Abstract
Dicentric chromosomes undergo breakage in mitosis, resulting in chromosome deletions, duplications, and translocations. In this study, we map chromosome break sites of dicentrics in Saccharomyces cerevisiae by a mitotic recombination assay. The assay uses a diploid strain in which one homolog has a conditional centromere in addition to a wild-type centromere, and the other homolog has only the wild-type centromere; the conditional centromere is inactive when cells are grown in galactose and is activated when the cells are switched to glucose. In addition, the two homologs are distinguishable by multiple single-nucleotide polymorphisms (SNPs). Under conditions in which the conditional centromere is activated, the functionally dicentric chromosome undergoes double-stranded DNA breaks (DSBs) that can be repaired by mitotic recombination with the homolog. Such recombination events often lead to loss of heterozygosity (LOH) of SNPs that are centromere distal to the crossover. Using a PCR-based assay, we determined the position of LOH in multiple independent recombination events to a resolution of ∼4 kb. This analysis shows that dicentric chromosomes have recombination breakpoints that are broadly distributed between the two centromeres, although there is a clustering of breakpoints within 10 kb of the conditional centromere.
Keywords: Saccharomyces cerevisiae, dicentric chromosomes, mitotic crossovers, loss of heterozygosity, break-induced replication
THE cells of solid tumors often have numerous chromosome alterations, both changes in chromosome number and structural alterations including deletions, duplications, and translocations (Cimini 2008). One mechanism that can contribute to the formation of chromosome alterations is the formation of dicentric chromosomes, followed by the breakage of the dicentric leading to secondary chromosome alterations (Stimpson et al. 2012).
In Saccharomyces cerevisiae, as well as in other eukaryotes, dicentric chromosomes can be formed in several ways. In yeast strains with certain telomeric defects, fusions between telomeres of different chromosomes or fusions between the telomere of one chromosome and an internal break on another chromosome can produce dicentric chromosomes (Myung et al. 2001; Craven et al. 2002; Mieczkowski et al. 2003; Pardo and Marcand 2005). Dicentrics also arise as a consequence of ectopic homologous recombination between retrotransposons on nonhomologous chromosomes (Mieczkowski et al. 2006) or as a consequence of nonhomologous end joining between two broken chromosomes (Myung et al. 2001; Craven et al. 2002). Intrachromosomal dicentrics can also be generated by processing or replication of inverted repeats (Lobachev et al., 2002; Narayanan et al. 2006; VanHulle et al. 2007). Lastly, dicentric plasmids and chromosomes have been generated by in vitro manipulations, followed by transformation of the resulting constructions into yeast (Mann and Davis 1983; Koshland et al. 1987).
Regardless of the mechanism by which they are formed, dicentric plasmids and chromosomes are unstable in S. cerevisae, undergoing various types of structural rearrangements to generate monocentric plasmids or chromosomes (Mann and Davis 1983; Koshland et al. 1987; Hill and Bloom 1989; Kramer et al. 1994; Narayanan et al. 2006; Pennaneach and Kolodner 2009). The instability is initiated when the two centromeres of a dicentric chromosome are pulled to different daughter cells during anaphase, resulting in the stretching of chromosomal sequences located between the two centromeres (Thrower and Bloom 2001). Although the subsequent double-stranded DNA breaks (DSBs) could simply reflect the mechanical forces applied by the spindle to the dicentric chromosome, breakage of the dicentric is observed when the chromosome experiences forces of ∼1 piconewton (pN) (Fisher et al. 2009), much less than the force required to mechanically break a double-stranded DNA molecule (∼480 pN; Bensimon et al. 1995). Based on these considerations and the observation that stretching and breakage of dicentric yeast chromosomes occur in anaphase (Thrower and Bloom 2001), it is likely that dicentric breakage requires the enzymatic production of DSBs in stretched chromatin. The enzymes involved in dicentric breakage, presumably endonucleases, have not yet been identified. Alternatively, or in addition, dicentric breakage could occur during nuclear fission or cytokinesis (Quevedo et al. 2012). For dicentric yeast chromosomes formed by telomere–telomere fusions, breakage often occurs at the telomere fusion junction, possibly involving resolution by enzymes that process cruciforms (Pobiega and Marcand 2010). Finally, it should be pointed out that the instability of dicentric chromosomes is not universal, since some dicentrics in human cells are relatively stable (Stimpson et al. 2012).
Following dicentric breakage, a stable karyotype in yeast requires the resulting DSBs to be repaired to generate telomere-capped monocentric chromosomes. There are a variety of mechanisms that can produce these products, depending on whether the strain is a haploid or diploid, the availability of repetitive sequences (for example, transposons) located near the break, the location of essential genes near the break, and other factors. One of the simplest mechanisms for repairing broken chromosome ends is by telomere addition (Haber and Thorburn 1984; Pennaneach and Kolodner 2009). Another common mechanism of generating a monocentric from a dicentric is a deletion that removes one of the two centromeres. Such deletions can occur by intrachromosomal homologous recombination between repeated genes that flank one of the two centromeres (Brock and Bloom 1994; Lemoine et al. 2005) or by nonhomologous end joining (NHEJ) following processing of the broken DNA ends (Kramer et al. 1994).
Alternatively, the repair process may involve recombination with a homolog or a nonhomologous chromosome. For example, a DSB resulting from breakage of a dicentric in a diploid strain could be repaired by homologous recombination with a homolog (Haber and Thorburn 1984), either by a crossover (Figure 1A) or by a break-induced replication (BIR) event (Figure 1B). If a broken chromosome contains a repetitive element (such as the retrotransposon Ty), it can also acquire a telomere by homologous recombination with a repetitive element on a nonhomologous chromosome; such events often involve BIRs (Umezu et al. 2002; Lemoine et al. 2005; Argueso et al. 2008; Vernon et al. 2008; Pennaneach and Kolodner 2009; Chan and Kolodner 2011). Alternatively, the broken chromosomes can be joined to the broken ends of nonhomologous chromosomes or to telomeres of other chromosomes by NHEJ, although homologous recombination events involving repeats are more common (Pennaneach and Kolodner 2009). Finally, it should be noted that repair of the broken chromosome may regenerate a dicentric chromosome, leading to additional chromosome rearrangements before a stable karyotype is generated (Admire et al. 2006; Narayanan et al. 2006; Pennaneach and Kolodner 2009).
Figure 1.
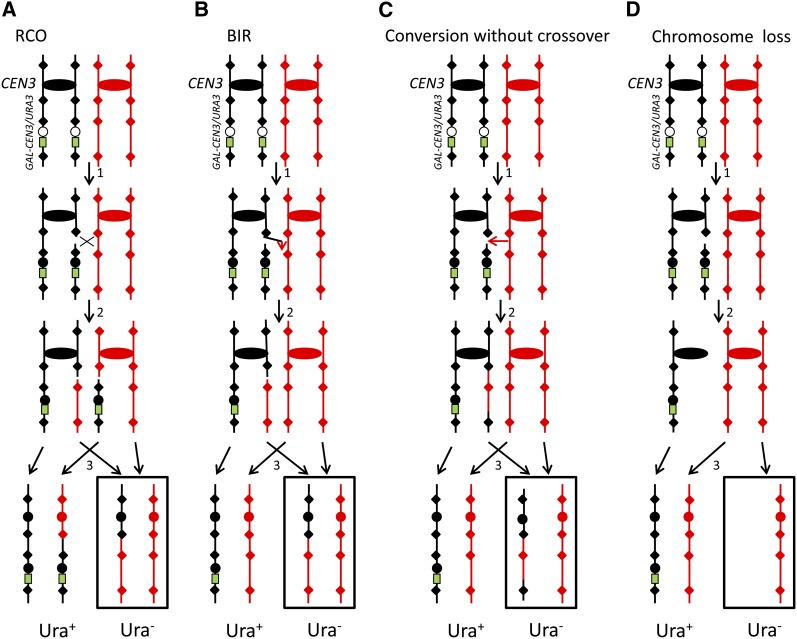
Mechanisms for loss of heterozygosity (LOH) resulting from breakage of a conditional dicentric chromosome. The two chromosome homologs are depicted in G2 with the duplicated chromatids held together at the centromere (shown as ovals or circles). The red and black colors signify polymorphisms that distinguish the two homologs: the black homolog derived from J178-#7-20 and the red homolog from PSL5. Red and black diamonds indicate representative SNPs that distinguish the two homologs: two located in the intercentromic region, one located near the telomere on the same chromosome arm at the conditional centromere, and one located on the opposite chromosome arm. The black homolog carries the GAL-CEN3/URA3 conditional centromere; the conditional centromere is shown as a circle next to the URA3 gene (shown as a green rectangle). When cells are grown on galactose-containing medium, the conditional centromere (white circle) is inactive as a consequence of transcription across the centromere, and the chromosome is functionally monocentric. When the cells are transferred to glucose-containing medium (step 1 in Figure 1, A–C), transcription across the centromere is repressed and the chromosome is functionally dicentric (indicated by black circles) (Hill and Bloom 1989). We show a DSB on only one of the two dicentric chromatids. Four pathways for the repair of the DSB are shown. (A) Reciprocal crossover (RCO). The broken chromosome is repaired by a reciprocal crossover with the homolog (step 2). If the recombined chromosomes segregate as shown by the arrows (step 3), one daughter cell (outlined in black) would be Ura− and the other cell would be Ura+. Only one of the two possible chromosome disjunction patterns is shown; the other pattern does not lead to the markers becoming homozygous. LOH is observed for markers distal to the crossover, but heterozygosity is maintained for the marker on the opposite chromosome arm. (B) Break-induced replication (BIR). In this pathway, one end of the broken black chromatid invades the red chromatid, duplicating all the sequences to the end of the chromatid. The net result of this process is one Ura− (ura3/ura3) cell and one Ura+ (ura3/URA3) cell. The pattern of marker segregation in the Ura− cell is indistinguishable from that shown for the crossover. (C) Conversion without crossover. A DSB occurring near the conditional centromere is processed to yield a conversion event that includes the conditional centromere. If this conversion event is unassociated with a crossover, an interstitial region of LOH would be formed, but heterozygosity would be maintained for the marker near the telomere and the one located on the opposite chromosome arm. (D) Chromosome loss. If the broken chromatid is not repaired, one Ura− monosomic daughter cell and one Ura+ (ura3/URA3) cell would be generated. LOH is observed for all markers.
Most studies of the behavior of dicentric yeast chromosomes focus on the chromosome aberrations generated during the repair of chromosome breaks. Since these aberrations are usually a consequence of homologous recombination between nonallelic Ty elements (as described above), these studies select for dicentric breaks that occur within a Ty element or breaks that occur near a Ty element that are processed to generate a Ty-containing broken end (Hoang et al. 2010). In our study, we examine the location of dicentric chromosome breaks by mapping homologous recombination events between a dicentric chromosome with one conditional centromere and a homologous monocentric chromosome. Our analysis shows that dicentric chromosomes break at sites distributed throughout the region between the two centromeres, but the region near the conditional centromere is particularly prone to breakage.
Materials and Methods
Genetic analysis and media
Standard yeast procedures were used for mating, sporulation, and tetrad dissection (Guthrie and Fink 1991). Rich growth medium, yeast extract, peptone, and dextrose (YPD), and omission media were made following standard protocols (Guthrie and Fink 1991; Lee et al. 2009). The growth media YPGal was identical to YPD, except 2% galactose was substituted for glucose. To select canavanine-resistant transformants, we used solid omission medium lacking arginine (SD −Arg) containing 120 μg/ml of canavanine.
Strains
The genotypes of all strains in this study are given in Supporting Information, Table S1. Three diploids were constructed (details in File S1). All diploids were isogenic except for the location of the heterozygous conditional centromere. The locations of the conditional centromeres in the three strains were on chromosome III near HIS4 (WS49), on chromosome V near CAN1 (WS83), and on chromosome V near coordinate 80 kb (WS92). In addition, the diploids were generated by crosses of one haploid parent isogenic with J178-1d (related to S288c; Brock and Bloom 1994) and one haploid isogenic with PSL5 (isogenic with YJM789; Lee et al. 2009). Since YJM789 has ∼60,000 SNPs distinguishing its genome from S288c (Wei et al. 2007), the diploids used in our study are heterozygous for markers distributed throughout the genome. The primers used in strain constructions are listed in Table S2.
The S288c strain in the Saccharomyces Genome Database (SGD) has a single Ty2 element on the left arm of chromosome III. By PCR and Southern analysis (described in File S1), we found that the left arm of chromosome III in J178-1d contains a Ty1 element closely linked to the Ty2 element. In addition, YJM789 lacks both the Ty1 and the Ty2 elements on the left arm. The primers used to diagnose the location of Ty elements are in Table S2.
Analysis of loss of heterozygosity
In yeast strains that are heterozygous for markers, mitotic crossovers or break-induced replication events can generate loss of heterozygosity (LOH) of markers centromere distal to the recombination event (Figure 1; Lee et al. 2009; St. Charles et al. 2012). We mapped the transition between heterozygous markers and homozygous markers using two techniques: a PCR-based method and an approach utilizing oligonucleotide-containing microarrays. Primers used for the PCR-based approach are listed in Table S3. The details of these methods are described in the Results section and in File S1.
Identification of strains with recombination events induced by dicentric chromosome breakage
The GAL-CEN3 conditional centromere is inactive in cells grown in medium containing galactose and active in cells grown in glucose (Hill and Bloom 1987). In our experiments, all diploid strains were grown from single cells to colonies on solid medium containing galactose (YPGal) at 30° for 2 days. Individual colonies were selected from these plates, and restreaked on plates containing glucose (YPD) and incubated at 30° for 2 days. The resulting colonies were then replica plated to solid galactose-containing omission medium lacking uracil (SGal −Ura) to identify derivatives that had lost the URA3 marker adjacent to the conditional centromere.
Ura− derivatives could be generated by recombination (crossovers or BIR; Figure 1A and 1B), gene conversion events unassociated with crossovers or intrachromosomal deletions that remove the GAL-CEN3/URA3 cassette (Figure 1C) or chromosome loss (Figure 1D). Since chromosome loss would result in LOH for markers on both centromeric arms (Figure 1D), we examined Ura− derivatives using a PCR-based analysis to detect LOH for markers on both the right and left arms of chromosomes III or V (details in File S1). Similarly, since strains with conversion events unassociated with crossovers or intrachromosomal deletions of the cassette containing the conditional centromere would retain heterozygosity for markers located near the end of the chromosome (Figure 1C), we excluded such events by a PCR-based analysis to examine heterozygosity for markers near the left telomeres of chromosomes III and V (details in File S1). The remaining Ura− derivatives were analyzed for the location of recombination breakpoints.
Statistical analysis
We performed two types of statistical tests. First, we determined whether the distributions of chromosome break sites were significantly different from a random distribution. Second, we looked for associations between recombination breakpoints and various chromosome elements (replication origins, tRNA genes, etc.). The details of these analyses are given in File S1.
Results
Experimental rationale
The diploid strain used to map breaks induced in a dicentric chromosome has two important features: (1) it is heterozygous for a conditional centromere and (2) it has multiple single-nucleotide polymorphisms (SNPs) that distinguish the homologs. The conditional centromere employed in our analysis is described by Hill and Bloom (1987). The activity of the centromere is regulated by the galactose-inducible GAL1 promoter located ∼100 bp from CEN3 sequences. When cells are grown on galactose-containing medium, transcription initiated at the GAL1 promoter inactivates the centromere; in glucose-containing medium, transcription is shut off and CEN3 is functional (Hill and Bloom 1987). Hill and Bloom (1989) inserted a cassette containing GAL-CEN3 and URA3 within the HIS4 gene on the left arm of chromosome III located ∼50 kb from the natural centromere. In strains with this insertion, chromosome III is unstable or stable depending on whether the strain is grown in glucose- or galactose-containing media, respectively. In conditions in which both centromeres are active, the intercentromeric region is stretched and, subsequently, broken (Thrower and Bloom 2001; Fisher et al. 2009). In haploid strains, these breaks result in chromosomes with deletions of either the conditional centromere or the natural centromere (Hill and Bloom 1989; Kramer et al. 1994).
Diploid cells have an option for the repair of DSBs that is unavailable to haploids, the use of the intact homologous chromosome as a template. Two of the expected pathways of repair that would result in loss of the conditional centromere are reciprocal crossovers (Figure 1A) and break-induced replication (Figure 1B). For both of these pathways, loss of the conditional centromere (detected as loss of the URA3 gene) would result in LOH of markers located centromere distal to the breakpoint. In experiments in which recombination events reflect DSBs formed at trinucleotide repeats (Tang et al. 2011) or at meganuclease-recognition sequences (Nickoloff et al. 1999), the site of the DSB maps at or near the transition to LOH. Thus, in our experiments, we mapped the transition between heterozygous and homozygous markers as a method of mapping dicentric breaks (discussed in the Mapping of LOH events section of Results below). It should be noted that our mapping of DSBs is limited to those DSBs that are repaired by recombination with the homolog. DSBs that are repaired by sister-chromatid exchange (Kadyk and Hartwell 1992) do not lead to LOH and are undetectable by our analysis.
Breakage of the dicentric chromosome was induced by restreaking cells from individual colonies grown on galactose-containing medium (inactive GAL-CEN) to glucose-containing medium (active GAL-CEN). The resulting colonies were then replica plated to medium lacking uracil. We expected most of the Ura− derivatives of the starting strain could be grouped into three classifications. In class 1 strains (Figure 1, A and B), a DSB resulting from breakage of the dicentric is repaired by a reciprocal crossover or a BIR event. Such events will result in LOH of all markers centromere distal to the position of the recombination event with retention of heterozygosity for markers located on the opposite chromosome arm. Since we analyze only the Ura− product, crossovers and BIR events are not distinguishable; in wild-type strains, however, crossovers are more common than BIR events (McMurray and Gottschling 2003; Ho et al. 2010). In class 2 strains (Figure 1C), a DSB located near the conditional centromere could be repaired to generate a gene conversion event unassociated with a crossover or could be repaired to generate an intrachromosomal deletion; both of these events result in an interstitial LOH event. For class 2 strains, we expect LOH for markers located near the conditional centromere and maintenance of heterozygosity for markers flanking the conversion event. Class 3 strains (Figure 1D) reflect chromosome loss and result in LOH for markers located on both arms of the chromosome.
To distinguish among these classes, our initial characterization of the strains utilized multiple markers located in the region between the conditional centromere and the native centromere, a marker located near the telomere, and a marker located on the opposite arm of the chromosome (details about coordinates of all markers in File S1). Three different dicentric strains were examined in our study: WS49 (conditional centromere located at SGD coordinate 67 kb of chromosome III), WS83 (conditional centromere located at SGD coordinate 32 kb of chromosome V), and WS92 (conditional centromere located at SGD coordinate 80 kb of chromosome V). For WS49, the telomere-associated marker was located at SGD coordinate 33 kb, ∼34 kb centromere distal to the conditional centromere. Since the median size of mitotic gene conversion tracts is 6–8 kb (Lee et al. 2009; St. Charles et al. 2012), most of the conversion tracts that include the conditional centromere should not include marker 33. The opposite-arm marker was at coordinate 116 kb. For the strains WS83 and WS92, the telomere-associated marker was at coordinate 7 kb, located ∼25 kb from the conditional centromere in WS83 and 73 kb from the conditional centromere in WS92; the markers on the opposite arm were located at coordinates 561 kb (WS83) and 152 kb (WS92). In summary, for class 1 events, we see a continuous region of LOH beginning at a marker located within the intercentromeric region and extending through the marker located near the telomere; the marker on the opposite chromosome arm retains heterozygosity. For class 2 events, we observe LOH for markers near the conditional centromere but retention of heterozygosity for the marker near the telomere and the marker on the opposite side of the centromere. Class 3 events are defined by strains in which LOH is observed for all markers tested, including the marker on the opposite side of the centromere. Some of the Ura− strains (∼10%) could not be classified into any of these three classes. In most of these strains (defined as class 4; complex events), there was more than one transition between heterozygous and homozygous markers. Such events may reflect the “patchy” repair of mismatches within a heteroduplex (St. Charles et al. 2012) or multiple repair events.
From the strains WS49, WS83, and WS92, we isolated 80, 61, and 62 Ura− derivatives, respectively. The percentages of each class of event in the four strains were: WS49 (class 1, 34%; class 2, 14%; class 3, 44%; and class 4, 9%), WS83 (class 1, 41%; class 2, 36%; class 3, 20%; and class 4, 3%), and WS92 (class 1, 39%; class 2, 39%; class 3, 11%; and class 4, 11%).
Mapping of LOH events
In all diploids used in our experiment, the conditional dicentric homolog had many SNPs distinguishing it from its monocentric homolog. Most of the diploids were generated by a cross of haploids isogenic with J178-1d (Hill and Bloom 1987) with a haploid (PSL5) isogenic with YJM789 (Wei et al. 2007). The J178-1d strain has a mixed pedigree, but most of the SNPs that we examined were identical to those of S288c. Two methods were used to diagnose LOH. The first method (described in detail in File S1) was that used in our previous study (Lee et al. 2009). In brief, by BLAST comparisons of the DNA sequences of S288c and YJM789 in the intercentromeric region, we identified SNPs that altered a restriction enzyme recognition site. For each such SNP, we designed primers flanking the SNP. Following PCR amplification of genomic DNA, the resulting DNA fragment was treated with the diagnostic restriction enzyme and analyzed by gel electrophoresis to determine whether the diploid with the recombination event was heterozygous or homozygous for the SNP.
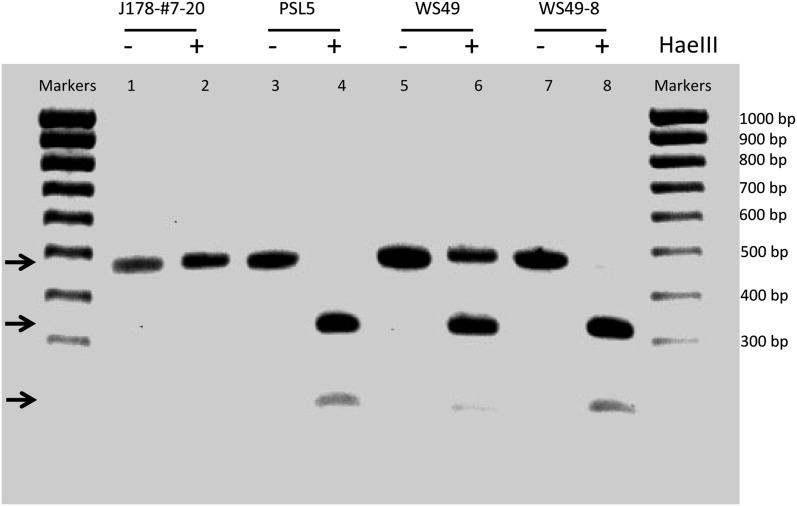
An example of this analysis is shown in Figure 2. In this example, we examined the Ura− derivative WS49-8 for LOH at a SNP located at SGD coordinate 91322 (marker 91 in Table S3). At this position, the haploid parental strain PSL5 (isogenic with YJM789) had a HaeIII site that was lacking in DNA from the other parental strain (J178-#7-20). The primers used to amplify the region containing this SNP resulted in a fragment of ∼500 bp. As shown in Figure 2, the fragment derived from the J178-#7-20 strain was not cut by HaeIII, whereas the fragment derived from PSL5 was cut into two fragments of ∼170 and 320 bp. The control diploid WS49, following treatment of the 500-bp fragment with HaeIII, had three bands of the appropriate size. The Ura− derivative WS49-8 had the pattern indicating LOH in favor of the PSL5-derived SNP. For WS49, we examined LOH in the intercentromeric region using 14 markers (listed in Table S3). Similar methods were used to map recombination events on chromosome V in diploids WS83 and WS92. For these two strains, we employed the same 34 SNPs previously described (Lee et al. 2009). Detailed mapping of recombination events was restricted to class 1 strains and our subsequent conclusions about dicentric breakpoints are based on only these strains.
Figure 2.
PCR-based method of detecting loss of heterozygosity (LOH). As described in the text, the two sequences of the two homologs differ at SGD coordinate 91,332 on chromosome III, resulting in a HaeIII restriction site in the PSL5 homolog that is missing in the J178-#7-20-derived homolog. Using primers that flank this polymorphism, we amplified genomic DNA from the haploid parental strains, the WS49 control Ura+ diploid and one of the Ura− WS49 derivatives (WS49-8). The resulting samples were either treated with HaeIII (lanes with the + sign) or were left untreated (lanes with the − sign) and then analyzed by agarose gel electrophoresis. In the untreated samples, all strains had a product of ∼500 bp. In the enzyme-treated samples, the fragment derived from J178-#7-20 genomic DNA had one fragment of ∼500 bp (lane 2), whereas the fragment derived from PSL5 had two fragments of ∼320 and 170 bp (lane 4). The Ura+ control diploid had three bands (lane 6), as expected, although the 170-bp fragment is difficult to visualize; the approximate positions of the fragments of 500, 320, and 170 bp are shown by arrows on the left side of the gel. In the Ura− derivative WS49-8 (lane 8), we observe two fragments of ∼320 and 170 bp, indicating that this diploid is homozygous for the PSL5-derived SNP at position 91,332. Using similar methods, we examined LOH for 17 markers on chromosome III and 37 markers on chromosome V. The two lanes at the left and right end of the gels are ladders of 100-bp size markers.
In addition to the PCR-based mapping method described above, seven of the events were also analyzed using oligonucleotide-containing microarrays (SNP arrays). Using 25-base oligonucleotides that are identical to PSL5- or J178-1d-derived SNPs, we detected LOH events in the intercentromeric regions as described in our previous studies (St. Charles et al. 2012). The results of the microarray analysis, in general, were in good agreement with those obtained by the PCR-based methods and will be described in more detail below.
Mapping break sites in a strain (WS49) with a dicentric chromosome III
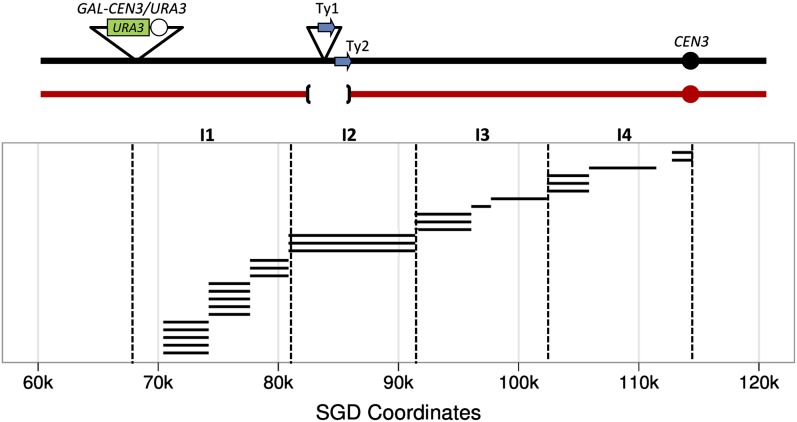
In the diploid WS49, the dicentric homolog has an insertion of the conditional centromere within the HIS4 locus at SGD coordinate 67 kb (Hill and Bloom 1989). In addition to SNPs that distinguish the two homologs, the J178-#7-20-derived homolog has a pair of Ty elements (one Ty1 and one Ty2) located centromere distal to LEU2 that are not present in the PSL5-derived homolog (File S1). We mapped recombination events in 27 class 1 Ura− derivatives (Figure 3). In this figure, each horizontal line denotes the distance between the markers at the transition between heterozygosity and homozygosity.
Figure 3.
Mapping of dicentric chromosome break sites on chromosome III. Recombination events in 27 class 1 Ura− derivatives of WS49 are shown. Using the PCR-based method described in Figure 2, we mapped recombination events between the conditional centromere located at SGD coordinate 68,096 and CEN3, a distance of ∼53 kb (including the heterozygous Tys). Each horizontal black line represents an independent event. The length of the line shows the distance separating the closest centromere-proximal heterozygous markers and the closest centromere-distal homozygous markers for each derivative. The J178-1d-derived homolog (shown in black) has two Ty elements (blue arrows) that are missing in the PSL5-derived homolog. Dotted lines show the intervals (I1–I4) used in the statistical analysis described in the text.
Although it is evident that the dicentric-induced breaks map throughout the intercentromeric region, the distribution of breaks is nonrandom. To examine the distribution, we divided the intercentromeric region into four intervals of similar size (markers at the ends of intervals shown in parentheses): interval 1 (68–81), interval 2 (81–91 plus 6 kb Ty element), interval 3 (91–102), and interval 4 (102–115). We compared the observed distribution of events in these intervals with the expected distribution if the events were random (details in File S1). The distribution was significantly different from random by chi-square analysis (P = 0.03). We also tested each of the four intervals separately and determined that interval 1 (near the conditional centromere) had a significant (P = 0.01) excess of breakpoints relative to the sum of the other intervals. This value remained significant (P < 0.05) after correcting for multiple comparisons using the method of Benjamini and Hochberg (1995).
Mapping break sites in strains (WS83 and WS92) that have a dicentric chromosome V
The elevated frequency of breakpoints near the conditional centromere in WS49 could reflect a property of the conditional centromere. Alternatively, this region of chromosome III could be intrinsically susceptible to chromosome breaks. To distinguish between these two possibilities, we constructed two diploid strains isogenic with WS49 where the conditional centromere was located on chromosome V instead of III. In WS83, the conditional centromere was inserted at the CAN1 locus ∼120 kb from the natural CEN5, and in WS92, the conditional centromere was located on the same chromosome arm ∼72 kb from CEN5.
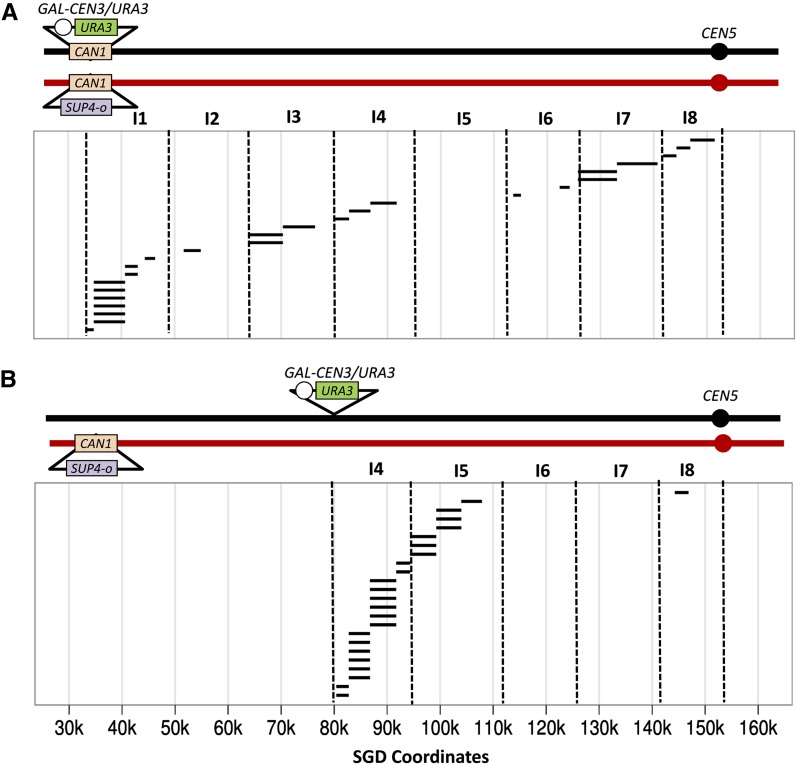
We previously mapped breakpoints for spontaneous mitotic recombination events between CAN1 and CEN5 using the PCR-based method (Lee et al. 2009). The mapping of recombination events reflecting dicentric breaks in WS83 was performed using the same 34 markers employed in our previous study. As shown in Figure 4A, the breakpoints cluster near the conditional centromere. For the statistical analysis, we divided the intercentromeric region into eight intervals: interval 1 (33–49), interval 2 (49–64), interval 3 (64–80), interval 4 (80–94), interval 5 (94–112), interval 6 (112–126), interval 7 (126–141), and interval 8 (141–152). The distribution of events in WS83 was significantly (P = 0.01) different from random. In addition, the number of events in interval 1 was significantly elevated relative to the sum of the events in the other intervals (P < 0.0001, corrected for multiple comparisons).
Figure 4.
Mapping dicentric chromosomal break sites on chromosome V. The same procedure described in Figure 3 was used to map recombination events in class 1 Ura− derivatives of WS83 and WS92. As in Figure 3, dotted lines show the intervals used in the statistical analysis. (A) Location of recombination breakpoints in WS83. A total of 25 independent events were mapped in a region of 120 kb between the conditional centromere and CEN5. The conditional centromere is inserted within the CAN1 on the J178-#7-20-derived homolog allelic to the insertion of SUP4-o on the PSL5-derived homolog. (B) Mapping of recombination breakpoints in WS92. The WS92 strain contains the conditional centromere near SGD coordinate 80 kb. We mapped 24 events in the 72-kb interval between the conditional centromere and CEN5.
In our previous analysis of spontaneous mitotic recombination events, the region near CAN1 had elevated frequencies of recombination (Lee et al. 2009). To determine whether the DNA sequences near the conditional centromere had elevated recombination in regions that were not intrinsically prone to exchange, we constructed the diploid WS92 in which the conditional centromere was inserted near SGD coordinate 80 kb. This region does not have elevated spontaneous mitotic recombination (Lee et al. 2009) and has a low frequency of dicentric breakpoints in WS83 (Figure 4A). As shown in Figure 4B, in WS92, the breakpoints of this new dicentric chromosome are clustered near the conditional centromere. The distribution is significantly (P < 0.0001) nonrandom, and the interval closest to the conditional centromere (interval 4) is significantly enriched for breakpoints relative to the sum of the other intervals (P = <0.01, corrected for multiple comparisons). In summary, these results demonstrate that the region near the conditional centromere is particularly susceptible to breakage in dicentric chromosomes.
Analysis of recombination breakpoints for overrepresentation of elements of chromosome structure or sequence
A number of factors or chromosome elements in yeast have been suggested to be associated with elevated levels of DNA breakage or mitotic recombination (Aguilera et al. 2000 and references in File S1). Using statistical tests described in File S1, we looked for significant overrepresentation of such elements within the dicentric recombination breakpoints including palindromic sequences (≥16 bp), tandem repeats (repeats between 2 and 213 bp with a minimum repeat tract of 24 bp), G4 DNA (four tracts of three G’s separated by spacers <25 bp), tRNA genes, ARS elements, triplet repeats (≥8 repeats), long terminal repeats, peaks of gamma-H2AX, Rrm3p pause sites, replication-termination regions, and highly transcribed genes. For this analysis, we excluded the 10-kb region adjacent to the conditional centromere, since this region has an elevated frequency of recombination breakpoints by a mechanism likely to be specific for the conditional centromere. After corrections for multiple comparisons, no significant correlations with any of these elements were observed in any of the three strains or in the combined data of all three strains.
Mapping of dicentric breakpoints using SNP microarrays
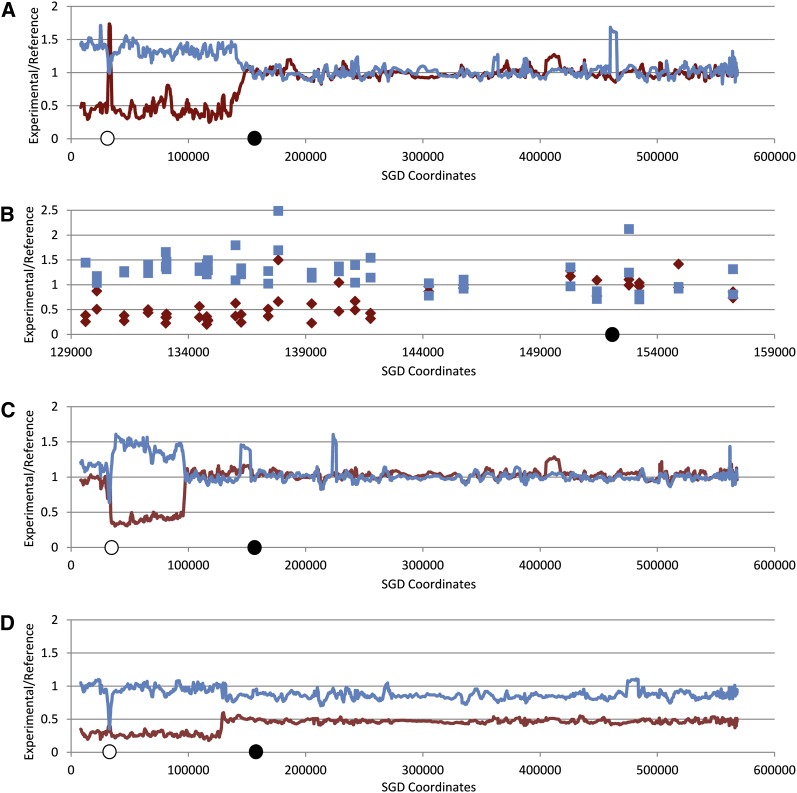
All of the events shown in Figures 3 and 4 were mapped using the PCR-based approach. We also used SNP microarrays to map seven events (WS49-16, WS49-37, WS49-39, WS83-12, WS83-30, WS83-39, and WS83-40) also examined by the PCR-based approach. This alternative method confirmed our mapping procedure and, in addition, mapped the events to higher resolution. We used oligonucleotide-containing microarrays (St. Charles et al. 2012) capable of distinguishing SNP heterozygosity and homozygosity on chromosomes III and V (details in File S1). In Figure 5, A and B, we show low- and high-resolution analysis of a recombination event on chromosome V (WS83-40). In this figure, the blue lines and squares indicate the normalized hybridization ratio (experimental to control) to the YJM789-specific oligonucleotides, and the red lines and diamonds show the normalized hybridization ratio to the J178-1d-specific oligonucleotides. The transition between heterozygosity and homozygosity for markers is between SNPs located at coordinates 141,779 and 144,265, in good agreement with our previous PCR-based mapping of the event between markers 141 and 144 kb. As expected, since the recombination is initiated by breakage of the dicentric chromosome, the homozygous region is derived from the YJM789-related parental strain PSL5. The “spike” of increased hybridization for the J178-1d-specific SNPs near SGD coordinate 30,000 in Figure 5A results from deletion of the YJM789 sequences associated with the insertion of the SUP4-o gene into the YJM789-derived chromosome. Similar patterns of hybridization, confirming our PCR-based mapping, were also observed for strains WS49-16, WS49-37, and WS49-39.
Figure 5.
Mapping of recombination events on the left arm of chromosome V by SNP microarrays. We examined three Ura− WS83 derivatives, previously analyzed by the PCR-based procedure, by SNP microarrays. The conditional centromere (white circles on x-axis) and CEN5 (black circles) are near SGD coordinates 32 kb and 152 kb, respectively, on the x-axis. A hybridization ratio of ∼1 indicates SNP heterozygosity. (A) Low-resolution depiction of the Ura− derivative WS83-40. The transition from heterozygous to homozygous SNP occurs at about SGD coordinate 142 kb. The pattern of hybridization is consistent with that expected for a crossover or BIR event. (B) High-resolution depiction of WS83-40. Blue squares and red diamonds represent hybridization to individual PSL5-specific SNPs and J1780-1d-specific SNPs, respectively. (C) Low-resolution depiction of the Ura− derivative WS83-12. In this sample, there is a transition between heterozygous and homozygous SNPs near SGD coordinate 96 kb, and a second transition between homozygous and heterozygous SNPs near SGD coordinate 31 kb. This pattern is consistent with a gene conversion event unassociated with a crossover or with a double crossover between the homologs. (D) Low-resolution depiction of the Ura− derivative WS83-30. The hybridization pattern in this sample indicates that a recombination event occurred near SGD coordinate 128 kb. Following the recombination event, the recombinant chromosome was lost in at least half of the cells in the culture.
The strain WS83-12 had a different pattern of hybridization than most of the other samples. By the PCR-based mapping method, this Ura− strain had a homozygous–heterozygous transition between markers 94 and 99. Marker 25, however, located centromere distal to the conditional centromere retained heterozygosity. The microarray analysis showed that this strain has two transitions, one located between SGD coordinates 96,550–97,221 and another located distal to the conditional centromere between coordinates 33,332 and 33,770 (Figure 5C). This pattern of hybridization suggests that WS83-12 resulted from a very long (60 kb) gene conversion event unassociated with a crossover, or a double crossover. Since the microarray analysis showed that SNPs derived from the YJM789-related homolog were duplicated, this hybridization profile is not consistent with a large heterozygous deletion. The breakpoints for WS83-12 or other strains that retained heterozygosity distal to the conditional centromere were not used in subsequent analyses.
In the strain WS83-30, the pattern of hybridization indicates that two events occurred. First, there was a crossover or BIR event near SGD coordinate 125 kb; high-resolution analysis shows a breakpoint between SNPs located at 127,038 and 128,941. Following the recombination event, the recombinant chromosome was lost in about half of the cells. A similar hybridization pattern was observed for WS83-39. Elevated frequencies of nondisjunction leading to loss (Campbell and Fogel 1977) or gain (Chua and Jinks-Robertson 1991) of recombinant chromosomes has been previously reported.
In summary, the mapping of recombination events by microarrays supports our previous analysis using the PCR-based method. These results demonstrate that mitotic homologous recombination is an important mechanism for the repair of DSBs generated by breakage of a dicentric chromosome. In diploid strains that are unable to repair DSBs by homologous recombination, the broken chromosome can be “capped” by addition of telomeric repeats (Kramer and Haber 1993), resulting in a large terminal heterozygous deletion. Since this process is much less efficient than homologous recombination (Kramer and Haber 1993), we did not expect it to contribute to the Ura− events in our experiments. The SNP arrays in Figure 5 are consistent with LOH resulting from mitotic recombination rather than deletion formation because the LOH regions have an elevated level of hybridization of the YJM789-specific SNPs associated with a reduced level of hybridization to the J178-1d-specific SNPs. A heterozygous deletion would have a reduced level of hybridization to the J178-1d-specific SNPs without an accompanying increase in the level of hybridization to the YJM789-specific SNPs. Our results, therefore, indicate most of our LOH events reflect mitotic recombination between homologs.
Discussion
Our analysis of recombination events associated with breakage of a dicentric chromosome shows two types of events. About half of the events are located near the conditional centromere, and the other half are randomly distributed in the intercentromeric region. Below, we discuss the interpretation of our mapping results in the context of previous studies of mitotic recombination and chromosome fragility.
Mitotic recombination and LOH
In S. cerevisiae, most DSBs are repaired by homologous recombination. In diploid cells in G2 of the cell cycle, the preferred substrate for the repair of a DSB is the sister chromatid, although the homologous chromosome is also used (Kadyk and Hartwell 1992). Of these two pathways, only exchange involving the homologs leads to LOH of markers distal to the event (Figure 1). In yeast, genetic evidence indicates that spontaneous recombination events are often initiated by a DSB in unreplicated chromosomes and that such breaks are preferentially repaired using the homolog as a substrate (Esposito 1978; Lee et al. 2009; Lee and Petes 2010, St. Charles et al. 2012). In the current experiments, although the DSBs are likely formed in anaphase, it is unclear whether dicentric-associated DSBs occur on one chromatid (as shown in Figure 1) or both chromatids simultaneously. If a DSB was induced in only one dicentric chromatid that was repaired using the sister chromatid as a template, the repaired chromosome would retain two centromeres and, therefore, would be likely to break in the subsequent cell cycle. The Ura− strains in our study are enriched for events in which the broken dicentric chromosome recombines with the intact monocentric homolog.
The repair of a DSB in one homolog using the intact homolog as a substrate can occur through a number of different pathways (Heyer et al. 2010). In the synthesis-dependent strand-annealing (SDSA) pathway, the broken DNA ends invade the intact homolog, synthesize DNA sequences that span the DNA break, dissociate from the template, and reanneal (Andersen and Sekelsky 2010). The net result of this process is a gene conversion event, the nonreciprocal transfer of sequences between the two chromosomes. This process would result in an interstitial region of LOH (Lee et al. 2009; St. Charles et al. 2012) rather than an LOH region extending to the telomere. Unless the conversion event removed the conditional centromere, it would not generate a Ura− derivative in our experiments and would not be detectable. Figure 5C shows an example of a possible gene conversion event.
Most of the Ura− strains examined in our experiments have the pattern of LOH that is consistent with repair of the dicentric by crossover or BIR events (Figure 1). We cannot distinguish between these two events in our analysis because we do not recover both daughter cells resulting from dicentric breakage. Since BIR events are less frequent than crossovers in wild-type strains (McMurray and Gottschling 2003; Barbera and Petes 2006; Ho et al. 2010), however, it is likely that most of the events represent crossovers.
An important issue is whether our observed recombination breakpoints accurately map the positions of the recombination-initiating DSBs. One possibility is that the broken ends undergo substantial nucleolytic degradation, forming large double-stranded gaps. By this mechanism, the position of the initiating DSB would be located telomere proximal to the location of the mapped LOH event. This scenario is unlikely for two reasons. First, degradation of the broken ends usually involves loss of one of the two DNA strands rather than formation of a double-stranded DNA gap (Symington and Gautier 2011). Second, events associated with a mitotic recombination hotspot (Tang et al. 2011) or an HO-induced DSB (Nickoloff et al. 1999) map close to the site of the DNA lesion. Another factor that affects the relationship between the site of the initiating DSB and LOH breakpoint is gene conversion. Most mitotic crossovers are associated with an adjacent tract of gene conversion (Pâques and Haber 1999; Lee et al. 2009; St. Charles et al. 2012). Since the conversion tracts have a median size of 6–8 kb, our mapping of the DSB sites is limited to that level of resolution. In addition, our analysis of LOH is also limited by the number of markers analyzed, with an average of ∼4 kb between markers on chromosomes III and V. Thus, our study yields only an approximate map position of the DSBs associated with breakage of a dicentric.
Clustering of DSBs near the conditional centromere
In all three dicentric strains examined, about half of the recombination breakpoints are within 10 kb of the conditional centromere. The comparison of the breakpoints in strains WS83 and WS92 demonstrate that clustering of events is a consequence of a property of the conditional centromere, since the region of chromosome V that has few events in WS83 (the interval between markers 81 and 91) becomes a hotspot for recombination when the conditional centromere is inserted nearby in WS92 (Figure 4, compare A and B). The mapping of breakpoints also suggests that the effect of the conditional centromere on DSBs extends at least 10 kb, since more than one of the marked intervals have elevated levels of events.
There are two related features of chromosome structure that extend about 10 kb from the yeast centromeres. First, cohesins are preferentially associated with yeast centromeres in a region extending 10–20 kb from the centromere (Blat and Kleckner 1999; Glynn et al. 2004). Second, the pericentric cohesins are bound intramolecularly, forming loops (Yeh et al. 2008). Several studies in Schizosaccharomyces pombe and S. cerevisiae have shown that the pericentric regions are more “stretchable” in preanaphase than other chromosomal regions (summarized by Thrower and Bloom 2001), although it is unclear whether this property is related to cohesin binding.
One explanation of our results is that the pericentric region located near the conditional centromere in the dicentric is more highly extended than other regions of the intercentromeric region, including the pericentric region located near the natural centromere. Why should the two pericentric regions behave differently? There is evidence that yeast centromere function is dependent on chromosome context. Megee et al. (1999) showed that centromere-associated loading of cohesins was more extensive in plasmids in which the centromeres were flanked by regions of low GC content. We measured the GC content in a 10-kb window centered on the natural and conditional centromeres in our strains. These percentages are: 34.6% (CEN3), 35.6% (CEN5), 45.1% (conditional CEN3 in WS49), 38.7% (conditional CEN3 in WS83), and 40.6% (conditional CEN3 in WS92). In all three dicentric strains, therefore, the conditional centromere is flanked by sequences that have significantly (P < 0.0001 by chi-square analysis) higher GC content than the natural centromere on the dicentric chromosome.
As discussed previously, although breakage of the dicentric chromosome requires the tension established by stretching the chromosome between different spindle poles, the mechanical force exerted by this tension is insufficient to break double-stranded DNA. One scenario is that the preferential extension of the chromosome near the conditional centromere results in increased access to cellular nucleases that generate recombinogenic DSBs. A related possibility is that stretching of the chromosome near the conditional chromosome increases the probability of DSBs formed by nucleases preloaded in the pericentric region. Top2p accumulates to high levels near the centromere during the S period (Bermejo et al. 2009). Although most Top2p binding is lost by G2/M, a small number of persisting Top2p molecules could be sufficient to generate DSBs in the pericentric region.
Another mechanism for chromosome breakage in the dicentric is scission of the chromosome during nuclear fission or cytokinesis (Quevedo et al. 2012). DNA breaks could be a direct consequence of the physical forces exerted during cell division or, perhaps more likely, an indirect consequence of stretching of the chromosome, followed by endonucleolytic cleavage. By this model, the conditional centromere would be located more closely to the cell cleavage plane than the natural centromere. Whatever the explanation for the elevated level of recombination breakpoints near the conditional centromere, our results argue that the conditional centromere has some properties that are different from the natural centromere.
Other evidence that the region near the conditional centromere is more susceptible to breakage than the region near the natural centromere is based on analysis of deletions associated with rad52 dicentric haploid strains (Kramer et al. 1994). In such strains, chromosomes become stabilized by deletion of one of the two centromeres. Kramer et al. (1994) found that deletion of the conditional centromere was about four times more common than deletion of the natural centromere. Since most of these deletions are likely to be initiated by a DSB, these results argue that the region near the conditional centromere is prone to breakage. We cannot, however, rule out the possibility that the preferential recovery of chromosomes with the natural centromere in the experiments of Kramer et al. (1994) was a consequence of a lower rate of nondisjunction of chromosomes with the wild-type centromere compared to chromosomes with only the conditional centromere.
In addition, studies in mammalian cells and plants have suggested that, in dicentric chromosomes, one centromere can be more prone to inactivation than the other (Sullivan and Schwartz 1995; Lamb et al. 2008; Stimpson et al. 2012); centromere inactivation in mammals involves both intracentromeric structural changes (deletions) and epigenetic mechanisms (Stimpson et al. 2010).
Dicentric breakpoints that are not associated with the conditional centromere
In addition to the recombination breakpoints located near the conditional centromere, we observed breakpoints that are widely distributed throughout the intercentromeric region. We were unable to associate these breakpoints with any single specific chromosome element. It should be pointed out, however, that the resolution of mapping the events and the relatively small number of mapped events makes the statistical analysis challenging. The wide distribution of events argues that the dicentric breaks occur at some common element of chromosome structure (for example, a promoter or other nucleosome-free region) or that the breaks are associated with multiple different types of chromosome elements. We also do not know the enzyme or enzymes associated with breakage of the dicentric, although the topoisomerases are obvious candidates. It is also important to mention that the pattern of recombination breakpoints produced in our system with one conditional centromere and one natural centromere may be different than recombination events induced in a chromosome with two natural centromeres.
Pobiega and Marcand (2010) created dicentric chromosomes with one conditional centromere and one natural centromere in which the dicentric was generated by a fusion between two telomeres. Upon activation of the conditional centromere, DSBs were observed. Although ∼40% of these breaks occurred within or near the telomere–telomere fusion (Pobiega and Marcand 2010), DSBs were also detected near the centromeres (S. Marcand, personal communication). The preference for breakage near the telomeric fusion may reflect a particular property of telomeric chromatin or the palindromic nature of telomere–telomere fusions.
In summary, there appear to be three types of chromosome breaks associated with dicentric chromosomes in yeast: breakage near (within 10 kb of) conditional centromeres, breakage at the junction of telomere–telomere fusions (Pobiega and Marcand 2010), and breaks that occur quasirandomly in the intercentromeric region.
Supplementary Material
Acknowledgments
We are grateful to Y. Yin, J. St. Charles, and J. Kelly for help in data analysis. We thank all members of the Petes and Jinks-Robertson labs for helpful advice, and S. Marcand, B. Sullivan, S. Covo, O. Quevedo, S. Andersen, E. Yeh, K. O’Connell, J. Sekelsky, and two reviewers for comments on the manuscript. The research was supported by National Institutes of Health grants GM24110 and GM52319 to T.D.P.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Admire A., Shanks L., Danzl N., Wang M., Weier U., et al. , 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20: 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Chavez S., Malagon F., 2000. Mitotic recombination in yeast: elements controlling its incidence. Yeast 16: 731–754. [DOI] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic and mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera M. A., Petes T. D., 2006. Selection and analysis of spontaneous reciprocal mitotic crossovers in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Stat. Soc. 57: 289–300. [Google Scholar]
- Bensimon D., Simon A. J., Croquette V., Bensimon A., 1995. Stretching DNA with a receding meniscus: experiments and models. Phys. Rev. 74: 4754–4757. [DOI] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Gonzalez-Huici V., Fachinetti D., Cocito A., et al. , 2009. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138: 870–884. [DOI] [PubMed] [Google Scholar]
- Blat Y., Kleckner N., 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms vs. the centric region. Cell 98: 249–259. [DOI] [PubMed] [Google Scholar]
- Brock J. A., Bloom K., 1994. A chromosome breakage assay to monitor mitotic forces in budding yeast. J. Cell Sci. 107: 891–902. [DOI] [PubMed] [Google Scholar]
- Campbell D. A., Fogel S., 1977. Association of chromosome loss with centromere-adjacent mitotic recombination in a yeast disomic haploid. Genetics 85: 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P., Jinks-Robertson S., 1991. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 1786: 32–40. [DOI] [PubMed] [Google Scholar]
- Craven R. J., Greenwell P. W., Dominska M., Petes T. D., 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M. S., 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75: 4436–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. K., Ballenger M., O’Brien E. T., Haase J., Superfine R., et al. , 2009. DNA relaxation dynamics as a probe for the intracellular environment. Proc. Natl. Acad. Sci. USA 106: 9250–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E. F., Megee P. C., Yu H. G., Mistrot C., Unal E., et al. , 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Microbiology, Academic Press, San Diego. [Google Scholar]
- Haber J. E., Thorburn P. C., 1984. Healing of broken linear dicentric chromosomes in yeast. Genetics 106: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.-D., Ehmsen K. T., Liu J., 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K., 1987. Genetic manipulation of centromere function. Mol. Cell. Biol. 7: 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K., 1989. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 1368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Mazon G., Lam A. F., Symington L. S., 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genetic integrity in budding yeast. Mol. Cell 40: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M. L., Tan F. J., Lai D. C., Celniker S. E., Hoskins R. A., et al. , 2010. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombination repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Rutledge L., Fitzgerald-Hayes M., Hartwell L. H., 1987. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell 48: 801–812. [DOI] [PubMed] [Google Scholar]
- Kramer K. M., Haber J. E., 1993. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 7: 2345–2356. [DOI] [PubMed] [Google Scholar]
- Kramer K. M., Brock J. A., Bloom K., Moore J. K., Haber J. E., 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. C., Yu W., Han F., Birchler J. A., 2008. Plant centromeres. Genome Dyn. 4: 95–107. [DOI] [PubMed] [Google Scholar]
- Lee P. S., Petes T. D., 2010. Mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. USA 107: 7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Greenwell P. W., Dominska M., Gawel M., Hamilton M., et al. , 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisae. PLoS Genet. 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosome translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Lobachev K. S., Gordenin D. A., Resnick M. A., 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevent of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Mann C., Davis R. W., 1983. Instability of dicentric plasmids in yeast. Proc. Natl. Acad. Sci. USA 80: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M. A., Gottschling D. E., 2003. An age-induced switch to a hyper-recombinational state. Science 301: 1908–1911. [DOI] [PubMed] [Google Scholar]
- Megee P. C., Mistrot C., Guacci V., Koshland D., 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4: 445–450. [DOI] [PubMed] [Google Scholar]
- Mieczkowski P. A., Mieczkowska J. O., Dominska M., Petes T. D., 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Lemoine F. J., Petes T. D., 2006. Review: recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Kolodner R. D., 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408. [DOI] [PubMed] [Google Scholar]
- Narayanan V., Mieczkowski P. A., Kim H.-M., Petes T. D., Lobachev K. S., 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Sweetser D. B., Clikeman J. A., Khalsa G. J., Wheeler S. L., 1999. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153: 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J. E., 1999. Multiple pathways of recombination induced by double strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Marcand S., 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24: 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V., Kolodner R. D., 2009. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS ONE 4: e6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S., Marcand S., 2010. Dicentric breakage at telomere fusions. Genes Dev. 24: 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevedo O., Garcia-Luis J., Matos-Perdomo E. A., Aragon L., Machin F., 2012. Nondisjunction of a single chromosome leads to breakage and activation of DNA damage checkpoint in G2. PLoS Genet. 8: e1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson K. M., Song I. Y., Jauch A., Holtgreve-Grez H., Hayden K. E., et al. , 2010. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 6: e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson K. M., Matheny J. E., Sullivan B. A., 2012. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosoma 20: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Schwartz S., 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4: 2189–2197. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Gautier J., 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Tang W., Dominska M., Greenwell P. W., Harvanek J., Lobachev K. S., et al. , 2011. Friedreich’s Ataxia (GAA)/(TTC) repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 7: e1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower D. A., Bloom K., 2001. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol. Biol. Cell 12: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHulle K., Lemoine F. J., Narayanan V., Downing B., Hull K., et al. , 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 27: 2601–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon M., Lobachev K., Petes T. D., 2008. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics 179: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., et al. , 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.