Abstract
Prezygotic barriers play a major role in the evolution of reproductive isolation, which is a prerequisite for speciation. However, despite considerable progress in identifying genes and mutations responsible for postzygotic isolation, little is known about the genetic and molecular basis underlying prezygotic barriers. The bumblebee-pollinated Mimulus lewisii and the hummingbird-pollinated M. cardinalis represent a classic example of pollinator-mediated prezygotic isolation between two sister species in sympatry. Flower color differences resulting from both carotenoid and anthocyanin pigments contribute to pollinator discrimination between the two species in nature. Through fine-scale genetic mapping, site-directed mutagenesis, and transgenic experiments, we demonstrate that a single-repeat R3 MYB repressor, ROSE INTENSITY1 (ROI1), is the causal gene underlying a major quantitative trait locus (QTL) with the largest effect on anthocyanin concentration and that cis-regulatory change rather than coding DNA mutations cause the allelic difference between M. lewisii and M. cardinalis. Together with the genomic resources and stable transgenic tools developed here, these results suggest that Mimulus is an excellent platform for studying the genetics of pollinator-mediated reproductive isolation and the molecular basis of morphological evolution at the most fundamental level—gene by gene, mutation by mutation.
Keywords: reproductive isolation, Mimulus, R3 MYB, anthocyanin concentration, flower color
UNDERSTANDING the origin of reproductive isolation barriers is a prerequisite for understanding speciation (Coyne and Orr 2004). Reproductive isolation barriers are usually partitioned into prezygotic factors (e.g., mate choice behavior, habitat adaptation, timing of reproduction, pollinator preference) and postzygotic factors (e.g., hybrid sterility, hybrid inviability) (Dobzhansky 1937; Mayr 1942; Coyne and Orr 2004). Prezygotic barriers are often thought to play a major role in the evolution of reproductive isolation (Dobzhansky 1937; Mayr 1942; Schluter 2001; Coyne and Orr 2004; Rieseberg and Willis 2007; Lowry et al. 2008) because they act early in the interaction between potential mates. However, despite considerable progress in identifying genes and mutations responsible for postzygotic isolation (Presgraves 2010; Rieseberg and Blackman 2010), little is known about the genetic and molecular basis underlying prezygotic barriers.
The pollinator-mediated prezygotic isolation between two sister species of monkeyflowers, Mimulus lewisii and M. cardinalis, represents a classic example for studying reproductive isolation and speciation in a natural ecological setting (Hiesey et al. 1971; Bradshaw et al. 1995; Schemske and Bradshaw 1999; Bradshaw and Schemske 2003; Ramsey et al. 2003). The bumblebee-pollinated M. lewisii has pale pink flowers with broad, flat petals and contrasting yellow nectar guides (Figure 1A). The hummingbird-pollinated M. cardinalis has red flowers with reflexed petals that form a tubular corolla (Figure 1B). Although the two species are interfertile with hand pollination, they rarely hybridize in wild sympatric populations due to pollinator preference (Hiesey et al. 1971; Schemske and Bradshaw 1999; Bradshaw and Schemske 2003; Ramsey et al. 2003).
Figure 1.
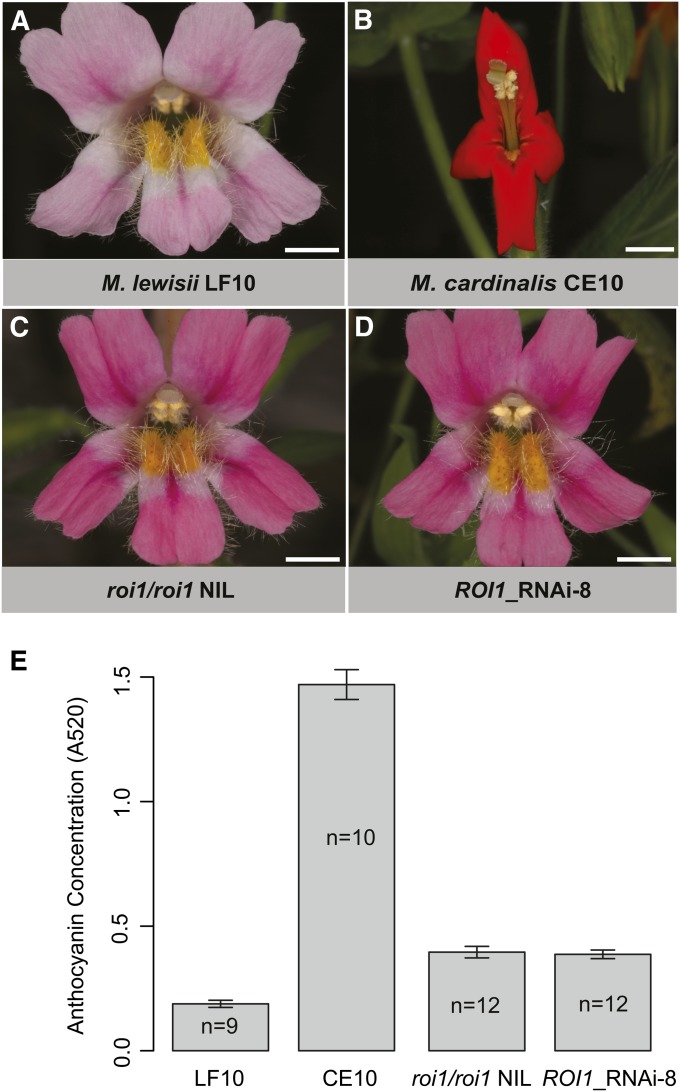
Flower phenotype and anthocyanin concentration. Bars, 5 mm. (A) M. lewisii inbred line LF10. (B) M. cardinalis inbred line CE10. (C) A roi1/roi1 NIL (17F9) in the LF10 background. (D) An RNAi transgenic line (ROI1_RNAi-8) in the LF10 background. (E) Anthocyanin concentration in the petal lobe. One disk was punched from each dorsal petal with a 1.5-ml Eppendorf tube, anthocyanins were extracted with 0.4 ml of methanol/0.1% HCl, and absorbance was recorded at 520 nm. CE10, the roi1/roi1 NIL, and the RNAi transgenic line are ∼7 times, 2 times, and 2 times higher than LF10, respectively. Sample sizes are shown in the bars. Bars are 1 SE.
The pale pink color of M. lewisii flowers results from a low concentration of anthocyanins and the absence of carotenoids (except in the nectar guides) (Figure 1A). The red color of M. cardinalis flowers is produced by a combination of high concentrations of both pigments. Both anthocyanin and carotenoid concentrations contribute to pollinator discrimination between the two species in their natural habitat (Schemske and Bradshaw 1999). A major quantitative trait locus (QTL) underlying floral anthocyanin concentration was identified as a dominant repressor in M. lewisii in a previous QTL mapping experiment (Bradshaw et al. 1995). The recessive allele in M. cardinalis leads to higher anthocyanin concentration in homozygotes. Here we generate genome sequences of these two species to facilitate fine-scale genetic dissection of this major anthocyanin QTL and to establish an efficient in planta transformation system for M. lewisii to perform functional characterization of target genes by transgenic experiments.
Through fine-scale genetic mapping, site-directed mutagenesis, and transgenic experiments, we demonstrate that a single-repeat R3 MYB repressor, ROSE INTENSITY1 (ROI1), is the causal gene underlying the major floral anthocyanin QTL and that cis-regulatory change rather than coding DNA mutations causes the allelic difference between M. lewisii and M. cardinalis.
Materials and Methods
Plant materials and growth conditions
Seeds of wild M. lewisii and M. cardinalis were collected by Amy Angert from a zone of sympatry along the South Fork of the Tuolumne River near Carlon, California, at ∼1400 m elevation. Inbred lines were developed from M. lewisii (LF10) and M. cardinalis (CE10) by self-pollination and single-seed descent for >10 generations.
All plants were grown with Sunshine soil mix #4 (Sun Gro Horticulture, Bellevue, WA) in the University of Washington Botany Greenhouse under natural light supplemented with sodium vapor lamps to provide a 16-hr day length, with daytime temperature of ∼20° and nighttime temperature of ∼15°. Plants were watered by sub-irrigation and fertilized as needed.
Genome sequencing and assembly
Genomic DNA of LF10 and CE10 were isolated using the BIO 101 System FastDNA kit (Qbiogene, Inc., Carlsbad, CA). A small-insert library (200–400 bp) was prepared for each line at the University of North Carolina High-Throughput Sequencing Facility. A total of 125 and 136 million 75-bp paired-end reads were generated by an Illumina Genome Analyzer II for LF10 (NCBI SRA056513: SRX198550) and CE10 (NCBI SRA056513: SRX200121), respectively. The resulting sequences were assembled into 80,139 contigs (N50 = 3598 bp) for LF10 and 80,737 contigs (N50 = 3794 bp) for CE10, using CLC Genomics Workbench. These draft genome assemblies are available for BLASTing or other analyses at http://target.iplantcollaborative.org/ (Han et al. 2009).
QTL mapping of flower petal anthocyanin concentration
Because M. lewisii is homozygous for the dominant allele of the major QTL that controls flower petal anthocyanin concentration (Bradshaw et al. 1995), a backcross (BC1) to M. cardinalis [(LF10 × CE10) × CE10; family #577; N = 192] was made for the initial coarse QTL mapping. Petal anthocyanin concentration was estimated from photographs taken at a standardized distance with a ring flash, using ImageJ (http://rsbweb.nih.gov/ij/) to determine the proportion of red (R), green (G), and blue (B) pixels in a rectangular area of the upper surface of a lateral petal. The concentration of anthocyanin (pink) pigment was estimated as [(R + B)/2] − G.
QTL affecting anthocyanin concentration in the backcross population were coarsely mapped with 65 SNP (AcycloPrime-FP, Perkin-Elmer) and indel markers at an average spacing of 7 cM (range: 0–32 cM) across the genome (Supporting Information, File S1; marker information is available at http://www.mimulusevolution.org/viewmap.php), using MAPMAKER/QTL 3.0 (Lincoln et al. 1992). The QTL with the largest effect on anthocyanin concentration (LOD = 17; 41% of BC1 phenotypic variance explained) was located near SNP marker MlSTS5209 (Figure S1). This QTL was named ROSE INTENSITY1 (ROI1), following the phenotypic nomenclature of Hiesey et al. (1971).
Refinement of the ROI1 QTL
A single BC1 plant (577-H12) heterozygous at ROI1 but homozygous at most other unlinked loci—a crude near-isogenic line (NIL)—was backcrossed to CE10 to produce a BC2 mapping population in which roi1/ROI1 heterozygotes (N = 177) could be identified unambiguously. Markers known to be linked to MlSTS5209 were used to map ROI1 to a 7.4-cM interval between MlSTS5467 and MgSTS305 (File S2). The 13 recombinants between MlSTS5467 and MgSTS305 were then screened with additional STS markers, resulting in ROI1 being mapped to a 1.7-cM interval between MgSTS55 and MgSTS280. Screening roi1/roi1 homozygotes recombinant between MlSTS5467 and MgSTS305 (N = 24) further located ROI1 between MgSTS158 and MgSTS55 (File S3).
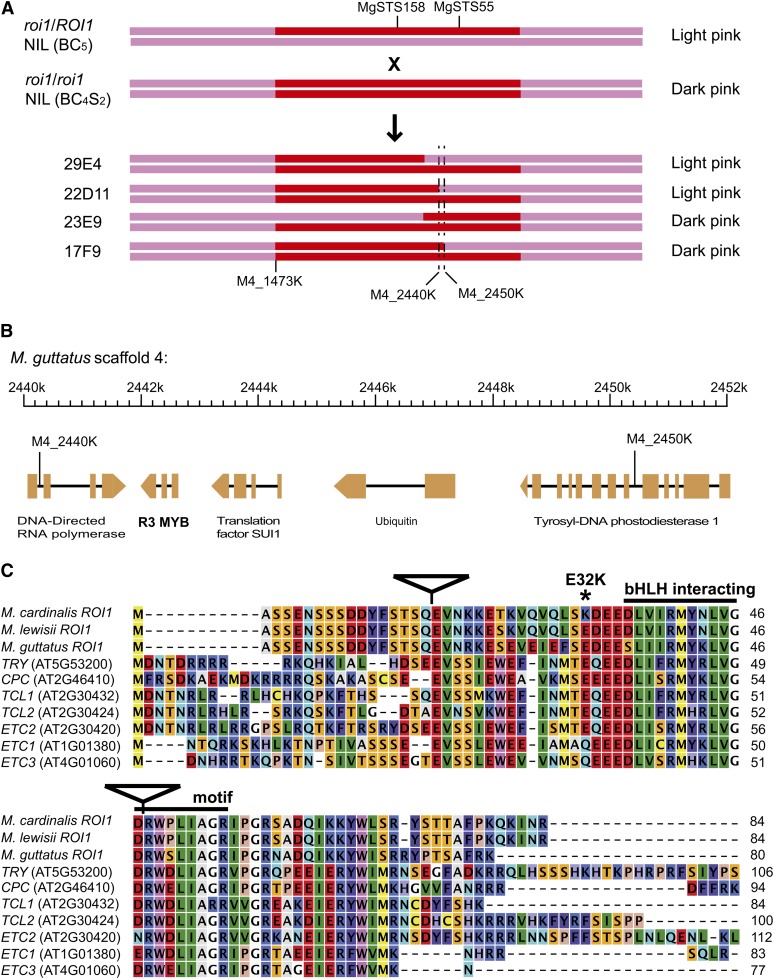
Fine-scale mapping of ROI1 was carried out in a high-resolution NIL in the M. lewisii LF10 genetic background. The recessive CE10 roi1 allele was introgressed into LF10 by four rounds of backcrossing and marker-assisted selection for loci linked to roi1 and against unlinked CE10 alleles. A selected backcross (BC4) plant was self-pollinated to produce a BC4S1 segregant with the dark-pink flower phenotype characteristic of roi1/roi1 homozygotes (Figure 1C). The roi1/roi1 BC4S1 plant was backcrossed to LF10 to produce a BC5 roi1/ROI1 heterozygote, and this BC5 heterozygote was crossed with a roi1/roi1 BC4S2 homozygote (Figure 2A) to produce a fine-mapping population segregating 1:1 for the dominant (light pink) and recessive (dark pink) flower phenotypes. Very young seedlings (two to four true leaves ∼1 cm tall) from the fine-mapping population (N = 2714) were genotyped at the flanking markers (MgSTS158 and MgSTS55, Figure 2A) by direct PCR (Phire, Finnzyme) of 0.5-mm leaf punches. Twenty-four individuals had recombinations between MgSTS158 and MgSTS55 and were transplanted and grown to flowering to determine their phenotype.
Figure 2.
Fine-scale mapping of the ROI1 gene. (A) Cross design to generate the fine-scale mapping population and the four most informative recombinants that reduced the candidate interval to a 10-kb region between markers M4_2440K and M4_2450K. Flower color phenotypes of the parental and recombinant lines are shown on the right. (B) The annotated genes in the 10-kb candidate interval. (C) Alignment of the single-repeat R3 MYB amino acid sequences of Mimulus and their homologs in Arabidopsis. Triangles represent intron positions. The signature motif [(DE)Lx2(RK)x3Lx6Lx3R] that interacts with bHLH proteins is marked above the alignment. The only nonconservative amino acid replacement (E32K) in M. cardinalis relative to M. lewisii is marked by an asterisk.
MgSTS158 and MgSTS55 lie 295 kb apart on scaffold 4 of the M. guttatus genome assembly (http://www.phytozome.net/cgi-bin/gbrowse/mimulus/). The 24 recombinants were further genotyped with an array of SNP and indel markers developed from assembled LF10 and CE10 contigs that contain orthologous genes in the 295-kb interval (File S4). The recombination breakpoints on either side of ROI1 reduced the candidate interval to a 10-kb region containing just five genes (Figure 2, A and B), one of which was a single-repeat R3 MYB-related gene and was considered the best candidate for ROI1 (see Results).
Plasmid construction
RNA interference (RNAi) plasmids were constructed to knock down the expression of the ROI1 gene in wild-type LF10. A 192-bp fragment containing part of the third exon and 3′ UTR of ROI1 was amplified with Phusion High-Fidelity DNA Polymerase (New England Biolabs) using a pair of primers with added restriction sites (underlined in the primer sequences) for cloning: ROI1_RNAi_F (GTTCTAGACCATGGCAAGAAATACTGGTTATCGCGA) and ROI1_RNAi_R (GTGGATCCGGCGCGCCTGAGCAGCATAACGCTTTGATTC). This fragment was BLASTed (Altschul et al. 1997) against the LF10 genome assembly to ensure target specificity. The amplified fragment was inserted into the binary vector pFGC5941 (Kerschen et al. 2004; Arabidopsis Biological Resource Center, CD3-447) by a two-step procedure. First, we cloned the PCR product into the original pFGC5941 vector in the sense orientation as an AscI/NcoI fragment. Then we cloned the same PCR product in the antisense orientation into the BamHI/XbaI site of the plasmid containing the sense fragment.
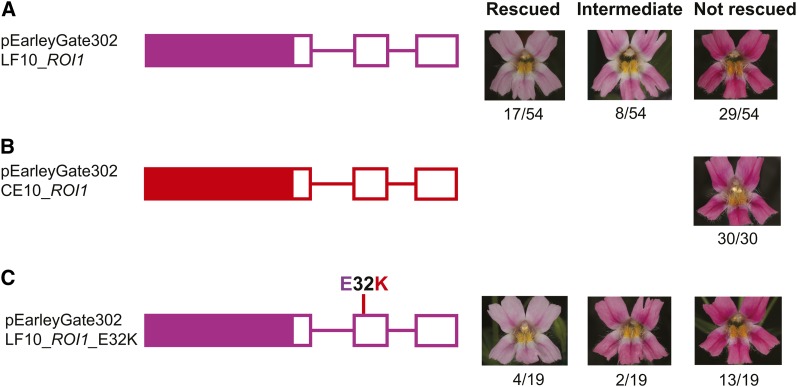
To test whether the wild-type LF10 ROI1 allele can rescue the flower color phenotype of the roi1/roi1 NIL, we generated a rescue plasmid using the pEarleyGate 302 vector (Earley et al. 2006; Arabidopsis Biological Resource Center, CD3-693) (Figure 3). We first amplified a 1944-bp genomic fragment, containing an ∼1.5-kb sequence upstream of the translation starting site and the entire ROI1 gene (including introns) from LF10 genomic DNA with Phusion polymerase and the primer pair ROI1_p302_F (CACCTGCCTTCTGCTGATTCTCAA) and ROI1_cdsR (ACGGTTTATCTTCTGTTTCGGA), and then cloned this genomic fragment into pEarleyGate 302 following Earley et al. (2006). Using the same procedure, we also built a rescue plasmid with the corresponding CE10 allele to serve as a negative control.
Figure 3.
Rescue experiments in the roi1/roi1 NIL background (dark-pink flower phenotype). (A) A construct containing the wild-type M. lewisii LF10 allele. (B) The wild-type M. cardinalis CE10 allele. (C) A mutated LF10 allele with the E32K replacement. All constructs were built from the pEarleyGate 302 vector. The LF10 and CE10 alleles are indicated by pink and red color, respectively. The open boxes represent exons; lines between exons represent introns; and solid boxes represent promoter regions. The mutated LF10 allele (LF10_ROI1_E32K) was created by site-directed mutagenesis to test the functional role of the E32K replacement. Numbers of completely rescued, partially rescued, and not rescued transgenic lines are indicated below the images that show the corresponding phenotypes.
To test whether the single nonconservative amino acid replacement (E32K; caused by a G-to A-nucleotide substitution) in the CE10 roi1 allele (Figure 2C) is functionally important, we mutated this “G” of the wild-type LF10 allele to an “A” by a PCR-based approach. To this end, the primers ROI1_p302_F and ROI1_p302E32K_R1 (TCTTTCGATAATTGAACTTGAACC) were used to amplify the first part of the 1944-bp fragment, and the primers ROI1_p302E32K_F2 (GGTTCAAGTTCAATTATCGAAAGA) and ROI1_cdsR were used to amplify the second part. These two fragments were then pooled together as template, amplified by bridge PCR using ROI1_p302_F and ROI1_cdsR to generate a full-length LF10 ROI1 allele with the G-to-A mutation. This mutated allele was then cloned to the pEarleyGate 302 vector. All plasmids were verified by sequencing before being transformed into Agrobacterium tumefaciens for subsequent plant transformation.
In planta transformation
Plant transformation was performed by a combination of the floral spray (Chung et al. 2000) and vacuum infiltration method (Bechtold et al. 1993). A. tumefaciens strain GV3101 was grown to stationary phase in liquid culture at 28°–30° in sterilized LB (tryptone 10 g/liter, yeast extract 5 g/liter, NaCl 5 g/liter) with kanamycin (50 mg/liter), rifampicin (25 mg/liter), and gentamicin (50 mg/liter). Pelleted bacteria cells from 400 ml of culture were resuspended in a 250-ml inoculation solution of 5% sucrose, 0.1 M acetosyringone, and 1 ml/liter surfactant Silwet L-77 (OSi Specialties, Inc., Danbury CT).
To prepare healthy plants for transformation, the apex was pinched from seedlings at about the eight-leaf stage to promote lateral growth and increase the number of flower buds. Inoculations were performed once the plants had developed lateral shoots that bore numerous flower buds. Only flower buds <5 mm were kept. Typically, each plant had 10–15 such small flower buds at the time of inoculation. The flower buds were sprayed with inoculation solution using a mist bottle until they were dripping. The plants were then placed in a vacuum chamber. The maximum vacuum (28–30 in Hg) was applied for 2–3 min and then released quickly. Inoculated plants were tented under plastic domes for 24 hr to maintain high humidity.
Flowers typically started opening 1 week after inoculation. Hand pollination was performed daily for 10–14 days. Pollen was obtained from untreated M. lewisii LF10 plants. The resulting capsular fruits were allowed to mature until beginning to yellow and then were harvested into seed envelopes where they continued to dry and dehisce.
The T-DNA in the pFGC5941 and pEarleyGate 302 vectors carries the BAR gene conferring resistance to the herbicide glufosinate (Finale, Farnam Companies, Inc., Phoenix, AZ). T1 seeds collected from inoculated plants were densely sown (∼8 seeds/cm2) in 25- × 50-cm flats and grown until cotyledons were fully expanded. Putative transformants were selected by spraying the seedlings with a 1:1000 dilution of Finale every 2 days. Glufosinate-resistant seedlings were transplanted for subsequent analysis. Transformation efficiency obtained by the current protocol is ∼0.02%; that is, one transformant per 5000 seeds or per five flowers, because each flower produces ∼1000 seeds.
Qualitative RT-PCR
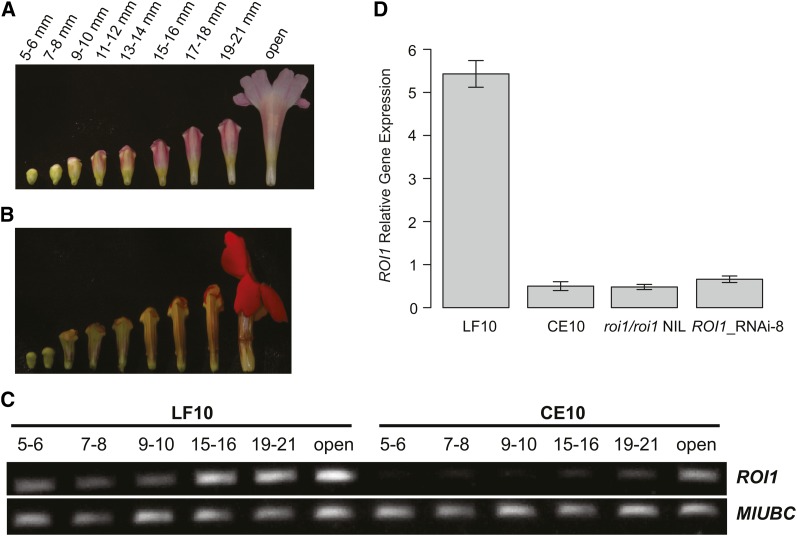
RT-PCR was performed to examine whether the ROI1 gene is expressed in different levels in LF10 and CE10 corollas and, if there is any difference, whether the difference is consistent across the multiple stages of corolla development. We divided LF10 corolla development into nine consecutive stages from the 5-mm flower bud to anthesis, each stage representing 1 day (Figure 4A), and divided CE10 corolla development into nine corresponding stages (Figure 4B). Total RNA was isolated from six of the nine stages of corolla development (Figure 4C) using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) and then treated with amplification grade DNaseI (Invitrogen). cDNA was synthesized from 1 μg of the resulting DNase-treated RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) and then diluted 40-fold before PCR. The gene-specific primers ROI1_SP2F (ATGGCCAGCTCAGAGAATTCTTC) and ROI1_cdsR (ACGGTTTATCTTCTGTTTCGGA) were used to amplify the 252-bp full-length coding DNA (CDS). The Mimulus ortholog of the Arabidopsis thaliana ubiquitin-conjugating enzyme gene At5g25760/UBC, MlUBC, was used as a reference gene for three reasons: (1) this gene was shown to have more stable constitutive expression than the traditional reference genes (e.g., Actin, UBQ10) (Czechowski et al. 2005); (2) this gene has a single copy in both Arabidopsis and Mimulus, so amplification is not confounded by paralogues in the same genome; (3) UBC is expressed at 5- to 20-fold lower levels than the traditional housekeeping genes, which is more comparable to typical transcription factors (Czechowski et al. 2005). The gene-specific primers MlUBC_SP3F (GGCTTGGACTCTGCAGTCTGT) and MlUBC_SP4R (TCTTCGGCATGGCAGCAAGTC) were used to amplify a 167-bp fragment spanning the last two exons.
Figure 4.
ROI1 expression in the corolla during flower development. (A) Corolla of M. lewisii LF10 at nine developmental stages over 9 consecutive days. (B) Corolla of M. cardinalis CE10 at nine stages corresponding to those of LF10. (C) RT-PCR of ROI1 and the reference gene MlUBC were examined at six of the nine stages. Note that the numbers shown for CE10 are not the absolute size of the corolla, but indicate corresponding stages as in LF10. ROI1 shows a substantial increase in expression after the 9- to 10-mm stage, when anthocyanins first become visible, and shows much lower expression in CE10 compared to LF10 in all corresponding stages. (D) qRT-PCR of ROI1 at the 19- to 21- mm corolla stage. Values are shown relative to a reference gene, MlUBC. ROI1 expression level in CE10, the roi1/roi1 NIL, and the RNAi transgenic line is 8.8%, 9.2%, and 12% of that in LF10, respectively. Bars represent 1 SE from three biological replicates.
Quantitative RT-PCR
The qualitative RT-PCR showed consistent difference between LF10 and CE10 in ROI1 expression across multiple developmental stages (Figure 4C). We selected the 19- to 21-mm corolla stage, when RNA is copious and the tissue sample is most convenient to handle, to quantify the ROI1 gene expression difference by quantitative RT-PCR (qRT-PCR) in LF10, CE10, the roi1/roi1 NIL, and the ROI1 RNAi line 8. RNA isolation and cDNA synthesis were as described above. Three independent biological replicates of each line were analyzed using iQ SYBR Green Supermix (Bio-Rad) run in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Samples were amplified for 40 cycles of 95o for 15 sec and 60o for 30 sec. Melting curves were examined to ensure that a single product had been amplified. The same ROI1 and MlUBC primers were used as in qualitative RT-PCR. We determined efficiencies for each primer pair using critical threshold values obtained from a dilution series (1:5, 1:20, 1:40, 1:80).
Results
QTL mapping of flower petal anthocyanin concentration
The QTL with the largest effect on anthocyanin concentration (41% of backcross phenotypic variance explained), ROI1, was initially mapped to an interval between marker MgSTS158 and MgSTS55 (Figure 2A). These two markers lie 295 kb apart on scaffold 4 of the M. guttatus genome assembly (http://www.phytozome.net/cgi-bin/gbrowse/mimulus/). Assuming that the M. guttatus, M. lewisii, and M. cardinalis genomes are collinear, the interval between these two markers contains 68 genes.
Fine-mapping of ROI1 was carried out in concert with constructing high-resolution NILs in the LF10 genetic background (see Materials and Methods), which will be used for future field experiments. Of 2714 individuals genotyped from the fine-mapping population (BC5 roi1/ROI1 × BC4S2 roi1/roi1 NIL; Figure 2A), 24 had recombinations between MgSTS158 and MgSTS55. Further genotyping of the 24 recombinants reduced the ROI1 candidate interval to a 10-kb region containing just five genes (Figure 2, A and B). One of the recombinants, 17F9 (Figure 2A), was selected to generate a roi1/roi1 homozygous NIL for subsequent experiments. The size of the chromosome fragment introgressed from CE10 to LF10 in this NIL was determined to be ∼1 Mb (Figure 2A, between M4_1473K and M4_2450K).
A single-repeat R3 MYB is the causal gene underlying ROI1
Of the five genes in the 10-kb interval, the single-repeat R3 MYB-related gene was considered the best candidate for ROI1 (Figure 2, B and C) because there is a group of R2R3 MYB transcription factors known to form a protein complex with basic helix-loop-helix (bHLH) and WD40 repeat proteins to activate anthocyanin biosynthesis in Arabidopsis, Antirrhinum, and other flowering plants (Koes et al. 2005; Ramsay and Glover 2005). Like the anthocyanin R2R3 MYB proteins, this R3 MYB has the signature motif [(DE)Lx2(RK)x3Lx6Lx3R] that interacts with the bHLH proteins (Figure 2C) (Zimmermann et al. 2004). The full length of this gene from the ATG initiation codon to the TAG stop codon, spanning three exons and two introns (Figure 2, B and C), is only 455 bp in LF10 (GenBank: JX992854) and 457 bp in CE10 (JX992855).
To test whether this candidate R3 MYB is the causal gene underlying the ROI1 QTL, we stably transformed the wild-type LF10 with an RNAi construct. Knocking down the expression of this R3 MYB in LF10 is expected to recapitulate the roi1/roi1 dark-pink flower phenotype (Figure 1C). We obtained nine independent RNAi lines, four of which show the dark-pink phenotype that is indistinguishable from the roi1/roi1 NIL (Figure 1, C and D). To further verify the gene identity, a rescue experiment was performed in the roi1/roi1 NIL background. We transformed the wild-type LF10 ROI1 allele (genomic DNA plus ∼1.5-kb upstream sequence from the ATG initiation codon) into the roi1/roi1 NIL. Seventeen of the 54 independent transgenic lines completely restored the wild-type light-pink phenotype, and eight lines showed an intermediate phenotype (Figure 3A). As a control, none of the 30 independent transgenics transformed with the corresponding CE10 roi1 allele rescues the NIL’s dark-pink phenotype (Figure 3B). Taken together, these results suggest that the R3 MYB is indeed the causal gene underlying the ROI1 QTL.
The CE10 petal lobe anthocyanin concentration is approximately sevenfold higher than LF10. Both the roi1/roi1 NIL and the RNAi transgenic line are approximately twofold higher than LF10 (Figure 1E). This suggests that the ROI1 gene alone accounts for ∼17% of the phenotypic difference between M. lewisii and M. cardinalis in petal lobe anthocyanin concentration and that anthocyanin concentration is a complex trait, which is affected by additional loci that remain to be identified (e.g., the minor QTL in Figure S1).
ROI1 homologs in Arabidopsis
To identify ROI1 homologs in A. thaliana, we performed BLASTP searches (Altschul et al. 1997) using the ROI1 amino acid sequence as query against the A. thaliana proteome (http://www.phytozome.net/search.php). The most closely related genes to ROI1 in the A. thaliana genome are a clade of seven R3 MYB repressors (Figure 2C), including CAPRICE (CPC) and TRIPTYCHON (TRY) (Wada et al. 1997; Schnittger et al. 1999). Like ROI1, these R3 MYBs are all small proteins (83–112 amino acids) with a single MYB domain that possesses the bHLH-interacting motif, but without an activation domain (Figure 2C). CPC and TRY function in root hair and trichome development in Arabidopsis as competitors with the R2R3 MYB transcriptional activators by sequestering the bHLH proteins into an inactive complex (Koes et al. 2005; Ramsay and Glover 2005). ROI1 is likely to negatively regulate anthocyanin biosynthesis through a similar mechanism by competing with the anthocyanin-activating R2R3 MYBs for a limited supply of bHLH proteins.
Cis-regulatory change causes ROI1 allelic difference
The ongoing debate over the relative importance of coding sequence polymorphisms and cis-regulatory mutations in generating phenotypic differences within and between species (Hoekstra and Coyne 2007; Stern and Orgogozo 2008) prompted us to determine the location of the causal mutation(s) underlying the allelic difference of ROI1 between M. lewisii and M. cardinalis.
There is a single nonconservative amino acid replacement (E32K) in the recessive CE10 allele (Figure 2C). This is a drastic substitution—from an acidic amino acid residue (E) to a basic residue (K)—in a fairly conserved site (Figure 2C) and, therefore, seemed likely to cause malfunction of the protein and to explain the recessiveness of the CE10 allele. To test this hypothesis, we transformed the roi1/roi1 NIL with a mutated LF10 allele (E32K) produced by site-directed mutagenesis (Figure 3C). If this mutation significantly interferes with the function of the protein, no complete rescue of the NIL’s dark-pink flower phenotype is expected. However, 4 of the 19 independent transgenic lines completely restored the M. lewisii light-pink phenotype, and 2 showed an intermediate phenotype (Figure 3C), similar to the experiment with the wild-type LF10 allele (Figure 3A). These results indicate that the E32K amino acid replacement is not responsible for the ROI1 allelic difference between M. lewisii and M. cardinalis.
Alternatively, if the recessive M. cardinalis allele is the result of one or more mutations in the cis-regulatory region of ROI1, the ROI1 mRNA level should be much lower in M. cardinalis than in M. lewisii flowers. RT-PCR indeed shows a substantially lower mRNA level in CE10 than in LF10 in all corresponding stages of corolla development (Figure 4C). Quantitative RT-PCR in the 19- to 21-mm flower bud stage (1 day before flower opening) reveals an ∼11-fold lower steady-state ROI1 mRNA abundance in CE10 than in LF10 (Figure 4 ).
Taking these results together, we conclude that altered gene expression of ROI1 is responsible for the natural variation of floral anthocyanin concentration between M. lewisii and M. cardinalis. Furthermore, the fine-mapping results ruled out trans-acting factors outside the 10-kb interval as potential causes of the gene expression change. Therefore, the causal mutation(s) should have occurred in the cis element of the ROI1 gene. There are numerous SNP and indel polymorphisms in the promoter region between the M. lewisii and M. cardinalis ROI1 alleles. Determining the specific causal cis-element change(s) will require more extensive promoter swapping and site-directed mutagenesis experiments in transgenic plants or flower protoplasts (Studer et al. 2011).
Discussion
Our results have established a causal link between a single-repeat R3 MYB gene and a flower color trait contributing to pollinator-mediated reproductive isolation between sister species of Mimulus, with experimental rigor (e.g., fine-scale genetic mapping, site-directed mutagensis, stable transgenics) that has rarely been achieved in ecological and evolutionary study systems (Hoballah et al. 2007; Chan et al. 2010).
ROI1 is among only a handful of genes that have been identified to underlie prezygotic isolation barriers in plants (reviewed in Rieseberg and Blackman 2010). It is noteworthy that none of the previously identified genes, except ANTHOCYANIN2 (AN2) in Petunia (Hoballah et al. 2007), has been subjected to a truly rigorous analysis, which should include genetic mapping and functional characterization of the causal gene and mutation(s), evaluation of its effect on reproductive isolation under field conditions, and molecular population genetic analysis to examine how allele variants spread within species (Lowry et al. 2008; Rieseberg and Blackman 2010). Identification and functional validation of the ROI1 gene has set the stage for future field experiments using the high-resolution ROI1 NIL or transgenic lines to test the effect of an individual locus on pollinator preference. A population-level survey of ROI1 allele sequences across the species range of M. lewisii and M. cardinalis is also underway to examine whether the differentially expressed alleles have been fixed in each species and to test for signatures of selection.
Identification of the ROI1 gene highlights the importance of fine-scale genetic mapping for understanding the genetic basis of a quantitative trait that underlies morphological differences between species (Orr 2001). Evolutionary transitions in floral anthocyanin intensity are thought to be most often due to mutations in the anthocyanin-activating R2R3 MYB genes (Streisfeld and Rausher 2011). In fact, these R2R3 MYBs have recently been associated with anthocyanin pigment variation in other Mimulus species (Cooley et al. 2011; Lowry et al. 2012), based on the commonly employed candidate gene approach. However, the candidate gene approach would not have predicted the single-repeat R3 MYB as the causal gene underlying the ROI1 QTL in our study; fine-scale genetic mapping was essential to discover novel genes underlying phenotypic difference.
Flower color variation between closely related species due to anthocyanin concentration is widespread in angiosperms (Rausher 2008). The ROI1 homologs are likely to play a role in generating flower color variation in many plant lineages, particularly in the Asteridae clade, where Mimulus and several other well-known study systems on plant–pollinator interactions (e.g., Antirrhinum, Petunia, Ipomoea) belong (Whibley et al. 2006; Hoballah et al. 2007; Des Marais and Rausher 2010).
The ROI1 example also illustrates the necessity for an efficient stable transformation system to distinguish the role of coding DNA vs. cis-regulatory change in generating morphological variation. Despite being a drastic substitution in a fairly conserved site, the E32K amino acid replacement was ruled out as a major factor in explaining the recessiveness of the M. cardinalis roi1 allele. We would not have been able to reach this conclusion without site-directed mutagenesis and transgenic experiments.
This study demonstrates that, in the M. lewisii/M. cardinalis system, we can now finely dissect the genetic basis of ecologically interesting traits with standards of experimental rigor formerly available in only a few genetic model organisms. The genomic resources developed here, in conjunction with community resources developed for M. guttatus, the other model species in the genus (Wu et al. 2008), greatly facilitate fine-scale mapping from phenotype to genotype. Stable transgenics allow rigorous testing of genetic hypotheses. In principle, the transgenic tools developed here will also permit detailed characterization of how gene products alter phenotypes during the developmental process, providing a mechanistic view of the evolution of phenotypic change, which is a major aim of evolutionary developmental biology (Sommer 2009). The capability of generating loss-of-function, gain-of-function, and alteration-of-function transgenics in Mimulus will enable us to study the genetics of pollinator-mediated reproductive isolation and the molecular basis of morphological evolution at the most fundamental level—gene by gene, mutation by mutation.
Supplementary Material
Acknowledgments
We thank Brian Watson, James Vela, Doug Ewing, Jeanette Milne, Paul Beeman, and Erin Forbush for plant care; Barbara Frewen and Dena Grosenbacher for conducting the preliminary trials of in planta transformation; Verónica Di Stilio, Richard Olmstead, Jennifer Nemhauser, Keiko Torii, and their lab members for sharing lab equipment and providing technical assistance; and Verónica Di Stilio for sharing her lab space and critically reading earlier versions of this manuscript. This work was supported by National Science Foundation Frontiers in Integrative Biological Research grant 0328636 and National Institutes of Health grant 5R01GM088805.
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J. H., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G., 1993. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. 316: 1194–1199. [Google Scholar]
- Bradshaw H. D., Schemske D. W., 2003. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178. [DOI] [PubMed] [Google Scholar]
- Bradshaw H. D., Wilbert S. M., Otto K. G., Schemske D. W., 1995. Genetic-mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus). Nature 376: 762–765. [Google Scholar]
- Chan Y. F., Marks M. E., Jones F. C., Villarreal G., Shapiro M. D., et al. , 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M. H., Chen M. K., Pan S. M., 2000. Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9: 471–476. [DOI] [PubMed] [Google Scholar]
- Cooley A. M., Modliszewski J. L., Rommel M. L., Willis J. H., 2011. Gene duplication in Mimulus underlies parallel floral evolution via independent trans-regulatory changes. Curr. Biol. 21: 700–704. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R., 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D. L., Rausher M. D., 2010. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution 64: 2044–2054. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species. Columbia University Press, New York. [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., et al. , 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Han Y. J., Burnette J. M., Wessler S. R., 2009. TARGeT: a web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Res. 37: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesey, W., M. Nobs, and O. Björkman, 1971 Experimental studies on the nature of species. V. Biosystematics, Genetics, and Physiological Ecology of the Erythranthe Section of Mimulus. Publ. 628, Carnegie Institute, Washington, DC.
- Hoballah M. E., Gubitz T., Stuurman J., Broger L., Barone M., et al. , 2007. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H. E., Coyne J. A., 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61: 995–1016. [DOI] [PubMed] [Google Scholar]
- Kerschen A., Napoli C. A., Jorgensen R. A., Muller A. E., 2004. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566: 223–228. [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F., 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Lincoln, S. E., M. J. Daly, and E. S. Lander, 1992 Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. Whitehead Institute Technical Report. Whitehead Institute, Cambridge, MA. [Google Scholar]
- Lowry D. B., Modliszewski J. L., Wright K. M., Wu C. A., Willis J. H., 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. B-Biol. Sci. 363: 3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Sheng C. C., Lasky J. R., Willis J. H., 2012. Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am. J. Bot. 99: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E., 1942. Systematics and the Origin of Species. Columbia University Press, New York. [Google Scholar]
- Orr H. A., 2001. The genetics of species differences. Trends Ecol. Evol. 16: 343–350. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Ramsay N. A., Glover B. J., 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Ramsey J., Bradshaw H. D., Schemske D. W., 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57: 1520–1534. [DOI] [PubMed] [Google Scholar]
- Rausher M. D., 2008. Evolutionary transitions in floral color. Int. J. Plant Sci. 169: 7–21. [Google Scholar]
- Rieseberg L. H., Blackman B. K., 2010. Speciation genes in plants. Ann. Bot. (Lond.) 106: 439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Willis J. H., 2007. Plant speciation. Science 317: 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemske D. W., Bradshaw H. D., 1999. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proc. Natl. Acad. Sci. USA 96: 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., 2001. Ecology and the origin of species. Trends Ecol. Evol. 16: 372–380. [DOI] [PubMed] [Google Scholar]
- Schnittger A., Folkers U., Schwab B., Jurgens G., Hulskamp M., 1999. Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11: 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., 2009. The future of evo-devo: model systems and evolutionary theory. Nat. Rev. Genet. 10: 416–422. [DOI] [PubMed] [Google Scholar]
- Stern D. L., Orgogozo V., 2008. The loci of evolution: How predictable is genetic evolution? Evolution 62: 2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisfeld M. A., Rausher M. D., 2011. Population genetics, pleiotropy, and the preferential fixation of mutations during adptive evolution. Evolution 65: 629–642. [DOI] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J., 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K., 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Whibley A. C., Langlade N. B., Andalo C., Hanna A. I., Bangham A., et al. , 2006. Evolutionary paths underlying flower color variation in Antirrhinum. Science 313: 963–966. [DOI] [PubMed] [Google Scholar]
- Wu C. A., Lowry D. B., Cooley A. M., Wright K. M., Lee Y. W., et al. , 2008. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100: 220–230. [DOI] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F., 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.