region boundaries. The two schematics also depict the graphed data for wild type (top) or unc-16 mutants mutants (bottom), with each red oval representing 0.1 lysosomes. Commissure data are from 320 commissures for each strain (54 animals each). (B) Graph depicting the number of lysosomes in each region of wild type as a percentage of the number of lysosomes in each region of an unc-16 null mutant. Note the sharp drop in the number of wild-type lysosomes distal to the axon initial segment.
Producing representative images:
After quantifying an image set we produced representative images for display by saving 8-bit versions of an image that was close to the mean ± standard error for the set. All images were scaled identically to the reference strain for each experiment. To crop images that show up as black due to low signal levels, we first produced an image scaled bright enough to see the subject and rotated and cropped the brightened image, noting the rotation and cropping coordinates. We then identically rotated and cropped the properly scaled version of the image.
Time lapse imaging and analysis of organelle movements
Mounting unanesthetized animals:
We produced 10% agarose pads by mixing 0.175 g of agarose and 1.75 ml of M9 buffer in a 2-ml screw-cap freezer tube (Sardstedt). We placed the tube with its cap tightened in 20 ml of water in a 50-ml beaker and autoclaved it for 5 min, after which we transferred it to a 95° hot block. After preheating two glass slides on the hot block for ∼1 min, we then arranged a taped slide (a slide with one layer of lab tape plus one layer of 3/4 inch Scotch tape) on each side of the mounting slide on the hot block. Using a small spatula we scooped agarose onto the slide and then compressed it with the other prewarmed slide, pressing down evenly to make a surface of even thickness. We removed the slide sandwich from the hot block and, after ∼1 min, removed the top slide, added a coverslip to the pad (to maintain a smooth surface), and stored it in a humidified chamber at 4° for up to 2 weeks. To mount animals, we prepicked ∼45 young adults from growing cultures to an unseeded culture plate. We lifted the protective coverslip from the agarose pad and added 5–7.5 μl (depending on pad diameter) of Polybead microspheres (Polysciences, 00876-15) to the center of the agarose pad. We used a pick with a gob of bacteria to transfer the animals to the Polybead suspension while viewing the transfer through the stereomicroscope. Shaking the pick vigorously in the drop promoted an even distribution of animals. We then used the pick to expand the area of the drop, approximately doubling its diameter. We used jewelers’ forceps to mount a no. 1 1/2 coverslip over the drop and sealed two corners with fingernail polish.
Time lapse imaging of lysosomes in commissures:
We collected time lapse images using the same microscope, objective, and illumination system described in the Fluorescence Imaging section, except that we lowered the light source power to 1.5 W (to reduce phototoxic effects) by adjusting the extent to which the light guide was pushed into the exit port of the illumination source, measuring the light power as described above. For the high-resolution movies of organelles transiting between cell somas or axons and commissures, we used 0.5–1.0 mm “rotational nudging” of the coverslip before applying nail polish to encourage orientations with the ventral or dorsal side of the animal facing the objective. On dorsally or ventrally oriented animals we then used Metamorph 6.3r4 to acquire 25- or 50-msec single plane exposures at 2-sec intervals for 4–20 min, making small adjustments to the fine focus manually during the 2-sec intervals to keep the region of interest in focus. To image large regions for representative movies for quantification of spot movement speeds and processivity, we used Metamorph 6.3r4 to acquire a 16-plane z-series (0.24 μm spacing) of 50-msec exposures centered on a commissure of interest every 30 or 60 sec in a 10-min time course. “Commissure of interest” was defined as the first commissure containing two or more CTNS-1-GFP spots when scanning the length of an animal. We collected only one time course from each animal with a commissure of interest, although in some cases the image contained two or three commissures of interest. We did not begin collecting a new time course if the slide was >45 min old. We then used “Review Multidimensional Data” in Metamorph 6.3r4 to make a maximum projection at each time point. We used “Track Points” in Metamorph 6.3r4 to measure distances of spot movements. “Movement” was defined as a spot translation of >0.5 μm. A movement ends when the spot pauses, changes direction, or exits the field of view or focal plane.
To measure spot movement speed, we examined each of the time-lapse stack files and identified time points containing “stuttered movement” that indicates that movement occurred during the z-series collection for that time point. We then used Metamorph to open the .stk file for the time point in which the stuttering occurred and determined the direction of movement (anterograde or retrograde) based on the location of the ventral somas. We then used the Metamorph Track Points tool to determine the distance traveled (until the spot left the focal plane). We noted the z-plane start and end of the movement and used an Excel spreadsheet to calculate the time elapsed as a fraction of the total time of the z-series. We experimentally measured the total time of a 16-plane z-series with 50-msec exposures as 9.39 sec ± 0.026 standard error of the mean (N = 7). This total time was divided by 15 (because the readout time of the final plane was not timed in this measurement) to give a “time per plane” of 626 msec.
Recording lysosome movements to and from the cell soma and axon initial segment:
We mounted animals as described above for Time Lapse Imaging of Lysosomes in Commissures, using 0.5–1.0 mm “rotational nudging” of the coverslip before applying nail polish to encourage orientations with the animal’s ventral side facing the objective. To prevent the excitation light from affecting other cell somas on the same animals, we closed the fluorescence diaphragm such that only the cell soma(s) of interest were illuminated, and we adjusted the light power as described above to 1.5 W to reduce photobleaching. We used the transgene’s soluble GFP signal to focus on a cell soma and commissure, and then collected 151 images in the Texas Red channel (for CTNS-1-mCherry) at 2-sec intervals, using a 50-msec exposure time. Every 10th frame we collected a 30-msec GFP image to allow us to identify the cell soma and commissure. In the 2-sec intervals between exposures we adjusted the focus if necessary. We sometimes imaged up to three cell somas and commissures on the same animal, but did not begin a new time course unless it could finish before 20 min had elapsed from the time the coverslip was applied. We prepared the time lapse movies as described above, except we loaded both colors, duplicating the missing GFP images to fill in the intervals between every 10th frame, thus providing a green cell soma and commissure background.
Antibody production
To produce the affinity-purified UNC-16 antibody KM37A-5.1, we prepared a recombinant maltose-binding protein (MBP)-UNC-16 (codons 1–462) fusion protein by transforming KG#657 [MBP-His10-UNC-16 (1–462)] into the bacterial expression host BL21-DE3(RIL). We then induced expression of the protein for 4 hr at 30° from a 5-liter culture at an A600 of 0.4 using 30 μM IPTG. We froze cell pellets in liquid nitrogen and rapidly thawed them before resuspending in 67 ml ice cold lysis buffer [20 mM HEPES, pH 8.0/300 mM NaCl/10 mM βme/10 mM imidizole/1 mM PMSF/0.3 mM EDTA/1 μg/ml pepstatin/1 μg/ml leupeptin/0.255 mg/ml lysozyme (Sigma L-6876)] per liter of culture. All steps hereafter were at 4°. To lyse, we stirred the suspension for 30 min and then added 5 mg of DNAse (Sigma; D-4527) and 400 μl of 1 M MgSO4 and incubated 30 min with stirring. After clearing the lysate with a 30-min 33,000 × g spin, we loaded it at 0.5 ml/min onto a 5-ml Ni-NTA agarose (Qiagen) column hooked to a Biologic (Bio-Rad) low-pressure chromatography system. We washed the column with 100 ml of lysis buffer and then eluted with 25 ml of lysis buffer + 250 mM imidizole while collecting 1-ml fractions. We dialyzed peak fractions containing the fusion protein for 20 hr at 6° in 4 liters of dialysis buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 5 mM βme) supplemented with 2% w/w His6-TEV [S219V] protease, which we purified from a pRK793 bacterial lysate as described (Kapust et al. 2001). We then dripped the dialysate over a fresh 5-ml Ni-NTA agarose column to rebind the His6-TEV protease, the cleaved MBP–His10 fusion partner, and any uncleaved fusion protein, chased with 15 ml of dialysis buffer, and concentrated the flow thru to 3.5 mg/ml using centrifugal concentrators (Millipore) for a total yield of 31.5 mg of UNC-16 (1-462; 53.3 kDa) with a purity of ∼90%.
We further purified 1000 μg of the protein on two SDS–PAGE gels, excised and homogenized the Coomassie-stained gel band in a 10-ml Kontes Potter-Elvehjem tissue grinder, using a drill press such that the final concentration was 250 μg/ml of gel suspension. We sent the suspension to Covance (Denver, PA) for injection into a rabbit (250 μg initial injection + four 125 μg boosts at 3-week intervals). To affinity purify UNC-16-specific antibodies we ran out 1000 μg of the purified protein on two SDS–PAGE gels and blotted to nitrocellulose. After staining the blots with Ponceau S, we excised the bands of interest and rinsed them in TBST (10 mM Tris, pH 8.0/150 mM NaCl/ 0.05% Tween-20). After blocking 30 min in TBST + 3% nonfat dry milk, we incubated 10 ml of the UNC-16 antiserum with the nitrocellulose strips in a 15-ml conical for 2 hr at room temperature with gentle shaking. After washing the strips four times for 5 min in TBST and once in PBS, we eluted bound antibodies with 1.0 ml of 0.1 M glycine, pH 2.5 for 5 min, and neutralized the solution with 100 μl of 1 M Tris, pH 8.0. We then eluted a second time with 0.2 M glycine, pH 2.3, neutralized with 79 μl of 1.5 M Tris base, combined the two eluants, and added 245 μl of 10% BSA as a cryoprotectant.
Immunostaining
We prepared freeze-cracked 4% paraformaldehyde fixed worms for immunostaining as described (Charlie et al. 2006a,b). For immunostaining UNC-16, we coimmunostained ceIs123 [unc-129::GFP] and unc-16(ce483); ceIs123 fixed animals using rabbit anti-UNC-16 (1/200; KM37A-5.1; this study) and mouse anti-GFP (1/500; mAb3E6; QBiogene). We labeled the UNC-16 primary antibody with a donkey anti-rabbit Dylight 550 secondary. We dual-labeled the GFP with donkey anti-mouse Dylight 488 and donkey anti-mouse Dylight 650 secondaries, which allowed us to easily find the signal using a GFP filter and to assess permeabilization on camera images using a Cy5 filter (since GFP can still fluoresce after fixation, a bright signal in the GFP channel does not necessarily indicate good permeabilization). For triple labeling of UNC-16, UNC-17 cholinergic synaptic vesicles, and UNC-13 active zones, we coimmunostained wild-type animals using rabbit anti-UNC-16 (1/200; KM37A-5.1; this study), mouse anti-UNC-17 (1/5000 from monoclonal ascites fluid) (Duerr et al. 2001), and goat anti-UNC-13L (1/800; KM21E-3.1) (Charlie et al. 2006b), using donkey anti-rabbit (Dylight 550), mouse (Dylight 488), and goat (Dylight 650) secondary antibodies. We used previously described methods for immunostaining of fixed animals (Charlie et al. 2006a,b), with the exception that we spread only 750 animals per slide at the final mounting step.
Results
goa-1(sa734) suppressor mutations disrupt UNC-16 (JIP3)
In C. elegans, an inhibitory GOA-1 (Gαo) pathway exerts its effects on locomotion by a mechanism that is dependent on the EGL-30 (Gαq) pathway (Perez-Mansilla and Nurrish 2009). To investigate how the GOA-1 and EGL-30 pathways regulate synaptic activity, we undertook a large forward genetic screen for mutations that suppress the slow growing, sickly phenotypes and hyperactive behaviors of goa-1 null mutants. The dual requirements of growth improvement and suppression of hyperactive behaviors produced a small number of prominent targets, including EGL-30 itself (Gαq, 34 mutants) and the Gαq effector UNC-73 (Trio RhoGEF domain; 8 mutants) (Williams et al. 2007). Here, we report our investigation of an unexpected target of this screen, UNC-16, which encodes the sole C. elegans ortholog of human JIP3 and fly Sunday Driver (SYD) (Bowman et al. 2000; Byrd et al. 2001). Our investigation has not yet revealed a direct connection between Gαo/Gαq signaling and UNC-16, so this study focuses on the striking organelle trafficking defects we observed in the unc-16 mutants.
The unc-16 mutations we isolated are early stop codons and are thus likely to be null or near-null mutants (Figure S1A). Although unc-16 mutant adults are slightly shorter than wild-type adults, they are healthy and fertile as homozygotes, having population growth rates of 72% ± 3% of wild type (n = 3 populations; ce421 allele). Quantitative locomotion assays revealed that the three unc-16 mutants have similarly sluggish locomotion rates that are ∼15% of wild type (Figure S1B).
unc-16 null mutants accumulate high levels of golgi and endosomal organelles, but not ER membranes, in their axons
To identify potential membrane trafficking defects in neurons that might contribute to the sluggish locomotion of unc-16 mutants, we used high-pressure freezing electron microscopy to analyze the organelle content of unc-16 mutant axons. The EM analysis revealed that unc-16 null mutants accumulate organelles in their ventral and dorsal axons at levels that are approximately nine- and sixfold higher than wild type, respectively (Figure 1, A and B). To identify the classes of organelles that accumulate in unc-16 mutant axons (i.e., the organelles in the electron micrographs) we produced a series of integrated transgenes that use the unc-129 promoter to express various organelle-specific fluorescently tagged markers in a subset of nine chlolinergic motor neurons. The nine motor neurons have their cell somas and dendrites on the ventral side of the animal and their synaptic axonal regions on the dorsal side. A long axonal commissure crosses the animal’s body to connect the cell soma to the dorsal synaptic region. After crossing the transgenes into an unc-16 null mutant background, we then imaged defined axonal and cell soma regions in young adult animals and quantified the steady-state fluorescence (total fluorescence/micrometer above background) of each organelle marker in those regions.
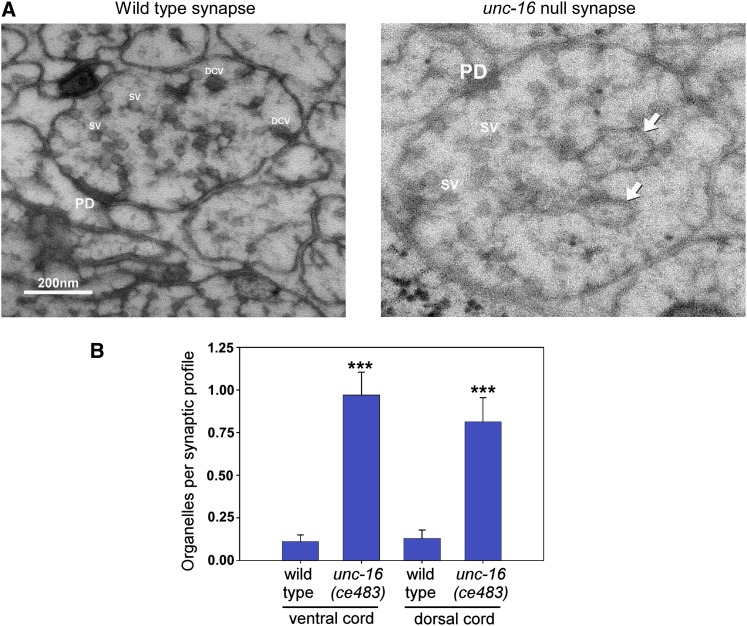
Figure 1.
unc-16 null mutants accumulate organelles in their axons. (A) Representative electron micrographs of synaptic profiles from the ventral nerve cord of wild type and the unc-16(ce483) null mutant as indicated. Arrows indicate representative organelles. PD, presynaptic density; SV, synaptic vesicle; DCV, dense core vesicle. (B) Graph quantifying the number of organelles in the dorsal and ventral nerve cords of wild type and an unc-16 null mutant. In this experiment we define “organelle” as any membrane-enclosed compartment that is larger than a dense core vesicle. Data are means and standard errors of the mean sampled from two young adults for each strain as follows (wild-type VNC, 72 profiles from 28 synapses; unc-16 VNC, 70 profiles from 25 synapses; wild-type DNC, 70 profiles from 22 synapses; and unc-16 DNC, 64 profiles from 17 synapses). ***P ≤ 0.0001.
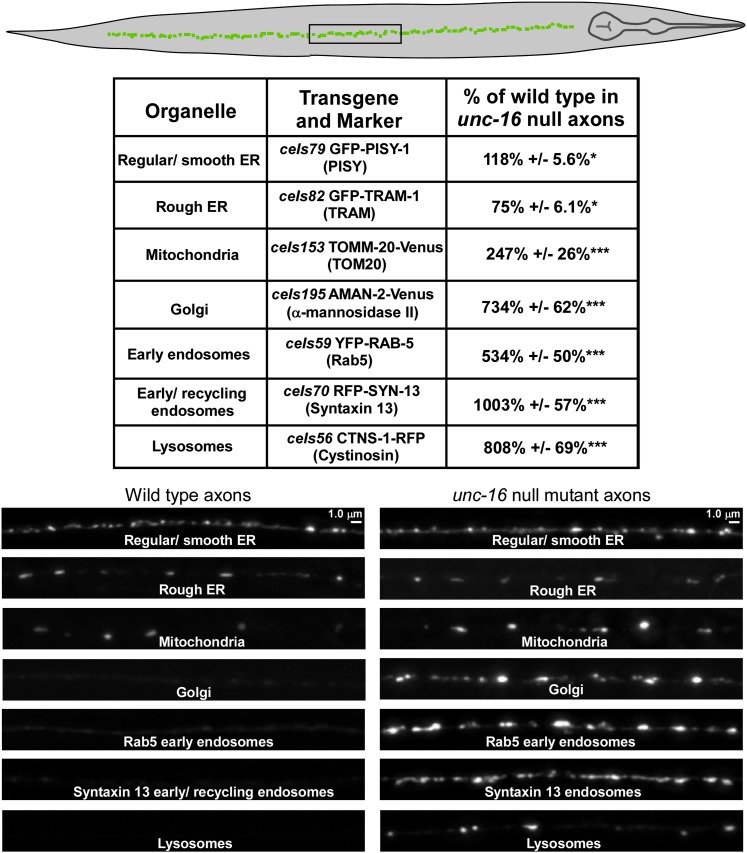
To quantify ER membranes we used the regular/smooth ER marker PISY-1 (a worm ortholog of phospatidylinositol synthase) and the rough ER marker TRAM-1 (a protein involved in translocation across the ER membrane). Both proteins have previously been shown to colocalize in a reticular pattern with each other and other ER markers when expressed from transgenes in C. elegans neuronal cell somas (Rolls et al. 2002). The axonal levels of the regular/smooth ER and rough ER membranes in unc-16 mutants were changed by <20% of wild type, and the differences were only marginally significant (Figure 2).
Figure 2.
unc-16 null mutants accumulate high levels of Golgi and endosomal membranes in their axons. The drawing shows an adult C. elegans oriented with its dorsal cord up and a box around the targeted image region. The table summarizes the levels of various organelle markers in unc-16(ce421) mutant axons expressed as percentages of the levels in wild-type animals. All data are derived from the indicated genomically integrated transgenes and are the means and standard errors of the background-adjusted integrated fluorescence per micrometer of axon length from 12–14 animals each. Asterisks indicate levels of statistical significance as follows: *P < 0.05; ***P < 0.0001. Each pair of wild-type and unc-16(ce421) representative images are scaled identically for brightness.
To quantify mitochondria, we expressed the mitochondrial membrane protein TOMM-20-Venus, previously shown to localize to mitochondria by fluorescence nanoscopy (Watanabe et al. 2011). The axonal levels of TOMM-20-Venus in unc-16 mutants were elevated to ∼2.5-fold higher than wild type (Figure 2); however, this difference was relatively small when compared to the remaining markers and the organelle levels we observed by EM.
The remaining markers tagged the Golgi, early/recycling endosomes, and lysosomes. We found that all of these markers accumulated in unc-16 mutant axons at high levels. AMAN-2-Venus (α-mannosidase II), which marks Golgi in C. elegans neurons (Orci et al. 2000; Rolls et al. 2002; Sumakovic et al. 2009); YFP-RAB-5, which marks early endosomes in C. elegans (Maxfield and McGraw 2004; Grosshans et al. 2006; Grill et al. 2007); RFP-SYN-13 (Syntaxin 13), which marks early/recycling endosomes in C. elegans neurons (Prekeris et al. 1998; Chun et al. 2008); and CTNS-1-RFP (Cystinosin, a lysosome-specific cysteine transporter), which marks lysosomes in C. elegans (Kalatzis et al. 2001; Mangahas et al. 2008) accumulated in unc-16 mutant axons at levels that were 7-, 5-, 10-, and 8-fold higher, respectively, than wild type (Figure 2). The fold increases in axons for the Golgi, lysosome, and endosome transgenic markers parallel the fold increases we observed for native organelle compartments in unc-16 mutant axons by electron microscopy, demonstrating that the accumulations of transgenic markers coincide with native membranes. As an important control, we found that expression from the transgenic promoter we used is not affected by an unc-16 null mutation (expression of soluble GFP in unc-16(ce483) cholinergic motor neurons was 118.4 ± 8% of wild type; N = 12 animals expressing the ceIs123 transgene).
To provide independent evidence that the accumulations represent Golgi, endosomes, and lysosomes, we tagged alternative organelle markers for each compartment using different color fluorescent proteins, separately integrated the resulting transgenes, and crossed them together. Imaging cholinergic motor neuron somas in these strains revealed that the CTNS-1-RFP marker colocalized with the alternative marker LMP-1-GFP (the C. elegans LAMP, or lysosomal membrane protein, ortholog; Figure S2A), and the two early endosome markers YFP-RAB-5 and RFP-SYN-13 colocalize (Figure S2B). As an alternative Golgi marker, we used PST-2-CFP. PST-2 is the C. elegans ortholog of a Golgi-resident PAPS (phosphosulfate) transporter that has been localized to Golgi stacks in C. elegans (Dejima et al. 2010). AMAN-2-Venus and PST-2-CFP generally colocalized, with some slight offset that could be due to the two proteins localizing to different regions of the Golgi (Figure S2C).
The high steady-state levels of organelles in unc-16 mutant axons result from loss of unc-16 in the same neurons because a transgene expressing the unc-16 cDNA in the same neurons completely rescued the hyperaccumulation of early endosomes (Figure S3A). Endosomes are known components of axons that are involved in synaptic vesicle recycling. Thus, it is not the presence of early endosomes in unc-16 mutant axons that is unusual; it is the high levels of this organelle. However, the presence of Golgi and lysosome membranes in adult C. elegans cholinergic motor neuron axons is highly abnormal. The brightness and relatively sparse distribution of the CTNS-1 lysosome marker made it ideal for counting as well as tracking by time lapse video microscopy, so we focused on this marker for the remaining study. All three unc-16 null mutants accumulated the CTNS-1-RFP lysosomal marker in their axons at eightfold higher levels than wild-type axons as measured by quantifying total CTNS-1-RFP fluorescence above background (Figure 3, A and B). Thresholding the data to count lysosomal puncta revealed only a diffuse, near-background level of the lysosomal marker and no lysosomal puncta in any of the 28 wild-type axons we analyzed. In contrast, unc-16 null mutants averaged a lysosomal punctum every 12 μm of axon length (Figure 3C).
unc-16 mutant cell somas are not depleted of the organelles that accumulate in axons
The unusually high levels of Golgi, endosomes, and lysosomes in unc-16 mutant axons were not associated with a corresponding depletion in the cell somas. The levels of the three organelle markers that were most elevated in unc-16 mutant axons (AMAN-2-Venus, RFP-SYN-13, and CTNS-1-RFP) were not statistically different from wild type with respect to their levels in cell somas, although all were slightly lower, and two of the three unc-16 nonsense mutants had slightly, but significantly lower numbers of lysosomal puncta in their cell somas (Figure 3, D–F) (S. L. Edwards and K. G. Miller, quantified data not shown for AMAN-2-Venus and RFP-SYN-13).
Lysosomes in unc-16 null mutants are less likely to re-enter the soma after they move into the axon
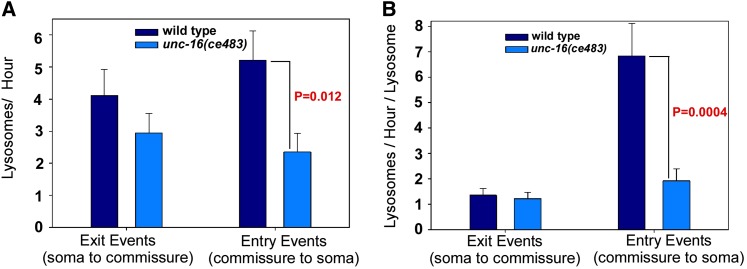
The quantitative data from static images led us to hypothesize that UNC-16 exerts an organelle gatekeeper function that inhibits organelle movements from the cell soma into the axon initial segment. The near normal organelle content in unc-16 mutant somas, as well as the near wild-type growth rate of unc-16 mutants, suggests that, despite heavy accumulations in axons over time, organelles move from unc-16 mutant somas to axons relatively infrequently, leaving time for them to be replaced as they leave the cell soma. To test these ideas, we constructed a transgenic array that coexpresses CTNS-1-RFP (to mark lysosomes) and soluble GFP (to mark cell somas and neuronal processes) in ventral cord cholinergic motor neurons. After integrating the array into the genome we crossed it into an unc-16 null mutant and used two-color time lapse imaging to track lysosome movements at the cell soma–axon boundary in living, unanesthetized animals. In time lapse data from >100 cell somas each for wild type and unc-16 mutants, we found that our initial gatekeeper hypothesis was wrong. Lysosomes in wild type and unc-16 null mutants were just as likely to exit the cell soma, doing so, on average, at a rate of one per 15–20 min (Figure 4A). However, the data also revealed that lysosomes in unc-16 mutants are less likely to re-enter the soma once they have moved into the axon initial segment (Figure 4A). Since the rate of lysosome movements into and out of the soma is affected by the number of lysosomes in the starting location, we also normalized the data to reflect the event rate per lysosome in the starting location. This enhanced the difference between wild type and the unc-16 mutant with respect to cell soma entry rates and more closely equalized the exit rates (Figure 4B).
Figure 4.
Lysosomes in unc-16 null mutants are less likely to re-enter the soma after they move into the axon initial segment. (A) Graph comparing the number of lysosomes per hour exiting the cell soma to the axon commissure or entering the cell soma from the axon commissure in wild type and unc-16 null mutants containing the ceIs134 integrated transgene, which expresses CTNS-1-RFP in cholinergic motor neurons. Data are from 114 (wild type) and 102 (unc-16 mutant) 5-min time lapse movies recorded from one to three cell somas per animal. (B) Graph normalizes the data from A to the average number of lysosomes at the starting location, which is the cell soma for exit events or the axon initial segment for entry events. See also Files S1 and S2, which each show sequential time lapses movies of four to five different somas from each genotype.
Files S1 and S2 each are a stack of four two-color clips from wild type and unc-16 mutants that are representative of the Figure 4 graph data. They show examples of lysosome movements into and out of the cell soma in each strain. The movies also show that, in some cases, the lysosomes transiting this boundary can be stretched into several micrometer long tubulovesicular structures as they enter the narrow axonal commissure. Thus, the lysosomal membranes moving out of cell somas include large organelle-size compartments.
UNC-16 exerts an organelle gatekeeper function at the axon initial segment
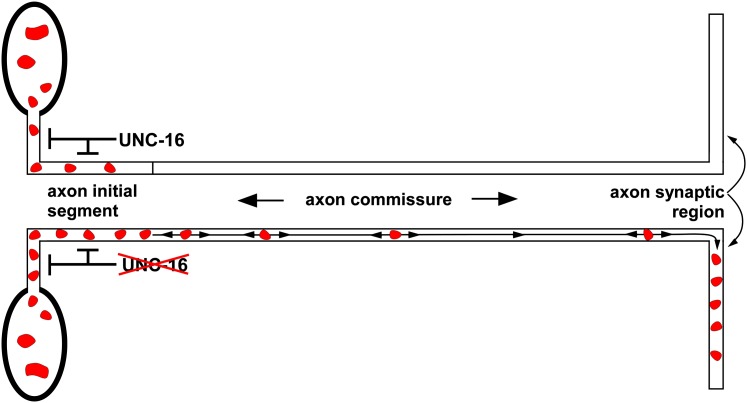
The above data suggest that UNC-16 is not acting at the cell soma–axon boundary as we initially hypothesized, but rather at some other location in the axon. To find the location of the gate region, we compared the distribution of lysosomes throughout the entire axon, from the cell soma to the synaptic region in wild type and unc-16 mutants. To accumulate in the dorsally located synaptic region of the axon, lysosomes in D-type cholinergic motor neurons must first pass through a long commissure that crosses the animal’s body to connect the ventral soma with the dorsal synaptic region. Although the commissure is technically part of the axon, it has distinctly different properties from the synaptic region of the axon as indicated by its lack of synapses and unique active transport properties highlighted in forthcoming data in this article. We divided the commissures into six different regions, starting with the axon initial segment (nearest to the soma) and finishing in the synaptic region on the other side of the animal. The data, obtained by counting lysosomes in the six regions of 320 commissures for each strain, revealed that lysosomes in wild type usually do not progress beyond the axon initial segment, whereas lysosomes in unc-16 mutants often escape this region and ultimately accumulate at high levels in the synaptic region (Figure 5, A and B). For example, segment 2 of wild type (the region just past the axon initial segment and possibly including part of the axon initial segment) had, on average <0.1 lysosomes per commissure, compared to 0.5 for the same region in the unc-16 null mutant. Beyond region 2, wild type had essentially no lysosomes in any of the 320 commissures, whereas these distal regions often contained a lysosome in the unc-16 mutant. The data thus suggest that UNC-16 exerts its gatekeeper function from the axon initial segment, not at the cell soma–axon boundary.
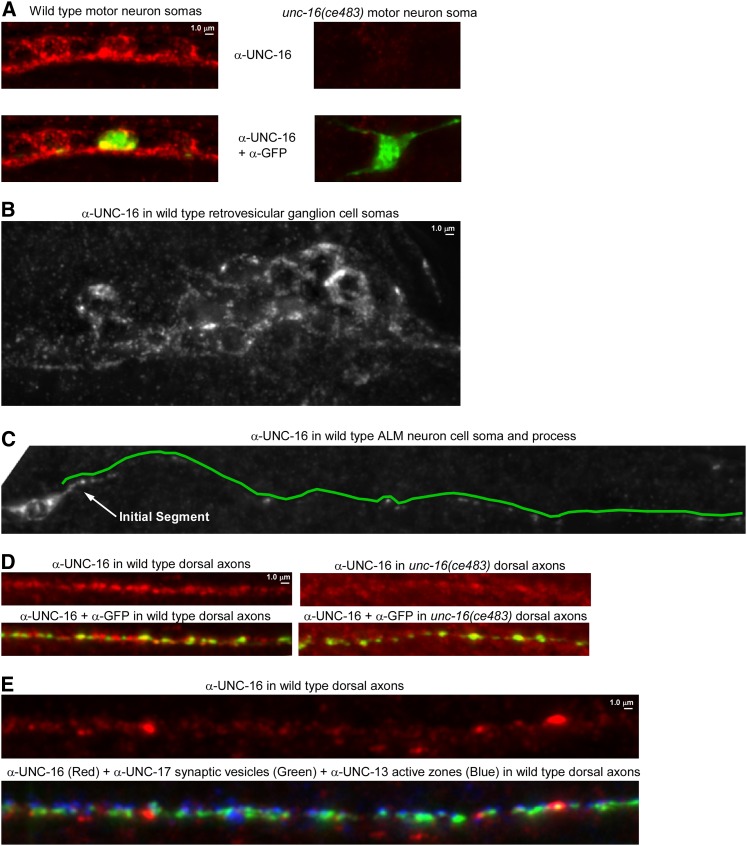
To determine whether native UNC-16 localizes to the axon initial segment we produced an affinity-purified antibody to UNC-16 and used it to immunostain C. elegans whole mounts. We saw UNC-16 signal broadly expressed throughout the nervous system (Figure 6, A and B shows examples from the ventral nerve cord in the animal’s body and the retrovesicular ganglion in the head). The UNC-16 signal was largely absent from the cell somas and axons of an unc-16 nonsense mutant (Figure 6, A and D). Due to the crowding of neuronal cell somas in the ventral cord and head ganglia, we were unable to determine whether UNC-16 has a special preference for the axon initial segment in these neurons. However, when we imaged the well-isolated ALM neuron, we observed that UNC-16 was equally concentrated in its cell soma and its initial segment (Figure 6C). The UNC-16 signal dropped off rapidly after the initial segment, but was still sparsely distributed throughout the remaining process and was also sparsely distributed in the dorsal axons of motor neurons in a pattern that did not align with synaptic sites (Figure 6, C–E). The localization of native UNC-16 is thus consistent with a function in the axon initial segment; however, it is also present on sparsely distributed structures distal to the initial segment and thus may also function at other locations.
Figure 6.
Native UNC-16 immunolocalization in C. elegans whole mounts. (A) Representative, identically scaled images of UNC-16 immunoreactivity (Red) in wild-type and unc-16(ce483) motor neuron cell somas in the ventral nerve cord. Note that the red signal is greatly diminished or absent in the unc-16 mutant, thus demonstrating that the red signal represents UNC-16. Each strain also contains the ceIs123 integrated transgene, which expresses soluble GFP in a subset of nine motor neurons. The top images show UNC-16 signal only. The bottom images show an overlay of the UNC-16 and GFP signals to demonstrate equal permeablization of wild-type and mutant animals. GFP protein (green) was detected using a Far Red secondary antibody to avoid native GFP fluorescence. (B) UNC-16 appears to be present in most or all neurons. The image shows UNC-16 protein in the retrovesicular ganglion in the animal’s head. Note that the staining shows up as small puncta, with some areas of heavy concentration. (C) UNC-16 protein in the isolated neuron ALM. Because this neuron is well separated from other neurons the initial segment of the process is visible. UNC-16 appears most heavily concentrated in the cell soma and the initial segment of the process, but it can also be found sparsely throughout the length of the process. A green line adjacent to the process marks its path. (D) The synaptic regions of axons also contain UNC-16. Representative, identically scaled images of UNC-16 protein (Red) in wild-type and unc-16(ce483) motor neuron axons in the dorsal nerve cord. Note that the red signal is greatly diminished or absent in the unc-16 mutant, thus demonstrating that the red signal represents UNC-16. Each strain also contains the ceIs123 integrated transgene, which expresses soluble GFP in a subset of nine motor neurons. Some UNC-16 signal remains in the mutant since the region used to produce the antibody contains sequences upstream of the nonsense mutation. The top images show UNC-16 signal only and is an area of exceptionally uniform concentration of UNC-16 in the center of the image. A more typical pattern is shown in E. The lower images show an overlay of the UNC-16 and GFP signals to demonstrate equal permeablization of wild-type and mutant animals. GFP protein (green) was detected using a Far Red secondary antibody to avoid native GFP fluorescence. (E) UNC-16 protein in the synaptic region of axons typically contains sparse areas of heavy concentration interspersed with large areas containing a low concentration of UNC-16. Shown is a 74-μm region of the dorsal cord. The top image shows UNC-16 signal only (red). The bottom image shows signals for synaptic vesicle clusters (green) and active zones (blue) overlaid on the UNC-16 signal.
Lysosomes in unc-16 mutants exhibit bidirectional active transport in the commissure, but are much less active in the synaptic region
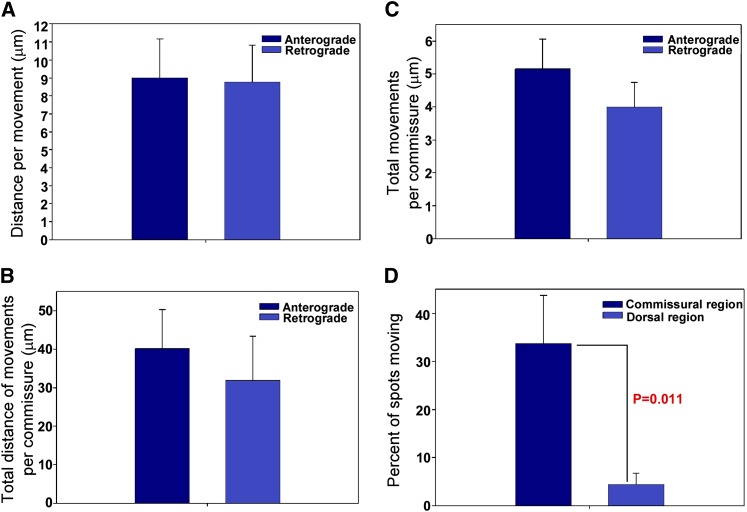
To determine how lysosomes reach the synaptic region in unc-16 mutants, and why they accumulate there, we produced time lapse movies of long regions of the commissure that allowed us to follow lysosome movements in living, unanesthetized animals by rapidly collecting a 16-plane z-series at each time point in physically immobilized animals. We observed that lysosomes in unc-16 null mutant axons moved vigorously and processively in both directions and sometimes stretched into long tubules as they moved through the narrow commissure (File S3). Anterograde and retrograde movements were not significantly different with respect to the overall distance per movement, the average distance of all movements in each commissure, or the average number of movements per commissure per unit time (Figure 7, A–C).
Figure 7.
Lysosomes in unc-16 mutants exhibit bidirectional active transport in the commissure, but are much less active in the synaptic region. (A–C) Graphs comparing anterograde and retrograde movements of lysosomes in unc-16 mutant commissures. Strain genotype is unc-16(ce483); ceIs83, which expresses CTNS-1-GFP in cholinergic motor neurons. Data are graphed as means ± standard errors. Sample sizes are as follows: (A) 45 anterograde and 36 retrograde movements from 7 adults; (B and C) 13 commissures from 10 adults. See also Files S3, S4, and S5. (D) Graph compares the fraction of CTNS-1-GFP lysosomes moving in commissural vs. dorsal axon regions in unc-16(ce483) mutants. Data are means ± standard errors of the mean. Commissural data are from 54 spots from 16 commissures in 14 animals; dorsal axon data are from 135 spots in 10 animals. See also File S4.
The movements were processive. The average distance before a pause, change in direction, or loss of the lysosome from the field of view was 9 μm for both retrograde and anterograde movements (Figure 7A). The maximum distance we observed for an anterograde movement was 62 μm (essentially the length of the commissure as it crosses the body), and the maximum retrograde distance was 49 μm.
The movements were vigorous and consistent with fast axonal transport velocities. The mean anterograde velocity was 1.21 ± 0.15 μm/sec (mean ± SEM; N = 14), while the mean retrograde velocity was 1.29 ± 0.16 μm/sec (mean ± SEM; N = 9). The maximum speeds we observed were 2.62 μm/sec (anterograde) and 2.28 μm/sec (retrograde).
Organelle active transport strongly decreases after deposition in the main axon
Since the bidirectional organelle movements in commissures appear balanced in each direction, we sought to determine why organelles accumulate in the synaptic region of unc-16 mutant axons. One possibility is that the bidirectional movements occasionally bring the organelle all the way to the synaptic region of the axons, and that organelle mobility decreases upon reaching this region. To test this, we quantified the number of lysosomal puncta moving in unc-16 mutant commissures vs. the synaptic region. Indeed, we found that lysosomal puncta in the synaptic region of unc-16 mutant axons were much less active than those in the commissural regions. File S4 shows simultaneous views of multiple lysosomes actively moving in the commissural region and not moving in the dorsal regions. However, as shown in the two clips in File S5, the bidirectional movements of lysosomes can persist for at least a few minutes even after reaching the synaptic region, and they can even move back into the commissure. The birectional movements into and out of the synaptic region are likely to be short-lived, however, because a quantitative analysis revealed that, on average, CTNS-1-GFP lysosomal puncta were nine times more likely to be moving when in the commissural region of the axon than when in the dorsal (synaptic) region (Figure 7D). Thus, if the vigorous bidirectional shuttling in the commissures successfully deposits a lysosome in the synaptic region, the lysosome would be less likely to return to the cell soma if it does not re-enter the commissure before its mobility decreases. This would cause lysosomes to accumulate in axons (at higher levels than commissures) without depleting them from the cell soma. These data are also significant because they suggest that the commissural regions of axons have specialized active transport properties compared to the synaptic regions.
Impairing UNC-116 (Kinesin) or dynein does not prevent axonal lysosome accumulation
Prior studies reported that the dynein intermediate light chain (a component of the dynein motor complex), the kinesin light chain (a cargo-binding subunit of the KIF5 kinesin motor), and KIF5 (kinesin heavy chain) directly bind to UNC-16 and/or its JIP3 orthologs (Bowman et al. 2000; Verhey et al. 2001; Kelkar et al. 2005; Nguyen et al. 2005; Sakamoto et al. 2005; Montagnac et al. 2009; Arimoto et al. 2011; Sun et al. 2011). Since these interactions involve active transport motors, we used mutations in these motors, along with the CTNS-1 lysosome marker to test the hypothesis that one or both motors carries organelles to axons in the absence of UNC-16. Eliminating DHC-1 or UNC-116 in C. elegans results in sterility or early embryonic lethality (Hamill et al. 2002; Koushika et al. 2004; Yang et al. 2005). Therefore, to produce adult animals with strongly impaired DHC-1 (dynein heavy chain) function, we grew strains with the temperature-sensitive allele dhc-1(or195) (Hamill et al. 2002; Koushika et al. 2004) at the restrictive temperature from hatching to adulthood, yielding 100% sterile adults. To impair UNC-116 (KIF5C) function in adults, we used the homozygous viable reduction-of-function mutation unc-116(e2310) (Patel et al. 1993).
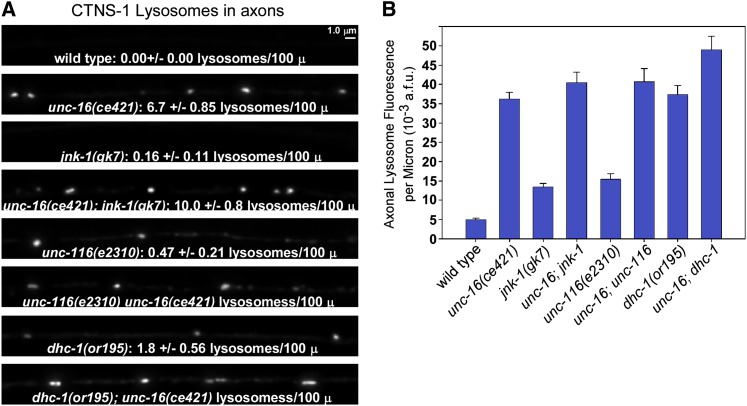
In mutants with strongly impaired DHC-1 function, axonal levels of CTNS-1-RFP (lysosomes) were 7.5-fold higher than wild type (Figure 8, A and B). Growing the dhc-1 mutants at the permissive temperature of 14° decreased CTNS-1-RFP levels in axons to 1.7 ± 0.19-fold higher than wild type, consistent with a small degree of impairment even at the permissive temperature (Figure S3B). To confirm that the dhc-1 mutant phenotype is caused by disruption of dynein function in motor neurons, we impaired dynein function by overexpressing DNC-2 (dynamitin) in the same neurons and observed a greater than fivefold increase in the axonal levels of CTNS-1-RFP lysosomes compared to wild type (Figure S3B). Overexpressing dynamitin dissociates the dynactin complex and causes phenotypes identical to dynein loss-of-function mutations (Burkhardt et al. 1997; Koushika et al. 2004). From these results we conclude that dynein is also required to inhibit the lysosomal marker from accumulating in axons.
Figure 8.
UNC-16 and the JNK-1 signaling kinase act in the same pathway to prevent lysosome accumulation in unc-16 mutant axons, but impairing KIF5 kinesin or dynein does not prevent the accumulation. (A and B) Representative, identically scaled images (A) and quantification (B) of CTNS-1-RFP lysosome fluorescence in defined dorsal and axonal regions of the indicated genotypes. All strains carry the genomically integrated transgene ceIs56. Graph shows the mean background-adjusted integrated fluorescence per micrometer of nerve cord length from images acquired from 11–12 animals each. Error bars represent standard errors. Intensity and scaling values for the unc-116; unc-16 and dhc-1; unc-16 double mutants have been reduced by 1.79- and 1.60-fold, respectively, to normalize for increased expression of the transgene in these two double mutants as determined by quantitatively imaging ceIs56 fluorescence in cell somas in each strain. The cell soma fluorescence of ceIs56 in all other strains is not significantly different from wild type. The means of all eight mutant strains are significantly different from wild type (P < 0.0001 using the unpaired t-test with Welch correction). The text data in A state the mean number of lysosomes/100 μm ± standard error of the mean, counted after defining a threshold level.
To test for a possible role for kinesin (KIF5) in the axonal accumulation of lysosomes, we crossed CTNS-1-RFP into the unc-116(e2310) mutant and observed a 3.1-fold increase relative to wild type (Figure 8, A and B). This phenotype results from a loss of UNC-116 (KIF5) function in motor neurons because transgenic, cell-specific unc-116 RNAi in the same neurons also caused a low, but significant axonal accumulation of the lysosomal marker (Figure S3C). Conversely, expressing the wild type unc-116 cDNA in unc-116 mutant motor neurons rescued, rather than enhanced, the lysosome accumulation phenotype (Figure S3C). The data thus show that conventional kinesin (KIF5) also inhibits the accumulation of the lysosome marker in axons.
An analysis of unc-16; dhc-1 and unc-16; unc-116 double mutants suggested that neither the dynein nor the kinesin motors are responsible for the axonal organelle accumulation that occurs in the absence of unc-16, since both double mutants accumulated lysosomes at levels slightly higher than unc-16 single mutants (Figure 8, A and B). This result could also suggest that the dhc-1 and unc-116 mutations disrupt the same organelle gatekeeper as the unc-16 null mutations, since the double mutants (unc-16; dhc-1 and unc-16; unc-116) did not strongly worsen the organelle accumulation phenotype of unc-16 mutants. However, this analysis quantified overall levels of the organelle markers in axons (total marker fluorescence above background). We also counted lysosomal puncta in the dorsal axons of the same strains. The punctal analysis revealed that unc-16 and unc-116 mutations do not have identical effects on lysosomal accumulation in axons. Most of the CTNS-1-RFP signal in unc-116 mutant axons was diffusely distributed or was associated with vesicles too small to be individually resolved by light microscopy. The number of lysosomal puncta in unc-116 mutants, though significantly higher than wild type, was only 7% of the number in unc-16 mutants (Figure 8A). Similarly, although the dhc-1 mutant accumulated the lysosomal marker in its axons at a high level that was not significantly different from unc-16 mutants, the number of lysosomal puncta in dhc-1 mutant axons was only 27% of the number in unc-16 mutants (Figure 8A). Thus, the lysosomal marker in unc-16 mutant axons appears more likely to be associated with sparsely distributed organelle size compartments, whereas the lysosomal marker in the unc-116 and dhc-1 mutants appears more likely to be associated with uniformly distributed membranes in the axon.
UNC-16 and the JNK-1 MAPK act in the same pathway to prevent axonal lysosome accumulation
Prior studies have shown that UNC-16 and its JIP3 orthologs directly interact with multiple MAP kinase signaling proteins, including C. elegans JNK-1 (a JNK-type MAP kinase) (Ito et al. 1999; Kelkar et al. 2000; Byrd et al. 2001). To test whether MAP kinase signaling contributes to UNC-16’s organelle gatekeeper function, we compared the axonal steady-state levels of CTNS-1-RFP lysosomes in wild type and jnk-1 null mutants. The axonal level of lysosomes in jnk-1 null mutants was 2.5-fold higher than wild type vs. 8.1-fold higher for unc-16 null mutants. unc-16; jnk-1 double mutants did not significantly differ from unc-16 single mutants in axonal lysosome accumulation, consistent with UNC-16 and JNK-1 acting as part of the same process to inhibit the axonal accumulation of lysosomes (Figure 8). The less severe effect of the jnk-1 null mutation compared to the unc-16 null could indicate that JNK-1 has a nonessential modulatory role in regulating the organelle gatekeeper. Alternatively, JNK-1 may act in parallel with one or more of the five other C. elegans MAP kinases in the JNK and/or p38 MAP kinase subgroups (Sakaguchi et al. 2004).
Discussion
Organelle gatekeeper model for UNC-16 (JIP3) function
A prior study proposed that the massive accumulation of organelles in Drosophila SYD (JIP3) mutants is a failure of axonal transport (Bowman et al. 2000) and that the function of JIP3 is to promote organelle transport into axons. Since that study used SYD homozygotes derived from heterozygous mothers, the explanation for why a failure of axonal transport would lead to a buildup rather than a loss of organelles in axons was that a gradual loss of maternally contributed SYD leaves organelles stalled and stranded in axons. We also observed a massive accumulation of organelles in C. elegans unc-16 (JIP3) mutant axons. Using dual, independent organelle markers, we found that the most strongly affected organelles originate from Golgi and endosomal membranes, including lysosomes. However, the C. elegans mutants are homozygous viable and thus have never been exposed to wild-type UNC-16. Time lapse observations of lysosome movements in living, unanesthetized animals and an analysis of lysosome distributions support a model in which UNC-16 functions as an organelle gatekeeper to inhibit the escape of organelles from the axon initial segment (Figure 9). Additional time lapse data showed that organelles escaping the initial segment in unc-16 mutants undergo processive, bidirectional, fast axonal transport in commissures. Thus, instead of promoting organelle movements into axons as previously proposed (Bowman et al. 2000), this new data suggest that UNC-16 (JIP3) and its orthologs normally function to inhibit the escape of organelles from the axon initial segment. Our data suggest that organelles accumulate in unc-16 mutant axons because bidirectional transport of organelles that have escaped the axon initial segment occasionally deposits them in the synaptic region, where their mobility decreases, causing them to accumulate at higher levels than in commissures.
Figure 9.
Organelle gatekeeper model for UNC-16 (JIP3) function. The data from the current study suggest that UNC-16 inhibits the escape of Golgi and endosomal organelles (endosomes and lyosomes) from the axon initial segment. In wild type (top), many lysosomes enter the axon initial segment but, because they tend to return to the cell soma, very few move beyond this region. In mutants lacking UNC-16 (bottom) even more lysosomes are present in the axon initial segment, because they tend not to return to the cell soma, and they are more likely to escape this region and move deeper into the commissure. When organelles escape the axon initial segment, bidirectional transport occasionally deposits them in the synaptic region, where their mobility decreases, causing them to accumulate at higher levels than in commissures.
Prior studies have shown that the axon initial segment has a barrier/filtering function that affects the movement of molecules along the plasma membrane (Winckler et al. 1999) as well as the movement of small transport vesicles carrying specific proteins (Song et al. 2009). However, a role for the axon initial segment in restricting the flow of organelles into axons has not been reported. Our study is the first to demonstrate that a gatekeeper function restricts the flow of Golgi and endosomal organelles, including lysosomes, through the axon initial segment.
Our data do not rule out the possibility that UNC-16 may have one or more other functions or sites of action in addition to its role at the axon initial segment. Indeed, our immunolocalization of native UNC-16 showed that it is present in the synaptic region of axons. Since our time lapse data suggest that UNC-16 promotes the return of organelles from the axon initial segment to the cell soma, we hypothesize that UNC-16 in the synaptic region may also promote the return of selected organelles to the soma. Consistent with UNC-16 targeting selected organelles in the synaptic region, our data showed that UNC-16 was heavily concentrated around large, sparsely distributed puncta in the synaptic region.
A prior study also reported a massive accumulation of the early endosome marker YFP-RAB-5 and unidentified membranous cisternae in unc-16 mutant axons (Brown et al. 2009). However, that study suggested that UNC-16 has a specific role in regulating the dynamics of a RAB-5-associated organelle compartment and, even more specifically, the activity state of the RAB-5 on that compartment. The authors proposed a connection between this RAB-5-specific function and the reduced number of synaptic vesicles at unc-16 mutant synapses (Brown et al. 2009). Our study revealed that UNC-16 has a much more general role as a gatekeeper that acts in the axon initial segment to restrict the flow of Golgi and endosomal organelles, including both early endosomes and lysosomes, into the synaptic region. While it is possible that UNC-16 has both the general organelle function that we discovered as well as an additional RAB-5-specific function that affects synaptic vesicle recycling, we believe that the more general UNC-16 gatekeeper model can also account for the reduced synaptic vesicle numbers at unc-16 mutant synapses, as discussed below.
What molecular motors contribute to the gatekeeper and the bidirectional movements in commissures?
Organelles that have escaped the axon initial segment undergo bidirectional movements that actively transport them within the commissure and occasionally deposit them in the synaptic region. The obvious candidates for the motors mediating these movements are kinesin (the main anterograde motor in axons) and dynein (the main retrograde motor in axons). As previously noted, subunits of both motors are known to directly interact with UNC-16 and its orthologs. However, reduction-of-function kinesin and dynein mutations failed to lessen the organelle accumulations in unc-16 mutant axons, suggesting that other motors mediate these movements. Indeed, we found that a nonnull kinesin reduction-of-function mutation, as well as kinesin RNAi (also likely to cause a mild reduction-of-function phenotype, since RNAi does not work efficiently in neurons) caused a low level of organelle accumulation in the synaptic region of axons, and that neither the kinesin nor the dynein mutations strongly worsened the axonal organelle accumulation in unc-16 null mutants. At face value, this could suggest that kinesin and dynein function with UNC-16 as part of the same organelle gatekeeper, although, as pointed out in the results, there appear to be differences in the organelle accumulation phenotypes caused by the motor protein mutations compared to the unc-16 null. These differences may or may not indicate that the motor protein mutations cause organelle accumulation by a different mechanism. The finding that a kinesin reduction-of-function mutation causes organelle marker accumulation in axons is unexpected because kinesin is an anterograde motor, and thus reducing its function should decrease the anterograde movements of organelles (movements directed away from the cell soma). However, this result is consistent with previous Drosophila studies, which reported massive accumulations of organelles in the axons of fly mutants lacking either Kinesin Heavy Chain or Kinesin Light Chain, as well as other phenotypes, such as larval tail flipping, that are similar to Sunday Driver mutants (Hurd and Saxton 1996; Gindhart et al. 1998; Bowman et al. 2000).
Why do neurons need an organelle gatekeeper?
There are at least three trafficking-related needs which, either singly or in combination, might have driven neurons to evolve an organelle gatekeeper function at the axon initial segment: (1) preserving the integrity and efficiency of synaptic vesicle biogenesis; (2) preventing organelle traffic jams or clogs in axons; and (3) regulating axon growth or repair.
First, neurons may use the gatekeeper to preserve the integrity of synaptic vesicles during their biogenesis or to maintain the efficiency of synaptic biogenesis. During synaptic vesicle biogenesis, synaptic proteins must traverse the Golgi and a portion of the early endosomal system as they come together with other synaptic vesicle proteins to form synaptic vesicle precursors in the cell soma (Bonanomi et al. 2006). In the absence of a gatekeeper, synaptic vesicle proteins might prematurely move from the cell soma into the synaptic region of axons as the cargo of Golgi or endosomal membranes, resulting in decreased synaptic vesicle production. Consistent with this prediction, a previous study found that unc-16 mutations allow a fraction of the synaptic vesicle proteins SNB-1-GFP and SNT-1 to move to axons by a method of transport that does not require the UNC-104 synaptic vesicle motor (Byrd et al. 2001). Since UNC-104 appears to specifically recognize mature synaptic vesicle precursors for transport (Hall and Hedgecock 1991; Okada et al. 1995; Pack-Chung et al. 2007), the UNC-104-independent transport of synaptic vesicle proteins in unc-16 mutants indicates that those proteins are not components of mature synaptic vesicle precursors. Further supporting the idea that unc-16 mutations impact synaptic vesicle biogenesis, synaptic vesicles numbers at motor neuron synapses are reduced in unc-16 mutants (Brown et al. 2009).
Second, neurons may require a gatekeeper function to prevent axonal clogging. Our time lapse experiments show that large organelles dramatically reshape themselves from spherical to elongated compartments as they move through the narrow commissures. The extent to which these large organelles may contribute to temporary or permanent organelle traffic jams in axons is unknown. However, fly mutants with phenotypes that are similar to SYD (JIP3) mutants are described as having organelle “clogs” or “jams” in their axons (Hurd and Saxton 1996; Gindhart et al. 1998). It is possible that this is a bigger problem in animals such as flies, mice, and humans, which live much longer than C. elegans and thus might have a greater chance of developing clogs that could compromise neuronal function. Our study does not address whether the organelle accumulations in the synaptic regions of unc-16 mutants increase linearly over time or whether they reach a steady state that is maintained by counteracting trafficking pathways; however, in nonquantitative observations, we saw similar accumulations in newly hatched animals and adults (K. G. Miller, data not shown), suggesting that the accumulation may reach a steady-state early in the animal’s life (or even embryonically) that is balanced by reentry of some organelles into the cell soma.
Third, wild-type animals may use the organelle gatekeeper to allow Golgi and/or endosomal organelles to flow to distal regions of axons in response to the need for axonal growth or repair after injury. Although our data suggest that Golgi membranes and lysosomes are largely excluded from wild-type motor neurons in healthy animals, it remains possible that specific stressors, such as damaged axons or the need for further growth, could produce a requirement for these organelles in the distal regions of axons. In this case the neuron would inhibit the UNC-16 gatekeeper function to allow the outflow of the required organelles. Thus, rather than acting as a permanent blockade of organelles at the initial segment, the UNC-16 gatekeeper may be regulated by signal transduction pathways to allow organelle outflow in response to specific signals or conditions. Consistent with this idea, we found that the MAP kinase JNK-1, a component of a signal transduction pathway, contributes to UNC-16’s organelle gatekeeper function.
Conclusion
Using the strengths of the genetic model C. elegans we uncovered a previously unrecognized function within neurons. UNC-16’s organelle gatekeeper function is the opposite of what is predicted by the untested stalled transport model, on which the prevailing hypothesis for the function of the fly and human orthologs of UNC-16 is based. The ability to maintain strong nervous system mutants as homozygotes in C. elegans and the ability to perform high-resolution, two-color time lapse imaging of organelles in the axons of living, unanesthetized animals were the key strengths of C. elegans that allowed us to test the stalled transport hypothesis. Although it remains possible that UNC-16 (JIP3) has opposite functions in flies and worms, this seems unlikely given its strong conservation throughout the animal kingdom and given the fact that both the fly and worm JIP3 mutants have massive accumulations of organelles in their axons. Future studies, including forward genetic screens for positive and negative regulators of the gatekeeper, may reveal the pathway components and signals that regulate this important function of neurons.
Supplementary Material
Acknowledgments
We thank Andrew Fire, Yishi Jin, Joshua Kaplan, Michael Nonet, James Rand, and Kang Shen for generously providing some of the plasmids and strains we used in this study (indicated in C. elegans Strains and Plasmids in File S6) and Sandhya Koushika for sharing unpublished data related to unc-16. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from the NIH (GM080765 to K.G.M; R01 MH073156 to J.E.R.) and a grant from the Oklahoma Center for the Advancement of Science to K.G.M. (HR06-078). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Note added in proof: See Zheng and Nonet 2013 (pp. 35–37) in this issue, for a related work.
Footnotes
Communicating editor: K. Kemphues
Literature Cited
- Arimoto M., Koushika S. P., Choudhary B. C., Li C., Matsumoto K., et al. , 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31: 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D., Benfenati F., Valtorta F., 2006. Protein sorting in the synaptic vesicle life cycle. Prog. Neurobiol. 80: 177–217. [DOI] [PubMed] [Google Scholar]
- Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., et al. , 2000. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103: 583–594. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of C. elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. M., Van Epps H. A., Goncharov A., Grant B. D., Jin Y., 2009. The JIP3 scaffold protein UNC-16 regulates RAB-5 dependent membrane trafficking at C. elegans synapses. Dev. Neurobiol. 69: 174–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B., 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139: 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd D. T., Kawasaki M., Walcoff M., Hisamoto N., Matsumoto K., et al. , 2001. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32: 787–800. [DOI] [PubMed] [Google Scholar]
- Charlie N. K., Schade M. A., Thomure A. M., Miller K. G., 2006a Presynaptic UNC-31 (CAPS) is required to activate the Gαs pathway of the synaptic signaling network. Genetics 172: 943–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie N. K., Thomure A. M., Schade M. A., Miller K. G., 2006b The dunce cAMP phosphodisterase PDE-4 negatively regulates Gαs-dependent and Gαs-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network. Genetics 173: 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun D. K., McEwen J. M., Burbea M., Kaplan J. M., 2008. UNC-108/Rab2 Regulates Post-endocytic Trafficking in C. elegans. Mol. Biol. Cell 19: 2682–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K., Murata D., Mizuguchi S., Nomura K. H., Izumikawa T., et al. , 2010. Two Golgi-resident 3′-phosphoadenosine 5′-phosphosulfate transporters play distinct roles in heparan sulfate modifications and embryonic and larval development in Caenorhabditis elegans. J. Biol. Chem. 285: 24717–24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr J. S., Gaskin J., Rand J. B., 2001. Identified neurons in C. elegans coexpress vesicular transporters for acetylcholine and monoamines. Am. J. Physiol. Cell Physiol. 280: C1616–C1622. [DOI] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Milfort M. C., Brown B. S., Gravlin C. N., et al. , 2008. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 6: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Richmond J. E., Hegermann J., Eimer S., et al. , 2009. Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J. Cell Biol. 186: 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Di Schiavi E., Bergamasco C., Bazzicalupo P., 2007. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene 395: 170–176. [DOI] [PubMed] [Google Scholar]
- Gindhart J. G., Jr, Desai C. J., Beushausen S., Zinn K., Goldstein L. S., 1998. Kinesin light chains are essential for axonal transport in Drosophila. J. Cell Biol. 141: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill B., Bienvenut W. V., Brown H. M., Ackley B. D., Quadroni M., et al. , 2007. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55: 587–601. [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Ortiz D., Novick P., 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103: 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb M. S., Burrone J., 2010. Building and maintaining the axon initial segment. Curr. Opin. Neurobiol. 20: 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Hedgecock E. M., 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65: 837–847. [DOI] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C., Bowerman B., 2002. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3: 673–684. [DOI] [PubMed] [Google Scholar]
- Hurd D. D., Saxton W. M., 1996. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144: 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Yoshioka K., Akechi M., Yamashita S., Takamatsu N., et al. , 1999. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol. Cell. Biol. 19: 7539–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatzis V., Cherqui S., Antignac C., Gasnier B., 2001. Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J. 20: 5940–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R. B., Tozser J., Fox J. D., Anderson D. E., Cherry S., et al. , 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14: 993–1000. [DOI] [PubMed] [Google Scholar]
- Kelkar N., Gupta S., Dickens M., Davis R. J., 2000. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol. Cell. Biol. 20: 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar N., Standen C. L., Davis R. J., 2005. Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol. Cell. Biol. 25: 2733–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S. P., Schaefer A. M., Vincent R., Willis J. H., Bowerman B., et al. , 2004. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J. Neurosci. 24: 3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas P. M., Yu X., Miller K. G., Zhou Z., 2008. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 180: 357–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F. R., McGraw T. E., 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5: 121–132. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10(12): 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Emerson M. D., Rand J. B., 1999. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G., Sibarita J. B., Loubery S., Daviet L., Romao M., et al. , 2009. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19: 184–195. [DOI] [PubMed] [Google Scholar]
- Nguyen Q., Lee C. M., Le A., Reddy E. P., 2005. JLP associates with kinesin light chain 1 through a novel leucine zipper-like domain. J. Biol. Chem. 280: 30185–30191. [DOI] [PubMed] [Google Scholar]
- Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N., 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81: 769–780. [DOI] [PubMed] [Google Scholar]
- Orci L., Amherdt M., Ravazzola M., Perrelet A., Rothman J. E., 2000. Exclusion of Golgi residents from transport vesicles budding from Golgi cisternae in intact cells. J. Cell Biol. 150: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E., Kurshan P. T., Dickman D. K., Schwarz T. L., 2007. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat. Neurosci. 10: 980–989. [DOI] [PubMed] [Google Scholar]
- Patel N., Thierry-Mieg D., Mancillas J. R., 1993. Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc. Natl. Acad. Sci. USA 90: 9181–9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mansilla B., Nurrish S., 2009. A network of G-protein signaling pathways control neuronal activity in C. elegans. Adv. Genet. 65: 145–192. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y. A., Scheller R. H., 1998. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143: 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N. K., Schade M. A., Miller K. G., 2005. Convergent, RIC-8 dependent Gα signaling pathways in the C. elegans synaptic signaling network. Genetics 169: 650–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M. M., Hall D. H., Victor M., Stelzer E. H., Rapoport T. A., 2002. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell 13: 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing P., Weimer R. M., Jorgensen E. M., Triller A., Bessereau J. L., 2004. Preservation of immunoreactivity and fine structure of adult C. elegans tissues using high-pressure freezing. J. Histochem. Cytochem. 52: 1–12. [DOI] [PubMed] [Google Scholar]
- Sakaguchi A., Matsumoto K., Hisamoto N., 2004. Roles of MAP kinase cascades in Caenorhabditis elegans. J. Biochem. 136: 7–11. [DOI] [PubMed] [Google Scholar]
- Sakamoto R., Byrd D. T., Brown H. M., Hisamoto N., Matsumoto K., et al. , 2005. The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Mol. Biol. Cell 16: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M. A., Reynolds N. K., Dollins C. M., Miller K. G., 2005. Mutations that rescue the paralysis of C. elegans ric-8 (Synembryn) mutants activate the Gαs pathway and define a third major branch of the synaptic signaling network. Genetics 169: 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D., Ch’ng Q., Dybbs M., Tavazoie M., Kennedy S., et al. , 2005. Systematic analysis of genes required for synapse structure and function. Nature 436: 510–517. [DOI] [PubMed] [Google Scholar]
- Song A. H., Wang D., Chen G., Li Y., Luo J., et al. , 2009. A selective filter for cytoplasmic transport at the axon initial segment. Cell 136: 1148–1160. [DOI] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988 Methods, pp. 596–597 in The Nematode Caenorhabditis elegans, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sumakovic M., Hegermann J., Luo L., Husson S. J., Schwarze K., et al. , 2009. UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J. Cell Biol. 186: 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhu C., Dixit R., Cavalli V., 2011. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 30: 3416–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K. J., Meyer D., Deehan R., Blenis J., Schnapp B. J., et al. , 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152: 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Punge A., Hollopeter G., Willig K. I., Hobson R. J., et al. , 2011. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods 8: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer R. M., Richmond J. E., 2005. Synaptic vesicle docking: a putative role for the Munc18/Sec1 protein family. Curr. Top. Dev. Biol. 65: 83–113. [DOI] [PubMed] [Google Scholar]
- Williams S. L., Lutz S., Charlie N. K., Vettel C., Ailion M., et al. , 2007. Trio’s Rho-specific GEF domain is the missing Gαq effector in C. elegans. Genes Dev. 21: 2731–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B., Forscher P., Mellman I., 1999. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature 397: 698–701. [DOI] [PubMed] [Google Scholar]
- Yang H. Y., Mains P. E., McNally F. J., 2005. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J. Cell Biol. 169: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.