Abstract
The deleterious effects of different X-chromosome dosage in males and females are buffered by a process called dosage compensation, which in Drosophila is achieved through a doubling of X-linked transcription in males. The male-specific lethal complex mediates this process, but is known to act only after gastrulation. Recent work has shown that the transcription of X-linked genes is also upregulated in males prior to gastrulation; whether it results in functional dosage compensation is not known. Absent or partial early dosage compensation raises the possibility of sex-biased expression of key developmental genes, such as the segmentation genes controlling anteroposterior patterning. We assess the functional output of early dosage compensation by measuring the expression of even-skipped (eve) with high spatiotemporal resolution in male and female embryos. We show that eve has a sexually dimorphic pattern, suggesting an interaction with either X-chromosome dose or the sex determination system. By manipulating the gene copy number of an X-linked transcription factor, giant (gt), we traced sex-biased eve patterning to gt dose, indicating that early dosage compensation is functionally incomplete. Despite sex-biased eve expression, the gene networks downstream of eve are able to produce sex-independent segmentation, a point that we establish by measuring the proportions of segments in elongated germ-band embryos. Finally, we use a whole-locus eve transgene with modified cis regulation to demonstrate that segment proportions have a sex-dependent sensitivity to subtle changes in Eve expression. The sex independence of downstream segmentation despite this sensitivity to Eve expression implies that additional autosomal gene- or pathway-specific mechanisms are required to ameliorate the effects of partial early dosage compensation.
Keywords: dosage compensation, Drosophila, segmentation, robustness
IN Drosophila, dosage compensation occurs by an upregulation of transcription from the X chromosome in males (Mukherjee and Beermann 1965; Belote and Lucchesi 1980; Straub et al. 2005). The best-characterized mechanism of upregulation depends on the male-specific activity of the male-specific lethal (MSL) complex (Gelbart and Kuroda 2009), which binds to sites on the X chromosome and enhances the rate of transcript elongation (Larschan et al. 2011). MSL activity is inhibited in females by the translational repression of Msl2 protein, which is necessary for complex formation, by the protein product of the sex-determination gene Sex-lethal (Sxl) (Kelley et al. 1995). The canonical MSL-dependent mechanism does not appear to be active prior to gastrulation as msl2 transcript is not detected until cleavage cycle 14 (Lott et al. 2011) and the earliest expression of Msl3 protein in male embryos is detected only at stage 6 (gastrulation) (Rastelli et al. 1995; Franke et al. 1996; Cline 2005).
Despite the lack of MSL-dependent dosage compensation during the blastoderm stage, the transcription of X-linked genes expressed before gastrulation is, in fact, elevated in males (Lott et al. 2011). With the exception of runt (run), which is known to be dosage-compensated in an MSL-independent but Sxl-dependent manner (Gergen 1987), the mechanism of the upregulation or dosage compensation of early expressed genes is not known.
Although the increased transcription of early expressed X-linked genes in males is suggestive of early dosage compensation, it has not yet been determined whether the observed upregulation results in functional dosage compensation. Two aspects of the pattern of upregulation of X-linked genes lead us to suspect that early dosage compensation can be incomplete: first, the pattern of upregulation is not uniform over the X chromosome, and many genes are upregulated less than twofold in males. Lott et al. (2011) found that 36 of 85 zygotically expressed X-linked genes had an average female/male ratio >1.5 and that X-linked gene expression was female biased. In general, the sex ratios of gene expression were smoothly distributed between 1 and 2, similar to later, MSL-mediated compensation. The second aspect of the pattern of upregulation suggesting incomplete compensation is that the female/male ratio of transcript abundance of X-linked genes varies over cycle 14 (Lott et al. 2011), implying that the efficacy of early dosage compensation can also vary in time during early development.
Many early expressed X-linked genes are transcriptional regulators of key developmental processes such as anteroposterior (AP) (e.g., giant) and dorsoventral (e.g., brinker) patterning. Autosomal targets of these X-linked regulators, such as even-skipped (eve), are expressed as early as cycle 8 (Pritchard and Schubiger 1996), making it possible to test whether early dosage compensation is functional by comparing the expression of target genes in the two sexes. If the observed upregulation of the early expression of X-linked genes is still insufficient for functional dosage compensation, their autosomal targets are expected to be expressed in sexually dimorphic patterns.
We investigate the possibility of incomplete functional dosage compensation and sex-biased pattern formation by characterizing the spatiotemporal expression of an autosomal AP patterning gene (Akam 1987; Surkova et al. 2008), eve, in male and female embryos. Pattern formation has been extensively studied in Drosophila, but no attempt has been made in previous studies to distinguish between the two sexes. Investigations of dosage compensation (Hamada et al. 2005; Lott et al. 2011), in contrast, have ignored potential spatial and/or temporal variation in sex-biased gene expression. eve is expressed in seven stripes during the latter half of cleavage cycle 14 of the Drosophila blastoderm (Macdonald et al. 1986; Frasch et al. 1987) (Figure 1A). Each stripe is about three to five nuclei wide and is established by localized repression of early broad eve expression over a period of ∼30 min (Surkova et al. 2008; Ludwig et al. 2011). Eve expression levels can be measured to an accuracy of ∼10% (Surkova et al. 2008; Ludwig et al. 2011) and provide a sensitive readout of potential sex differences over time and position. This level of precision is desirable since the magnitude of sex ratio variation of X-linked gene expression is relatively small—at most twofold. Here we demonstrate that Eve is expressed at different levels in male and female embryos in the region between stripes one and two (1–2 interstripe).
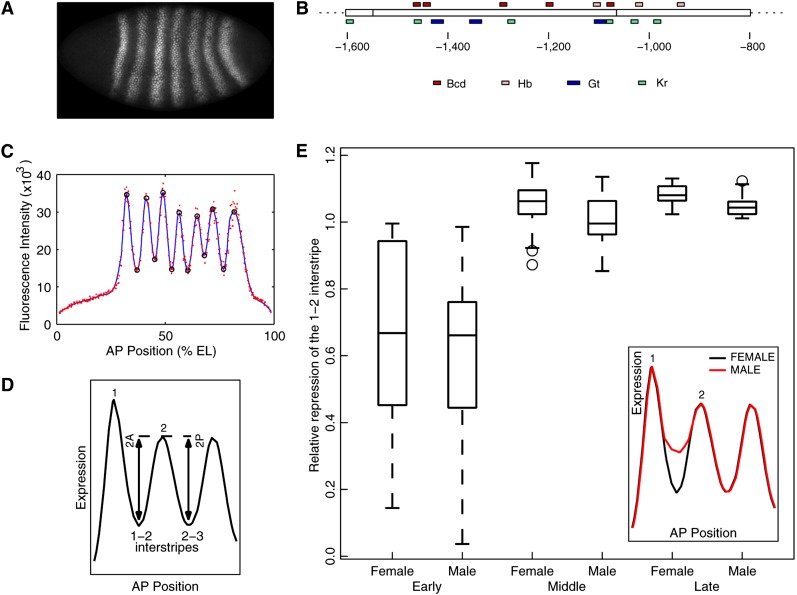
Figure 1.
Sexually dimorphic Eve expression in w1118. (A) The Eve expression pattern in middle–late cleavage cycle 14. Anterior is to the left and dorsal is above. (B) The 800-bp-long 5′ regulatory region that drives stripe 2 expression (Ludwig et al. 2005). Distance is relative to the transcription start site. The box in the middle is the minimal stripe 2 element. Footprinted binding sites (Stanojevic et al. 1989; Small et al. 1991) for the activators Bicoid (Bcd) and Hunchback (Hb) and for the repressors Giant (Gt) and Krüppel (Kr) are shown above and below, respectively. (C) Measurement of the features of the Eve expression pattern. The red points are average fluorescence intensities of Eve staining in nuclei lying in a strip that extends 10% dorsoventrally around the AP axis. The blue line is a smoothing cubic spline fit to data. Black circles are local maxima and minima of the spline. (D) Schematic of the measurement of stripe 2 border height. (E) Boxplot of relative repression of the 1–2 interstripe in w1118 embryos. In all boxplots shown, the box lines are the first quartile, median, and the third quartile. The whiskers extend to the most extreme values lying within 1.5 times the interquartile range, and any datapoints outside the whiskers are shown as circles. Embryos were classified into three 12-min long bins: early, middle, and late (see Materials and Methods). Relative repression was measured as the ratio of the height of the anterior border to that of the posterior border (D). n = 14, 13, 25, 31, 16, 15. The inset is a schematic illustration of the sex-specific expression of Eve.
One advantage of querying functional dosage compensation with a segmentation gene is that it is relatively easy to trace any observed difference both upstream and downstream in the well-established regulatory network (Schroeder et al. 2004; Jaeger 2011). It is known that the second stripe of eve, for example, is activated by Bicoid (Bcd) and Hunchback (Hb) and repressed in the anterior by Giant (Gt) and in the posterior by Krüppel (Kr) (Frasch and Levine 1987; Stanojevic et al. 1991; Small et al. 1992). We exploit this feature of the segmentation system to test the functional compensation of gt, an X-linked gene, by measuring the response of eve expression to gt dose.
Next, we determine whether the sex-biased expression of Eve also leads to sex-biased segmentation during later development. Eve is a regulator of the segment-polarity gene engrailed (en) (Macdonald et al. 1986; Fujioka et al. 1995; Fujioka et al. 2002), which, along with wingless (wg), establishes the molecular prepattern of the segmented embryo (Ingham et al. 1985; Baker 1987). en patterning and the proportions of parasegments have not yet been critically examined in the sexes. Using En expression as a marker, we measure the ratio of parasegments in extended germ-band embryos of each sex to establish that segmentation at a later developmental stage is sex-independent.
A potential explanation for the robustness of en patterning and parasegment proportions is that en expression is insensitive to Eve expression in the 1–2 interstripe region. In a final set of experiments, we test this hypothesis by characterizing the sensitivity of en expression to Eve expression in the 1–2 interstripe. For this purpose, we take advantage of an eve transgene that has a reduced level of Eve expression in the interstripe region. This transgene was derived from the endogenous eve locus by modifying the cis regulation of the stripe 2 enhancer (S2E) (Ludwig et al. 2011). S2E is ∼800 bp long and contains 17 binding sites, identified by in vitro DNase protection assays, for Bcd, Hb, Gt, and Kr (Figure 1B). A 480-bp fragment, called the minimal stripe 2 element, is, however, necessary and sufficient for stripe 2 expression (Small et al. 1992; Ludwig et al. 2005). The transgene MSE was derived from the endogenous locus by deleting 244 bp of sequence flanking the minimal stripe 2 element and tagging the C terminus of Eve with yellow fluorescent protein (YFP). We also constructed a control YFP-tagged transgene, “WT”, which has wild-type eve cis-regulatory sequences. Both WT and MSE provided healthy rescue of the lethality of eveR13, a null allele; Df(2R)eve, a small deletion covering eve; and eveΔMSE, a synthetic allele of eve in which the minimal stripe 2 element has been replaced by w+ sequence. Nevertheless, MSE drove defective stripe 2 formation: Eve expression was lower in both stripe 2 and the 1–2 interstripe region.
We measure Eve and En expression driven by the MSE transgene in each sex to show that (1) 1–2 interstripe Eve expression is lowered specifically in males and becomes indistinguishable from the female level and (2) En expression and parasegment proportions are perturbed in males to give sex-biased segmentation in MSE. Based on these results we argue that en regulation is sensitive to the level of Eve expression in the 1–2 interstripe and postulate that additional mechanisms must act on the segmentation genes to ensure sex-independent larval segmentation in the wild type.
Materials and Methods
Stocks and transgenic fly lines
The following laboratory stocks were used: w1118, y1,sc1,gtX11/FM6 (Bloomington 1529) and Df(1)JA27/FM7c,P[GAL4-Kr.C],P[UAS-GFP.S65T] (Bloomington 5193). The latter two stocks were crossed to produce y1,sc1,gtX11/ FM7c,P[GAL4-Kr.C],P[UAS-GFP.S65T] flies. eveΔMSE is a recessive lethal mutant of eve created by replacing the 480-bp fragment corresponding to the minimal stripe 2 element from the endogenous eve locus with the white+ gene using ends-out homologous recombination (Ludwig et al. 2011).
The construction of the transgenic strains is reported in detail elsewhere (Ludwig et al. 2011Briefly, WT is a 16.4-kb fragment from the eve locus created by recombineering with a Red/ET counterselection BAC Modification Kit (Gene Bridges GmBH, Heidelberg, Germany). A superfolding variant of YFP, SYFP2 (Ben Glick, University of Chicago), was added to the C terminus of the Eve peptide, allowing us to visualize transgenic expression independently of the endogenous locus. MSE was derived from WT by deleting 33 and 211 bp 5′ and 3′ of the minimal stripe 2 element, respectively (Figure 2A). Both constructs were integrated into the same site on the third chromosome (attP2) using phiC31 site-specific integration (Markstein et al. 2008). The gtYFP construct was made similarly, using a 26.2-kb region of the gt locus, tagged with SYFP2 employing Drosophila codon usage (Supporting Information, File S1), and provides healthy rescue of a null allele of gt, gtX11 (Table S2).
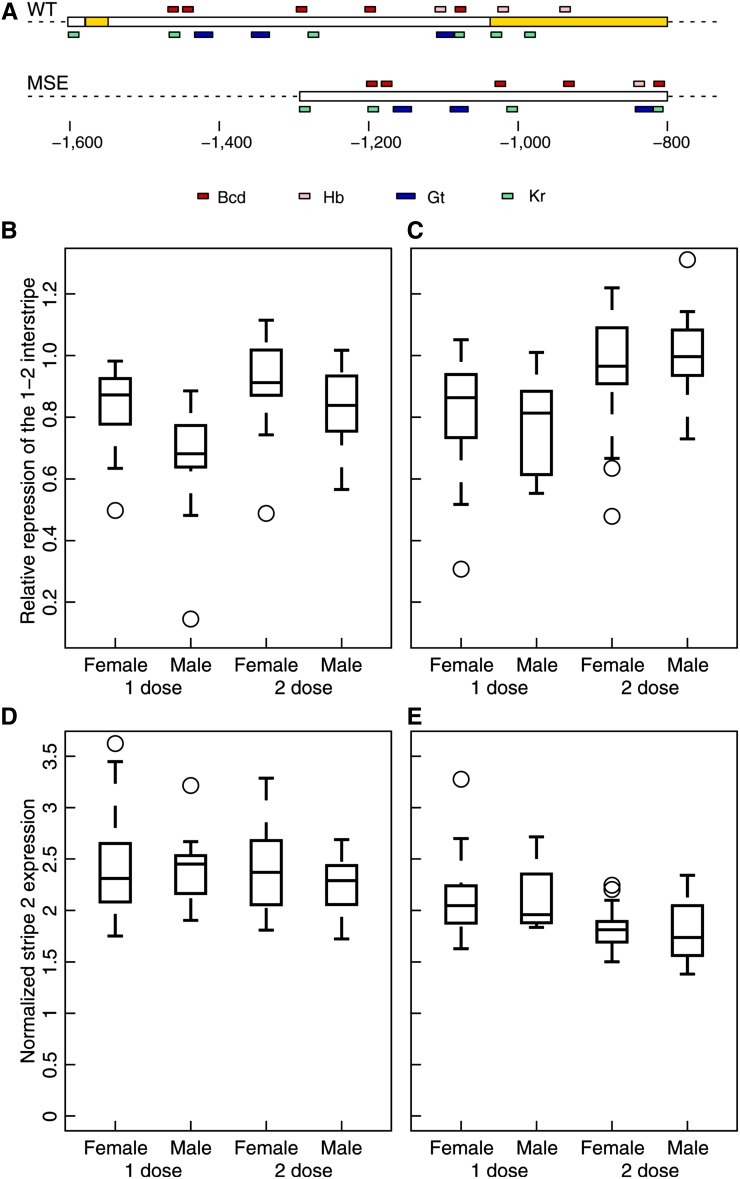
Figure 2.
Stripe 2 expression is sexually dimorphic in WT but lacks sex bias in MSE. Embryos had one copy of the transgene and were either null (eveR13;WT(MSE)/+) or hemizygous (eveR13/CyO,P[hb-Lacz];WT(MSE)/+) for endogenous eve, giving a total eve dose of either one or two, respectively. Data are from embryos in mid-cycle 14 stained for Eve-YFP with anti-GFP. (A) The stripe 2 regulatory region in the WT and MSE eve transgenes. Yellow regions were deleted in MSE. Binding sites are shown as in Figure 1B. (B and D) WT. (C and E) MSE. (B and C) Boxplots of relative repression of the 1–2 interstripe. Relative repression was measured as the ratio of the height of the anterior border to that of the posterior border (Figure 1D). (B) WT; n = 20, 15, 18, 20. (C) MSE; n = 25, 20, 23, 9. (D and E) Boxplots of expression at stripe 2 peak. Peak expression was normalized to mean Eve fluorescence in each embryo. Sample sizes are the same as in B and C.
Genetic crosses, staining, and genotyping for En and Eve patterning
Wild-type Eve patterning was assayed in w1118 embryos (Figure 1). Eve patterning driven by the WT and MSE transgenes (Figure 2) was assayed in embryos from a cross between eveR13/CyO,P[hb-Lacz] females and eveR13;WT(MSE) males, yielding the eveR13;WT(MSE)/+ or eveR13/CyO,P[hb-Lacz];WT(MSE)/+ genotypes. The gt dosage series (Figure 3) was constructed in two separate crosses. Genotypes with zero to two doses were the offspring of a cross between gtX11/FM7c,P[GAL4-Kr.C],P[UAS-GFP.S65T] females and w1118 (w−) males while the three- to four-dose genotypes were the offspring of the cross between gtYFP males and females. To measure the effects of the rescue transgenes on En pattern and segmentation (Figure 4), we crossed eveΔMSE/CyO,P[hb-LacZ] and eveR13;WT(MSE) flies and collected embryos for further analysis.
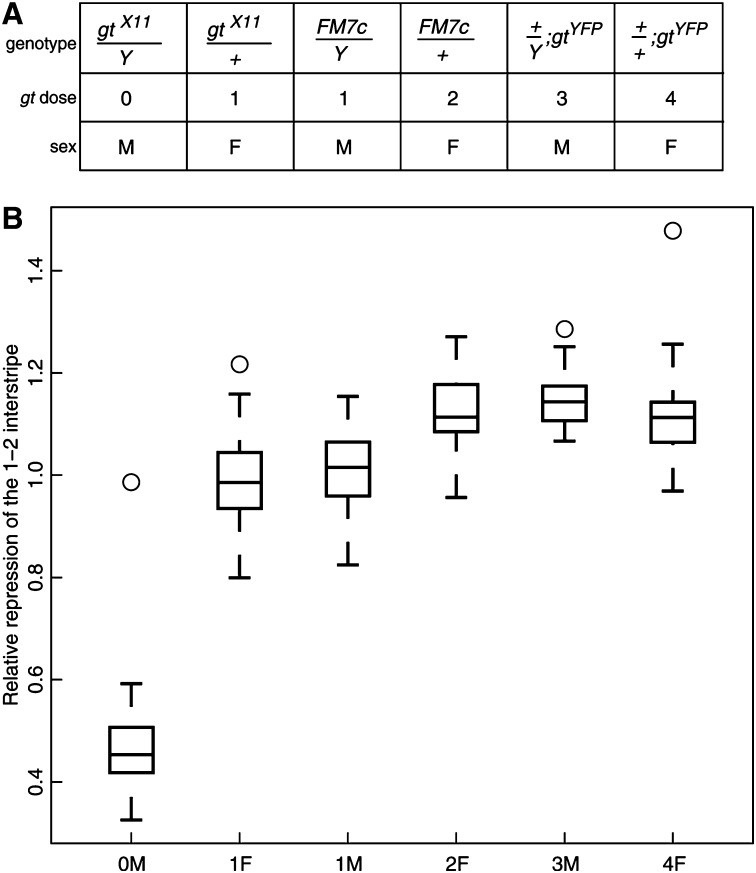
Figure 3.
The response of stripe 2 relative repression to gt dose. (A) The genotypes used to make a dosage series for gt. (B) Response of the relative repression of the 1–2 interstripe to gt dose. Relative repression was measured as before (Figure 1, D and E). Embryos were at mid-cycle 14. n = 13, 28, 16, 14, 22, 17.
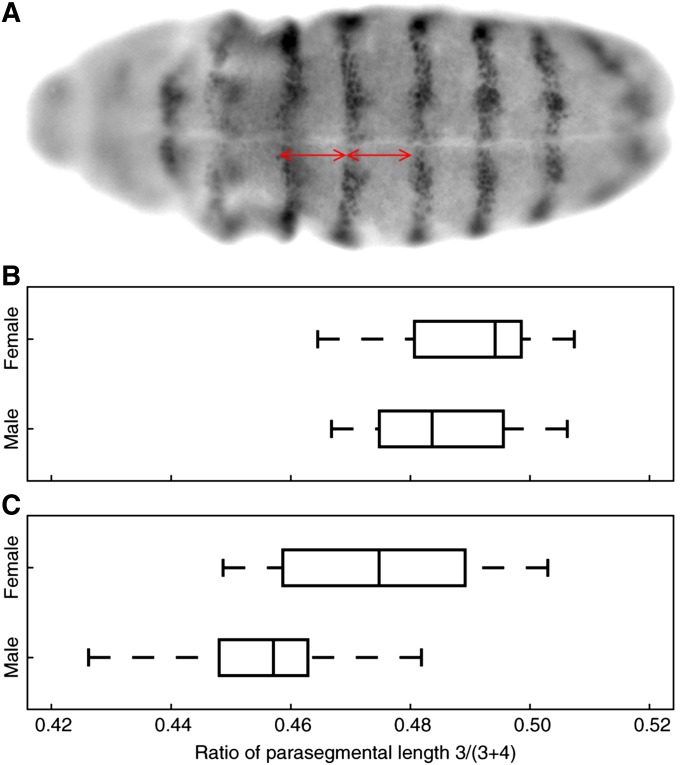
Figure 4.
Parasegmental proportions in the rescue of eveΔMSE/eveR13 lethality by the WT or MSE transgenes. The genotype of the rescued embryos is eveΔMSE/eveR13;WT(MSE)/+. (A) Ventral view of a stage 11 embryo stained for En. Red lines show the measurement of the length of parasegments 3 (left) and 4 (right). (B and C) Boxplot of the ratio of parasegment 3 length to the total length of 3 and 4. (B) WT; from top to bottom, n = 8, 13 embryos. (C) MSE; n = 14, 7 embryos.
Embryos were collected, fixed, and immunostained with antibodies against Eve (Figures 1E and 3), GFP (Figure 2), or En (Figure 4) as described (Ludwig et al. 2005, 2011). Embryos stained fluorescently for GFP and Eve were imaged with a confocal microscope (see below). The lengths of both parasegments 3 and 4 in En pattern were measured at the ventral site of early stage 11 embryos. The reported numbers are the mean of three repeated measurements by hand using ImageJ (National Institutes of Health). After imaging, individual embryos were genotyped by PCR with primers for the Y chromosome, P[hb-LacZ], and P[UAS-GFP.S65T] (File S1) to determine sex and eve or gt dosage. The protocol is available on request.
Confocal imaging and feature detection
Immunofluorescently stained embryos were imaged with a Vti Infinity 3 confocal (Visitech International, Sunderland, MA) and a Zeiss AxioPlan 2 microscope (Carl Zeiss, Inc.) using a 16× PLAN-NEOFLUAR objective. The 491-, 561-, 642-nm lasers, a 488/565/643 dichroic, 535/605/700 emission filter, and an EMCCD camera (Hamamatsu Photonics UK Ltd., Hertfordshire, UK) were employed to acquire images at a resolution of 512 × 512. To maximize dynamic range, camera gain and exposure were chosen so that the brightest embryos in an experiment had a few saturated pixels. All the embryos in an experiment, irrespective of genotype, were imaged with the same settings. Laterally oriented embryos were imaged in three planes 1 μm apart transecting the nuclei at the embryo surface flattened against the coverslip, and the images were averaged. Embryos were also imaged at the mid-sagittal plane to determine their length.
Embryos were staged according to a standard scheme (Surkova et al. 2008) using only membrane invagination as a marker; “early” cycle 14 corresponds to time classes T3–T4, the earliest time at which a nascent stripe 2 is detectable; “middle” is T5–T6; and “late” is T7–T8, ∼9 min prior to gastrulation. The images were segmented using described methods (Janssens et al. 2005), and the location of and the mean fluorescence intensity in each nucleus were saved for further processing.
The positions of and expression values at the extrema were estimated using a smoothing cubic spline (CSAPS function of MATLAB, MathWorks Inc.) fit to data extracted from the nuclei lying in a 10%-wide dorsoventral strip along the anteroposterior axis. The level of smoothing was chosen to minimize the error made by the spline approximation without detecting spurious extrema (Ludwig et al. 2011). The same level of smoothing was applied to all embryos in the same time class irrespective of experiment or genotype. The error made by the spline approximation at the extrema was ∼5%.
Results
Sex-specific eve expression in wild type
To investigate early dosage compensation, we measured the spatiotemporal dynamics of eve expression separately in male and female embryos. We fluorescently immunostained embryos of a standard laboratory stock, w1118, with anti-Eve antibody. These embryos were mounted one per slide and imaged with a confocal microscope. After imaging, the embryo was removed and genotyped for sex using PCR. The images were staged into three classes according to the scheme of Surkova et al. (2008), using only nuclear morphology and membrane invagination as markers. Each class is ∼12 min long, spanning the period from early cycle 14 when Eve is expressed in a triband pattern, to just before gastrulation, when Eve is expressed in seven fully mature saw-tooth-shaped stripes (Frasch et al. 1987; Surkova et al. 2008). We processed the confocal images using an image-processing algorithm (Janssens et al. 2005) to estimate Eve expression in individual nuclei. Finally, we extracted data from a narrow strip lying along the AP axis of the embryo and fit a cubic spline to these data to measure several features of the pattern (Figure 1C).
In all, we measured 43 features of the Eve pattern. We estimated the expression level at the peak of each stripe and at the trough between two stripe peaks, henceforth referred to as the “interstripe.” These measurements were normalized to the mean of Eve expression in all nuclei of the embryo to correct for experimental image intensity variation. We also computed the positions of the stripe peaks, interstripe troughs, and the stripe borders along the AP axis. Since Eve stripes form by localized repression in the interstripe nuclei (Stanojevic et al. 1991; Small et al. 1992), we also define a measure for the strength of repression in an interstripe. Let the height of a stripe border be the difference in the expression level of the stripe peak and the interstripe trough (for example, 2A in Figure 1D). We take the ratio of the border heights of the two borders of a stripe, for example, 2A and 2P in Figure 1D, to obtain a normalized measure of the repression of a border, which we refer to as “relative repression.”
We found evidence for sex-specific patterning differences in the relative repression of three of the seven Eve stripes: stripe 2 (Figure 1E; P = 0.0538 mid-cycle 14 and P = 0.0096 late cycle 14; the Wilcoxon rank-sum test was used here and in all subsequent statistical tests), stripe 3 (Figure S1C, P = 0.0312 late cycle 14), and stripe 4 (Figure S1C, P = 0.0345 late cycle 14). No sex bias was detectable in any of the other features (Figure S1 and Figure S2).
As we show below, the relative repression of stripe 2 (Figure 1E) is the most consistent sex-biased patterning feature, and we focused our subsequent analysis on this feature. There is no detectable difference in early cycle 14, possibly due to the large variability of relative repression during the early stages of stripe 2 formation (Ludwig et al. 2011) (Figure 1E). A measurable difference of ∼7% appears in middle cycle 14 (Figure 1E), with the male having a lower value of relative repression than the female. The difference reduces to ∼4% in late cycle 14, but is still statistically significant due to the precise measurement allowed by the low variability of the late Eve pattern; the standard deviation of relative repression is ∼3%.
Lower values of relative repression imply that the male embryo has a shorter—that is, derepressed—stripe 2 anterior border compared to the female, which is shown schematically in the inset of Figure 1E. We could detect greater Eve expression in the 1–2 interstripe in males during late cycle 14 (Figure S3A, P = 0.0604), whereas no sex difference was detectable in Eve expression in the 2–3 interstripe (Figure S3B, P > 0.2), suggesting that sex-biased relative repression is attributable to higher Eve expression in the 1–2 interstripe.
Sex-specific features of Eve patterning were rechecked in independent experiments using the WT eve transgene, a 16.4-kb region from the eve locus, tagged with YFP and integrated on the third chromosome (Table S1). We crossed eveR13/CyO,P[hb-Lacz] females with eveR13;WT males. Embryos collected from the cross were mounted, imaged, and genotyped in the same manner as the w1118 embryos, with two differences. First, we detected the CyO balancer by costaining with β-galactosidase antibody to identify embryos with one (eveR13;WT/+) or two (eveR13/CyO,P[hb-Lacz];WT/+) eve doses. Second, we focused on middle cycle 14 to maximize sample size since this cross produced many more genotypes than the w1118 experiment.
This experiment confirmed the sex-dependent patterning of stripe 2. Males have a derepressed anterior border compared to females in WT (Figure 2B; P = 0.0012, one eve dose and P = 0.0346, two eve doses). Relative repression and 1–2 interstripe expression were the only sex-biased stripe 2 features in WT (Figure S4); Eve expression in the 1–2 interstripe was elevated in males relative to females (Figure S5; P = 0.0021, one eve dose and P = 0.2395, two eve doses), replicating w1118 results. The derepression of the 1–2 interstripe is eve-dose-dependent since relative repression differs between the sexes by ∼22 and ∼8% in the one- and two-dose rescues, respectively (Figure 2B). The other sex differences—in the relative repression of stripes 3 and 4 in w1118 (Figure S1C) and a new one found in this experiment in the relative repression of stripe 5 (Figure S6C)—were inconsistent between the two experiments.
Incomplete dosage compensation of gt
The sex-specific features of eve expression in wild type implies that there is an as-yet-unappreciated interaction between the sex determination or dosage compensation systems and the segmentation genes. One possibility is that sex-determination genes, such as Sxl, regulate eve’s expression in the 1–2 interstripe. Another possibility is that one or more X-linked genes regulating eve are not dosage-compensated and have different levels of expression in the two sexes, resulting in eve’s sex-dependent features. To regulate eve specifically in the 1–2 interstripe, the sex-determination genes would need to be expressed in an AP-position-dependent manner, but are, in fact, expressed uniformly throughout the embryo (Erickson and Cline 1993). Although uniform expression does not completely preclude an interaction with eve, we first checked the latter possibility—that eve’s sex-specific expression originates in the lack of dosage compensation for one or more X-linked loci—by varying the dosage of a candidate X-linked locus, gt, and measuring the response of the eve interstripe phenotype.
gt is a promising candidate as a source of dimorphic eve expression in the 1–2 interstripe because Gt protein is known to repress eve expression in that region. The minimal stripe 2 enhancer contains three footprinted binding sites for Gt (Stanojevic et al. 1991; Small et al. 1992). Deletion of these sites leads to a derepression of the anterior border in a reporter assay (Stanojevic et al. 1991; Small et al. 1992). Stripes 1 and 2 are fused in gt− embryos (Frasch et al. 1987), suggesting that the phenotype is responsive to gt dose. Given the known role of Gt as a repressor of eve, the observed derepression of eve expression in the 1–2 interstripe is consistent with a lower level of Gt protein expression in male embryos.
Our strategy was to vary the gene dose of gt in the two sexes and to measure the response of the eve 1–2 interstripe phenotype. The reasons to measure the response of eve instead of Gt expression levels directly are twofold. First, the difference between male and female gt expression is potentially quite small (Lott et al. 2011), and the precise and dynamic spatiotemporal expression of eve allows for much greater sensitivity of detection. Second, this experiment would test not only whether gt is dosage-compensated but also whether it drives the observed sex-specific features of eve expression.
We crossed gtX11/FM7c flies (a null allele; Table S1) to w1118 to generate gt− male embryos (0M; the number is gt dose and the letter signifies sex), 1-gt-dose female embryos (1F), 1-gt-dose male embryos (1M), and 2-gt-dose female embryos (2F) (Figure 3A). Using the same experimental procedures used to establish the sex-specific expression of Eve, we imaged Eve expression and measured its features quantitatively in these genotypes. In agreement with our earlier results, the 1M genotype has a derepressed 1–2 interstripe relative to 2F in middle cycle 14 (Figure 3B, P = 0.0011).
The 1F genotype is hemizygous for gt but homozygous for all other X-linked loci. 1F also produces a derepressed interstripe (Figure 3B, P = 0.0002), showing that a lower gt dose leads to a derepression of the 1–2 interstripe in female embryos. In fact, 1F and 1M are indistinguishable from each other (P = 0.3864), strongly suggesting that gt accounts for all of the effect.
Because gt regulates the expression of eve 5–6 and 6–7 interstripes (Frasch and Levine 1987) in addition to that of interstripe 1–2, their expression should have a similar response to gt dose. Figure S7 shows the P-values of the rank-sum test for relative interstripe expression between 2F and 0M, 1F, or 1M. 0M (gt−) differs from 2F in the interstripes 1–2, 5–6, and 6–7. Both 1M and 1F differ from 2F in the 1–2 and 6–7 interstripes but not elsewhere.
We also used a whole-locus transgene for gt, gt-YFP (Table S1), which provides healthy rescue of gt− (Table S2) to boost gt dose to 3 in males (3M) and 4 in females (4F) (Figure 3A). As expected, repression of the 1–2 interstripe is an increasing function of gt dose (Figure 3B). The dose–response curve saturates after two doses, however, suggesting that two gt doses are sufficient to completely turn off eve transcription in 1–2 interstripe nuclei.
Late segmentation is sex-independent
Do the sex-dependent features of Eve expression lead to sexually dimorphic segmentation of the embryo? Although no gross differences in the segmentation of embryos between males and females have been observed to date, we conducted a quantitative analysis to detect more subtle differences in the proportions of segments. We measured the proportions of parasegments 3 and 4 in extended germ-band-stage embryos using En expression as a marker. en is expressed in the anterior part of parasegments, and its anterior margin marks parasegmental grooves (Ingham et al. 1985). Parasegment 3 is under the control of Eve expression in stripe 2 since embryos homozygous for eveΔMSE (Table S1), the endogenous eve locus in which the stripe 2 enhancer has been replaced by w+ sequence, lack eve stripe 2 expression and have defective third parasegments (Ludwig et al. 2005).
We crossed eveΔMSE/CyO,P[hb-LacZ] and eveR13;WT flies to specifically rescue the lethality of stripe 2 ablation with one dose of the WT eve transgene. Stage 11 embryos were costained with β-galactosidase to identify the eveΔMSE/eveR13 genotype and En antibodies. We measured the number of pixels between the anterior margins of En stripes 3–4 and 4–5 on the ventral side to estimate of the length of parasegments 3 and 4, respectively (Figure 4A). The length of parasegment 3 was normalized to that of the total length of parasegments 3 and 4 to correct for variation in embryo size. This comparison between male and female embryos revealed no evidence for a difference in the median ratio of the parasegmental length of 3 to 3 + 4 (Figure 4B, P = 0.2937). The sex-specific features of Eve stripe 2 expression do not, therefore, appear to be carried forward developmentally to segmentation in extended germ-band-stage embryos.
Sex-independent segmentation in WT requires compensatory mechanisms
The sex-dependent features of eve expression pose potential challenges for its downstream targets, such as the pair-rule genes fushi-tarazu, runt, and hairy and the segment polarity genes en and wg. For late segmentation to be the same in the two sexes, are downstream genes indifferent to sex-specific eve expression? Or are there mechanisms interceding to attenuate the sex-dependent differences in eve expression?
The relative repression in males differs by a small amount, ∼10%, from the female level. One possible explanation for the apparent robustness is simply that en or other eve targets are insensitive to the relative repression of stripe 2. In this section, we test the sensitivity of en regulation to sex-specific Eve expression by assaying Eve and En expression in male and female embryos of a transgenic line having altered levels of relative repression.
In earlier work investigating the robustness of the cis regulation of eve stripe 2 (Ludwig et al. 2011), we modified the WT eve transgene to create another transgene, MSE (Table S1). MSE was derived from the WT transgene by deleting 244 bp of regulatory sequence flanking the minimal stripe 2 element (Figure 2A). Stripe 2 expression driven by the MSE transgene is weaker than WT expression, has greater variability than WT, and lacks temperature compensation.
Of more direct interest to us, MSE also has a different level of interstripe repression than WT, which offers an opportunity to test the potential functional impact of interstripe derepression in males. Time series data of live Eve-YFP expression showed that the relative repression of the 1–2 interstripe was greater in MSE relative to WT (Ludwig et al. 2011). If the ∼10% difference between males and females is functionally relevant, modulation of the level of repression should result in downstream segmentation phenotypes.
The live-imaging experiments (Ludwig et al. 2011) had not distinguished between the sexes; male or female embryos, or both, might have greater relative repression than their WT counterparts. We therefore measured the features of Eve expression driven by the MSE transgene separately in fixed male and female embryos that had either one (eveR13;MSE/+) or two (eveR13/CyO,P[hb-Lacz];MSE/+) doses of eve. Confocal imaging and image processing of MSE were carried out with settings identical to WT.
MSE males lose the derepression of the 1–2 interstripe and have a level of relative repression indistinguishable from MSE females (Figure 2C; P = 0.3145, one dose, and P = 0.5686, two doses). The increase in relative repression in MSE relative to WT is restricted to males (Figure 2, B and C; P = 0.3711, one dose, and P = 0.0002, two doses), and relative repression is not altered in females (Figure 2, B and C; P = 0.7609, one dose, and P = 0.2423, two doses); the sequences removed in MSE appear to affect eve expression specifically in males. One possible explanation for the insensitivity of relative repression in females is that two gt copies are sufficient to completely turn transcription off in the interstripe (Figure 3B). We were not able to detect sex-specific expression in any other stripe 2 feature in MSE (Figure S4). In particular, although peak stripe 2 expression is lower in MSE relative to WT [Figure 2, D and E; P(female) = 0.0126 and P(male) = 0.0950 in one dose, P(female) = 4.9548E-6 and P(male) = 0.0001 in two doses], peak expression does not differ between males and females in either line (P > 0.4, WT, and P > 0.6, MSE). The loss of 1–2 interstripe derepression in MSE males allows us to test its functional impact on downstream segmentation.
We assayed the proportions of parasegments 3 and 4 in eveΔMSE embryos rescued by one dose of the MSE transgene in the same manner as WT. We found that eve misregulation by MSE induces sex-specific en patterning. In contrast to WT (Figure 4B), the median ratio of the parasegmental length of 3 to 3 + 4 differs significantly between MSE males and females (Figure 4C; P = 0.0571). Assuming that each parasegment is 20 cells long (Ingham et al. 1985), the difference in MSE males and females (3.7%) corresponds to a shift of about one cell. This result indicates that the proportions of parasegments 3 and 4 are sensitive to subtle alteration of Eve expression. Even though both 1–2 interstripe and peak stripe 2 expression are altered in MSE, only the former is sex-dependent (Figure 2). For this reason, we tentatively conclude that the sex bias in En patterning results specifically from the sensitivity of en regulation to Eve 1-2 interstripe repression. It is not the case that sexually dimorphic eve expression is simply below the detection limit of downstream gene regulation. This in turn implies that as-yet-unknown mechanisms must compensate for sexually dimorphic eve expression in WT to produce sex-independent segmentation.
Discussion
Genomic transcriptional profiling of individual embryos established that transcription from the X chromosome is upregulated in males prior to gastrulation (Lott et al. 2011). It was not known whether the upregulation of transcription in males leads to functional dosage compensation. We suspected that early dosage compensation might be incomplete since the female/male ratio of gene expression is not uniform but rather varies between 1 (perfect compensation) and 2 (no compensation) (Lott et al. 2011). Incomplete early dosage compensation could lead to sex-biased pattern formation in the blastoderm.
We confirmed this suspicion by measuring the spatiotemporal expression of eve in wild-type male and female embryos. Whereas studies of spatiotemporally resolved patterning have ignored sex (Fowlkes et al. 2008; Surkova et al. 2008), genome-wide studies of dosage compensation (Hamada et al. 2005; Lott et al. 2011) have relied on whole-embryo extracts and hence have ignored differential spatial regulation between the two sexes. Eve expression differs between males and females in the 1–2 interstripe (Figures 1E and 2B) and perhaps in the 5–6 and 6–7 interstripes (Figure S7), which altogether comprise only ∼15% of embryo length. Dimorphic patterning of eve is remarkable since the robustness of eve expression has been extensively documented (Lucchetta et al. 2005; Holloway et al. 2006; Lott et al. 2007; Surkova et al. 2008; Ludwig et al. 2011); this is perhaps the first example of the lack of robustness among the segmentation genes.
Genetic analysis showed that the derepression phenotype is under the specific control of gt dose (Figure 3B). Despite empirical evidence for the upregulation of gt transcription in males (Lott et al. 2011), our results suggest that gt dosage compensation is partial and that incomplete compensation has functional consequences. It is notable that 1-gt-dose males and females are indistinguishable with respect to 1–2 interstripe repression (Figure 3B). The equivalence of gt functional output in males and hemizygous females implies that the mechanism compensating for gt dose operates independently of sex. run was found to rely on Sxl for its dosage compensation (Gergen 1987), and its messenger RNA (mRNA) contains Sxl-binding sites (Kelley et al. 1995). Consistent with a non-sex-dependent mode of compensation, gt mRNA lacks Sxl-binding sites (Lott et al. 2011).
Despite eve’s sex-biased expression, we could not detect any sex bias in the expression of En in extended germ-band eveΔMSE/eveR13;WT/+ embryos (Figure 4B). It is possible that the shift in the placement of parasegments is smaller than our measurement precision. However, given that we can detect a sex-specific shift of about one cell in eveΔMSE/eveR13;MSE/+ embryos, the shift in WT, if any, is likely too small to be biologically meaningful.
The differential expression of Eve in WT and MSE suggests two alternative hypotheses for the sex-specific shift in parasegment 3 En expression in MSE. Stripe 2 expression differs between WT and MSE in two aspects (Ludwig et al. 2011): (1) 1–2 interstripe repression is greater in MSE (Figure 2, B and C) and (2) peak expression is lower in MSE (Figure 2, D and E). According to the first hypothesis, the shift of parasegment 3 En expression in MSE males is specifically attributable to the first Eve phenotype—i.e., greater repression of the 1–2 interstripe. The alternative hypothesis proposes that the shift in En expression is due to lowered Eve stripe 2 expression in MSE, whereas its sex dependence arises for unrelated and as-yet-undefined reasons. We favor the first explanation for two reasons: (1) the increased repression of the 1–2 interstripe is, in fact, a sex-dependent Eve phenotype (Figure 2 and Figure S4) and (2) it is unlikely that the sex dependence of the En phenotype is unrelated to Eve expression because WT and MSE are co-isogenic strains, differing only in their eve transgene sequence. Nevertheless, experiments selectively disrupting either 1–2 interstripe expression or peak stripe 2 expression will be required to conclusively distinguish between these two hypotheses.
We propose that additional mechanisms are required to compensate for the effects of dimorphic Eve expression and to produce sex-independent En patterning. Our results allow us to infer some general properties of such buffering. First, it must be independent of known dosage compensation mechanisms since it would have to act on autosomal gene expression in a tissue-specific manner. Second, the mechanism appears to be specifically targeted to the derepression of the 1–2 interstripe in wild-type males since the MSE line, which has lost the derepression, has defective En patterning. One hypothesis for a downstream buffering mechanism is eve autoregulation (Harding et al. 1989) because we observe a smaller median shift of relative repression between males and females with two eve doses (Figure 2B, 8%) than with one eve dose (22%). Alternatively, the differential expression of gt may induce sex-specific expression in other pair-rule genes, which in turn could compensate for sex-specific eve expression in the regulation of the segment polarity genes. This might explain why en expression becomes sex-biased (Figure 4C) when eve is misregulated in MSE to have sex-independent expression (Figure 2C). The MSE transgene also shows that a functional stripe 2 enhancer architecture that suppresses sex-specific gt expression is possible even though the WT enhancer itself transduces differential gt expression. Cis-regulatory architecture, therefore, may be playing an integral role in fine-tuning sex-biased gene expression and perhaps has evolutionarily conserved features.
In previous work (Ludwig et al. 2011), we noted that the modifications to eve’s cis-regulation in MSE sensitize the strain to X-dosage perturbations. Even though both WT and MSE rescue eveR13 lethality with high efficiency, the male/female ratio is much lower in rescue by MSE [P = 0.01, homozygote; P = 2.3E-23, hemizygote; figure 1B and figure S1 of Ludwig et al. (2011), respectively]. Sensitivity to X dosage appears to specifically originate in stripe 2 regulatory sequences: while both WT and MSE rescue the lethality of eveΔMSE (Ludwig et al. 2005, 2011), an eve locus lacking the minimal stripe 2 element, only the MSE rescue genotype exhibits a reduced male/female ratio [P = 2.1E-5; figure S3 of Ludwig et al. (2011)].
Based on available evidence, we argue that the modulation of the repression of the 1–2 interstripe by gt dosage and stripe 2 cis-regulatory interactions underlies the sensitivity of the MSE strain to X dosage, although a definitive proof of causality will have to await further experiments. First, the loss of adult male viability in MSE is not peculiar to any mutant chromosome; the rescue of all tested chromosomes—eveR13/eveR13, eveR13/Df(2R)eve, and eveΔMSE/eveR13—shows loss of male viability (Ludwig et al. 2011). Second, in hemizygous rescue, MSE has significantly greater lethality than WT during embryogenesis, a point established in our earlier study (Ludwig et al. 2011). Third, as we noted above, the loss of male viability originates specifically in stripe 2 sequences; the WT and MSE transgenes are co-isogenic except for 244 bp of sequence flanking the minimal stripe 2 element. Notwithstanding these arguments, it is possible that the mutated stripe 2 sequences cause lethality not via parasegment 3 en misregulation, but by their pleiotropic effects on eve regulation in other tissues, such as the CNS (Fujioka et al. 1999). Although enhancers have been regarded as largely autonomous so far, long-range interactions between enhancers have been reported recently (Dunipace et al. 2011; Perry et al. 2011). Inspecting eve’s neuronal expression for sex bias might help to affirm or eliminate this possibility.
The incomplete dosage compensation of gt induces differences that are able to propagate through the segmentation hierarchy to the pair-rule genes in a sex-specific manner, but are attenuated before reaching the segment-polarity genes. We speculate that the propagation of these differences and mechanisms for their correction must be widespread since incomplete dosage compensation itself is common in other organisms and in evolution. In Drosophila, MSL-mediated compensation, like early dosage compensation, is also non-uniform (Hamada et al. 2005). In humans, a total of 25% of X-linked genes escape or show variable patterns of inactivation (Carrel and Willard 2005), and, as in Drosophila, canonical dosage compensation is inactive early in embryonic development (Steele 1970). Incomplete dosage compensation also arises during the evolution of neo-X chromosomes, and in ZW sex-determination systems, such as birds, global dosage compensation is replaced by a variable system of tissue-specific compensation of individual genes (Mank and Ellegren 2009). Phenotypic robustness is achieved not only through intrinsic or emergent mechanisms acting in gene networks (Manu et al. 2009; Acar et al. 2010) but also through specific adaptations involving individual genes, as exemplified by the ubiquity of microRNAs and redundant enhancers (Li et al. 2009; Frankel et al. 2010; Perry et al. 2010). Buffering of incomplete dosage compensation is likewise expected to be target-specific and therefore common, but subject to continual adaptive pressure to track changes in the efficacy of dosage compensation of continually evolving X-linked genes.
Supplementary Material
Acknowledgments
We thank J. Jaeger for providing MATLAB segmentation code; S. Wang, C. Williams, and A. Victorsen for technical assistance; J. Gavin-Smith for comments on the manuscript; and C. Miles, B. He, S. Lott, A. B. Carvalho, and K. P. White for discussion. This work was supported by award no. 1R01GM078381 (M.Z.L. and M.K.) from the National Institutes of Health and grant no. 5P50GM081892 from the National Institute of General Medical Sciences.
Footnotes
Communicating editor: T. Schüpbach
Literature Cited
- Acar M., Pando B. F., Arnold F. H., Elowitz M. B., Van Oudenaarden A., 2010. A general mechanism for network-dosage compensation in gene circuits. Science 329: 1656–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M., 1987. The molecular basis for metameric pattern in the Drosophila embryo. Development 101: 1–22. [PubMed] [Google Scholar]
- Baker N. E., 1987. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 6: 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote J. M., Lucchesi J. C., 1980. Control of X chromosome transcription by the maleless gene in Drosophila. Nature 285: 573–575. [DOI] [PubMed] [Google Scholar]
- Carrel L., Willard H. F., 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404. [DOI] [PubMed] [Google Scholar]
- Cline T. W., 2005. Reflections on a path to sexual commitment. Genetics 169: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L., Ozdemir A., Stathopoulos A., 2011. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development 138: 4075–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Cline T. W., 1993. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 7: 1688–1702. [DOI] [PubMed] [Google Scholar]
- Fowlkes C. C., Hendriks C. L. L., Keränen S. V. E., Weber G. H., Rübel O., et al. , 2008. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell 133: 364–374. [DOI] [PubMed] [Google Scholar]
- Franke A., Dernburg A., Bashaw G. J., Baker B. S., 1996. Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development 122: 2751–2760. [DOI] [PubMed] [Google Scholar]
- Frankel N., Davis G. K., Vargas D., Wang S., Payre F., et al. , 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Levine M., 1987. Complementary patterns of even-skipped and fushi-tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1: 981–995. [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H. J., Levine M., 1987. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6: 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Jaynes J. B., Goto T., 1995. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development 121: 4371–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Emi-Sarker Y., Yusibova G. L., Goto T., Jaynes J. B., 1999. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126: 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Yusibova G. L., Patel N. H., Brown S. J., Jaynes J. B., 2002. The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development 129: 4411–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J. P., 1987. Dosage compensation in Drosophila: evidence that daughterless and Sex-lethal control X chromosome activity at the blastoderm stage of embryogenesis. Genetics 117: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N., Park P. J., Gordadze P. R., Kuroda M. I., 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K., Hoey T., Warrior R., Levine M., 1989. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 8: 1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway D. M., Harrison L. G., Kosman D., Vanario-Alonso C. E., Spirov A. V., 2006. Analysis of pattern precision shows that Drosophila segmentation develops substantial independence from gradients of maternal gene products. Dev. Dyn. 235: 2949–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., Martinez-Arias A., Lawrence P. A., Howard K. R., 1985. Expression of engrailed in the parasegment of Drosophila. Nature 317: 634–636. [Google Scholar]
- Jaeger J., 2011. The gap gene network. Cell. Mol. Life Sci. 68: 243–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H., Kosman D., Vanario-Alonso C. E., Jaeger J., Samsonova M., et al. , 2005. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Dev. Genes Evol. 215: 374–381. [DOI] [PubMed] [Google Scholar]
- Kelley R. L., Solovyeva I., Lyman L. M., Richman R., Solovyev V., et al. , 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877. [DOI] [PubMed] [Google Scholar]
- Larschan E., Bishop E. P., Kharchenko P. V., Core L. J., Lis J. T., et al. , 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W., 2009. A microRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Kreitman M., Palsson A., Alekseeva E., Ludwig M. Z., 2007. Canalization of segmentation and its evolution in Drosophila. Proc. Natl. Acad. Sci. USA 104: 10926–10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A., et al. , 2011. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-Seq. PLoS Biol. 9: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta E. M., Lee J. H., Fu L. A., Patel N. H., Ismagilov R. F., 2005. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434: 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., Palsson A., Alekseeva E., Bergman C. M., Nathan J., et al. , 2005. Functional evolution of a cis-regulatory module. PLoS Biol. 3: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., Manu , Kittler R., White K. P., Kreitman M., 2011. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 7(11): e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G., 1986. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell 47: 721–734. [DOI] [PubMed] [Google Scholar]
- Mank J. E., Ellegren H., 2009. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102: 312–320. [DOI] [PubMed] [Google Scholar]
- Manu S. Surkova, A. V. Spirov, V. Gursky, H. Janssens et al, 2009. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 7: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. S., Beermann W., 1965. Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 207: 785–786. [DOI] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Bothma J. P., Levine M., 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20: 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Levine M., 2011. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 108: 13570–13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard D. K., Schubiger G., 1996. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10: 1131–1142. [DOI] [PubMed] [Google Scholar]
- Rastelli L., Richman R., Kuroda M. I., 1995. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev. 53: 223–233. [DOI] [PubMed] [Google Scholar]
- Schroeder M. D., Pearce M., Fak J., Fan H., Unnerstall U., et al. , 2004. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2: E271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Kraut R., Hoey T., Warrior R., Levine M., 1991. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 5: 827–839. [DOI] [PubMed] [Google Scholar]
- Small S., Blair A., Levine M., 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11: 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D., Hoey T., Levine M., 1989. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature 341: 331–335. [DOI] [PubMed] [Google Scholar]
- Stanojevic D., Small S., Levine M., 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254: 1385–1387. [DOI] [PubMed] [Google Scholar]
- Steele M. W., 1970. Incomplete dosage compensation for glucose-6-phosphate dehydrogenase in human embryos and newborns. Nature 227: 496–498. [DOI] [PubMed] [Google Scholar]
- Straub T., Gilfillan G. D., Maier V. K., Becker P. B., 2005. The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 19: 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu E. Myasnikova et al, 2008. Characterization of the Drosophila segment determination morphome. Dev. Biol. 313: 844–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.