Abstract
Examples of meiotic drive, the non-Mendelian segregation of a specific genomic region, have been identified in several eukaryotic species. Maize contains the abnormal chromosome 10 (Ab10) drive system that transforms typically inert heterochromatic knobs into centromere-like domains (neocentromeres) that move rapidly poleward along the spindle during meiosis. Knobs can be made of two different tandem repeat sequences (TR-1 and 180-bp repeat), and both repeats have become widespread in Zea species. Here we describe detailed studies of a large knob on chromosome 10 called K10L2. We show that the knob is composed entirely of the TR-1 repeat and is linked to a strong activator of TR-1 neocentromere activity. K10L2 shows weak meiotic drive when paired with N10 but significantly reduces the meiotic drive exhibited by Ab10 (types I or II) in Ab10/K10L2 heterozygotes. These and other data confirm that (1) there are two separate and independent neocentromere activities in maize, (2) that both the TR-1 and knob 180 repeats exhibit meiotic drive (in the presence of other drive genes), and (3) that the two repeats can operate in competition with each other. Our results support the general concept that tandem repeat arrays can engage in arms-race-like struggles and proliferate as an outcome.
Keywords: abnormal chromosome 10, genomic conflict, meiotic drive, satellite repeats, selfish DNA
AS a general rule eukaryotic chromosome movement is mediated by kinetochore proteins, which bridge the interaction between the centromeric DNA and the spindle microtubules. As cell division proceeds, the centromeres move toward spindle poles, while the chromosome arms drag behind. However, genes on a chromosome 10 variant in maize known as Abnormal chromosome 10 (Ab10) change this dynamic by providing heterochromatic regions called knobs the means to move poleward along the microtubule lattice during meiosis. Each knob may be composed of thousands of tandem repeats that are clearly separated into two distinct homology groups, the 350-bp TR-1 repeat and the 180-bp repeat (Peacock et al. 1981; Ananiev et al. 1998). When Ab10 confers activity to knobs they are referred to as neocentromeres (Rhoades 1952). Ab10 contains the genes that activate neocentromeres and other unknown functions that together cause the preferential transmission of knobbed chromosomes to progeny in a process known as meiotic drive (Longley 1945; Kikudome 1959; Rhoades and Dempsey 1988a).
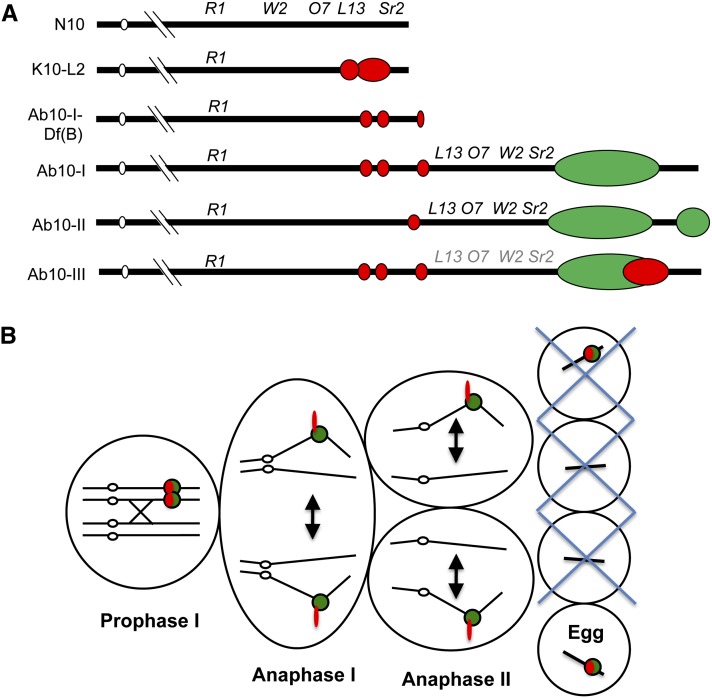
Structurally, Ab10 is similar to the canonical chromosome 10 (N10), but contains a large haplotype on the end of its long arm (Mroczek et al. 2006) (Figure 1A). The Ab10 haplotype contains the genes required for neocentromere activity and meiotic drive, as well as long arrays of both types of knob repeats (Hiatt and Dawe 2003a). Hundreds of other genes are present within the haplotype and are allelic with similar genes on N10; however, Ab10 and N10 do not recombine in this area due to the presence of multiple rearrangements (Mroczek et al. 2006). There are two well-studied cytological variants of the Ab10 haplotype that differ primarily by the size and repeat content of their knobs (as well as a third lesser known haplotype; Figure 1A). Previous work demonstrated that on Ab10-I, the region containing TR-1 rich knobs is linked to a gene(s) that selectively activates TR-1 repeats as neocentromeres, while the large 180-based knob is linked to a different gene(s) that moves knobs containing the 180 repeat (Hiatt et al. 2002). The two types of neocentromeres have visibly different cytological phenotypes at meiotic anaphase, such that TR-1-based neocentromeres tend to stretch out along spindle fibers, whereas knobs with the 180-bp repeat retain their knob-shaped appearance (Hiatt et al. 2002).
Figure 1.
Variants of maize chromosome 10 and meiotic drive model. (A) Six variations of maize chromosome 10. Only the end of the long arm is shown in detail. There are four classical markers on normal chromosome 10 (N10): Colored1 (R1), White Seedling2 (W2), Opaque Endosperm7 (O7), Luteus13 (L13), and Striate Leaves2 (Sr2). On abnormal chromosome 10 (Ab10) three of these loci lie within a large inversion that is characteristic of the Ab10 types. Ab10-III is a newly discovered form (Kanizay et al. 2012) and the arrangement of the four classical markers has not been verified (shown in gray, not black lettering). All variants of chromosome 10 (except N10) contain different amounts of TR-1 repeats (red) and 180-bp repeats (green). (B) The Rhoades model for meiotic drive. The process begins with a recombination event between centromere (open circles) and knobs (red and green). In anaphase I, knobs move laterally along the spindle poles ahead of the centromere, creating an outward orientation that is maintained through anaphase II. TR-1 repeats (red) advance to the pole faster than 180-bp repeats (green) and appear to stretch out along the spindle fibers. These events place knobbed chromatids in the top- and bottommost cells of the linear tetrad. The bottom cell normally develops into the egg.
Rhoades initially proposed that neocentromere activity is the key to the observed preferential transmission of Ab10 (Rhoades 1952). Further genetic analyses demonstrated that neocentromere activity is required but not sufficient for meiotic drive (Dawe and Cande 1996; Hiatt and Dawe 2003a). According to Rhoades model for meiotic drive (Figure 1B), a crossover event must first occur in the region between the centromere and knob of a heterozygous Ab10/N10 pair. This creates a heteromorphic dyad where each homologous chromosome contains one knobbed and one nonknobbed sister chromatid. As anaphase begins, the knobs on all chromosomes are activated as neocentromeres and begin their dramatic poleward movement. The speed and efficiency of neocentromere activity enable knobs to reach the spindle poles ahead of the centromeres (Yu et al. 1997), creating an outward orientation that ultimately delivers knobbed chromatids to the upper and lower cells of the linear tetrad. Since only the bottom cell develops into the egg in female flowers, Ab10 and other knobs are preferentially transmitted. Ab10 can be transmitted to female progeny at rates of up to 83% by this mechanism (Buckler et al. 1999) but in practice it is transmitted at levels that vary from 60 to 80%, apparently depending on environmental variables (Hiatt and Dawe 2003a). Meiotic drive does not occur in male meiosis because all four products of meiosis survive to produce pollen.
Since the major meiotic genes encoded on Ab10 are trans-acting (Dawe and Hiatt 2004), the Rhoades model accommodates the observation that other knobs also show meiotic drive when Ab10 is present (Rhoades and Dempsey 1966). Given that knobs are composed of tandem repeats, they presumably evolve by unequal recombination and replication strand slippage, both of which provide an equal chance for knobs to expand or contract in size (Charlesworth et al. 1994; Gemayel et al. 2010). However, Ab10 selects for larger knobs, and this has led to the spread and expansion of knobs across the Zea lineage. Cytologically visible knobs have been documented at multiple locations on the arms of all 10 maize chromosomes (McClintock et al. 1981; Buckler et al. 1999; Albert et al. 2010) and knob repeats can account for upwards of 8% of the maize genome (Dennis and Peacock 1984). It is clear that the 180-bp repeat is more abundant than TR-1 (Ananiev et al. 1998; Albert et al. 2010); however, little else is known about the selective forces operating on the two forms of knob repeat.
Given the fact that Ab10-mediated meiotic drive is based on a race-to-the-pole mechanism, it is theoretically possible that different forms of Ab10 haplotypes are actively competing with each other. A hypothetical example is the case where two different Ab10 haplotypes occur together in a heterozygous state. If they were identical in their efficiency of meiotic drive, the net result should be a 50/50 segregation. However, this is unlikely, as the three known haplotypes differ both structurally and genetically; one is likely to prevail. The haplotypes may differ in any number of ways, but one of the most obvious is the relative quantities of the 180-bp and TR-1 repeats. Here we provide evidence, based on extensive studies of a chromosome 10 variant called K10L2, that the 180-bp and TR-1 knob repeats behave as competitors within the Ab10-mediated meiotic drive system. These data help to explain the existence of two forms of knob repeats in maize and support the view that other repeat arrays, such as those in centromeres, may also be the outcome of an evolutionary arms race (Henikoff et al. 2001).
Materials and Methods
Cytology and fluorescent in situ hybridization
Root tip spreads were prepared from 3- or 4-day-old primary roots from the CI66 inbred line (PI 587148) and 2–23 individuals (302 total) from each of 37 landraces as previously published (Kanizay et al. 2012). Landraces were ordered from the National Plant Germplasm System (http://www.ars-grin.gov/npgs) (Supporting Information, Table S1). The knobs in each individual were counted and classified by fluorescent in situ hybridization (FISH) as TR-1 containing, 180-bp containing, or mixed (containing both repeats).
All tassels from CI66, landrace individuals that contained K10L2, and other heterozygous lines [Ab10-I/Ab10-II, K10L2/Ab10-I, K10L2/N10, and K10L2/Ab10-I-Df(B)] were staged under a dissecting microscope. Anthers of the correct stages (pachytene through anaphase II) were fixed in 4% paraformaldehyde and processed for FISH as previously described (Shi and Dawe 2006).
Testing the linkage of K10L2 to a neocentromere-activating factor
The R1 gene has many alleles that produce different patterns of kernel (aleurone) pigmentation. In this study we used four different alleles of R1: The recessive r1, which confers a colorless aleurone; the dominant R1, which confers a completely purple aleurone; R1-nj, which produces a purple cap and a purple embryo; and R1-st, which causes a purple-spotted aluerone.
Sixteen progeny from the testcross of a heterozygous K10L2 plant (r1_K10L2/R1_N10 × r1_N10/r1_N10) were examined for the presence of neocentromere activity at meiosis. We assayed nine individuals carrying K10L2 and seven carrying N10. One of the K10L2 individuals carried the dominant R1 allele, indicating there had been a recombination event between R1 and the K10L2 knob (this plant showed TR-1 neocentromeres like all other plants carrying K10L2). Twenty-five anaphase-II cells were scored from each individual.
Recombination mapping using polymerase chain reaction markers
Recombination between haplotypes was measured using two-point testcrosses, where one marker was the R1 gene that lies just proximal to the Ab10 haplotype and the second marker was one of three dominant polymerase chain reaction (PCR) markers derived from Ab10-I (D6, G8, and C2RJJM2 (Kanizay et al. 2012). For the cross R1_Ab10-I/r1_N10, R1_Ab10-I/r1_K10L2, and R1_Ab10-II/r1_K10L2 recombination was measured between the R1 locus and the G8 marker (G8 is present on Ab10 and absent on the others). For the cross r1_K10L2/R1-nj_N10 recombination was measured between R1-nj and the C2RJJM2 marker (C2RJJM2 is present on K10L2 and absent on N10). For the cross R1_Ab10-I/r1_Ab10-II, recombination was measured between R1 and the D6 marker (D6 is present on Ab10-I but absent on Ab10-II). DNA was extracted using a CTAB method (Clarke 2009) and PCR was performed as described previously (Kanizay et al. 2012).
Meiotic drive of chromosome 10 in different heterozygous plants
Progeny heterozygous for all possible combinations of Ab10-I, Ab10-II, K10L2, and N10 chromosomes (linked to different R1 alleles) were testcrossed so as to visualize segregation ratios. Crosses were made over the period of three seasons. Crosses in the first season (summer 2010) were performed on University of Georgia farmland. The testcrosses for the second (winter 2010/2011) and third seasons (summer 2011) were performed in Molokai, Hawaii. In seasons two and three, we examined the results when K10L2 was paired with three different N10 chromosomes, all linked to different R1 alleles. The results from each season varied such that there was a significant deviation from Mendelian segregation for some crosses in some seasons, but not in others. We then combined all the K10L2 testcross data as a single large data set, after first determining that the data could be pooled using a chi-squared test for independence.
Testing meiotic drive of the TR-1 rich knob on chromosome K6L
To test whether Ab10-I can drive TR-1 repeats independently of knob-180 repeats we crossed an Ab10-I individual to the Ki3 inbred (Ames 27123), which contains a TR-1 knob on the long arm of chromosome six (K6L). This created progeny that were heterozygous for both the Ab10-I chromosome and the TR-1 knob on chromosome 6. Two F1 individuals were testcrossed to knobless tama flint (Ames 21969) and progeny were scored for Ab10 and K6L using FISH.
Correlating knob abundance with variants of chromosome 10
Knobs in individuals from 37 landraces (Kanizay et al. 2012) were scored for repeat type using FISH. The average number of TR-1 only, 180-bp only, and mixed knobs was compared across three different groupings (individuals with Ab10, individuals with N10, and individuals with K10L2) by a one-way analysis of variance. The resulting groupings of means were compared with Tukey’s honestly significant difference test. The statistical analyses were performed using JMP (SAS Institute, Cary, NC; 27513).
Results
Classification of K10L2 as a TR-1-only knob
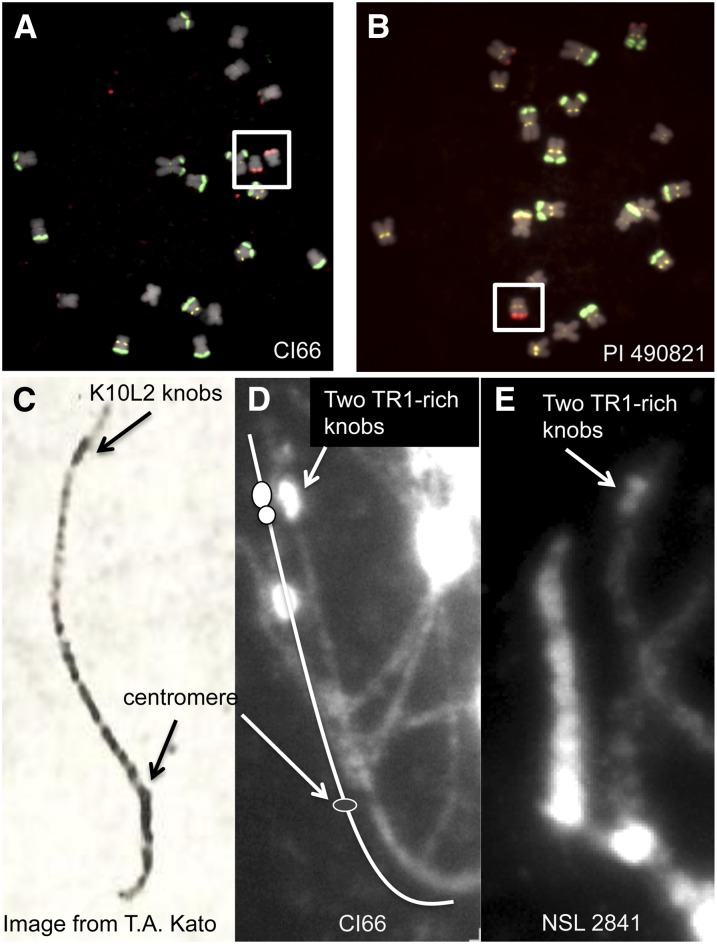
As a part of a large-scale mitotic karyotyping project, we observed that the inbred CI66 contains a variant of chromosome 10 with a large TR-1-rich knob on 10L (Albert et al. 2010). There are no 180-bp repeats in this knob as judged by our standard FISH assay (Figure 2A). A higher-resolution analysis of CI66 pachytene chromosomes revealed that there are two TR-1 knobs of different sizes followed by a distal portion of chromatin (Figure 2D). The overall structure is very similar to a knob called K10L2 that was previously reported in multiple landraces (McClintock et al. 1981) (Figure 2C). Although the particular landraces noted to contain K10L2 by McClintock and coworkers are no longer available, we requested 37 landraces from similar localities. Twelve of these races were segregating an apparently similar TR-1-rich knob on chromosome 10L as assayed by mitotic FISH (Figure 2B, Table S1). Individuals from four of the landraces were grown to meiosis and confirmed to show at pachytene a knob of the same structure (two TR-1 knobs) (Figure 2E). These data identify the large TR-1 knob in CI66 as K10L2 and confirm that it is prevalent in multiple landraces.
Figure 2.
CI66 contains the K10L2 TR-1-rich knob. (A) Mitotic chromosomes from the inbred CI66. This line contains two copies of a variant of chromosome 10 with a large TR-1-rich knob on the long arm (boxed). TR-1 repeats are in red, 180-bp repeats are in green, and centromeres (the CentC repeat) are in yellow. (B) The landrace PI 490831 segregates for a chromosome that appears similar to chromosome 10 in CI66. The individual shown was heterozygous for the chromosome. (C) A meiotic chromosome in the pachytene substage of meiosis showing the knob previously described as K10L2 (McClintock et al. 1981). Note that the knob appears to be composed of two smaller knobs. This image was prepared using classical acetocarmine methods. (D) Pachytene chromosome 10 from CI66 showing a structure similar to K10L2. (E) Pachytene chromosome from the landrace NSL 2841 showing a similar structure. The images in D and E show DNA staining only (DAPI), but FISH analysis of similar slides confirmed that K10L2 contains TR-1 and no evidence of the knob-180 repeat.
Lines with K10L2 show extreme TR-1 neocentromere activity
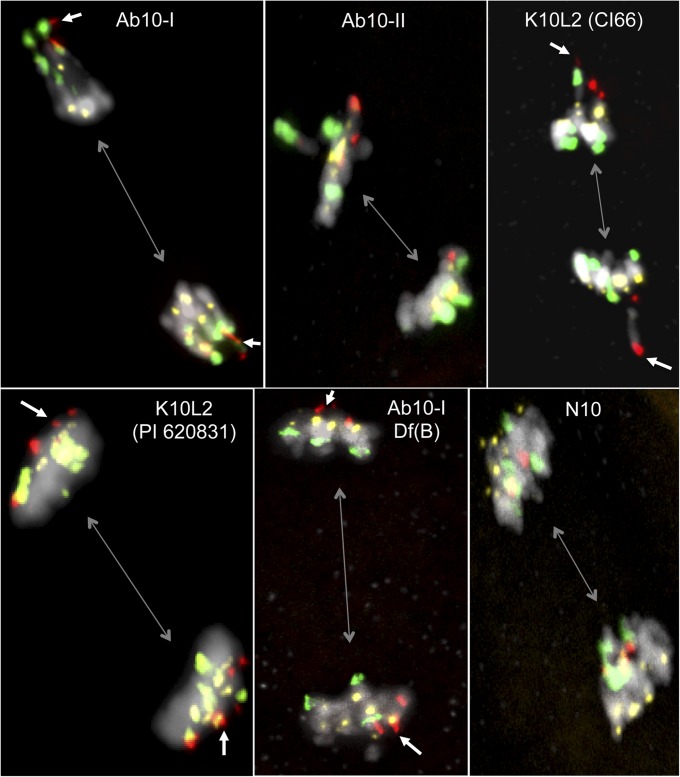
We wondered whether K10L2 might be either a new form of Ab10 or a deletion derivative of one of the known Ab10 chromosomes. Given that neocentromere activity is a hallmark of Ab10, we examined male meiocytes undergoing anaphase from the CI66 inbred where K10L2 was identified. We also processed control meiocytes from lines containing N10, Ab10-I, Ab10-II, and a terminal deficiency of Ab10-I-Df(B). Ab10-I is known to show neocentromere activity at both repeats (Hiatt et al. 2002), Ab10-II shows neocentromere activity at 180-bp repeats but not TR-1 repeats (Mroczek et al. 2006), and the Ab10-I deficiency Df(B) shows neocentromeres at TR-1 repeats but not 180-bp repeats (Figure 3).
Figure 3.
Lines with K10L2 exhibit strong neocentromere activity at TR-1 knobs. Anaphase II cells from six different genotypes are shown, highlighting the direction of the spindle axis (gray bidirectional arrows), TR-1 repeats (red), 180-bp repeats (green), and centromeres (yellow). White arrows indicate TR-1 neocentromere activity. Ab10-I exhibits neocentromere activity at both TR-I and 180-bp repeats. Ab10-II shows neocentromeres at 180-bp repeats (but not TR-1 repeats), while CI66 shows neocentromeres at TR-1 repeats (but not 180-bp repeats). The landrace PI 620831 has a phenotype similar to CI66. Ab10-I-Df(B) is a deletion derivative that lacks the capacity to induce neocentromeres at 180-bp repeats but retains TR-1 neocentromere activity. In N10 lines, the centromeres lead the chromosomes to spindle poles and knobs drag behind.
Individuals from the CI66 inbred showed extreme neocentromere activity of all knobs containing the TR-1 repeat (including TR-1-only and mixed knobs), but no neocentromeres on knobs that were composed exclusively of 180-bp repeats (Figure 3C). We also assayed one landrace (PI 620831) containing K10L2 and observed similar TR-1-based neocentromere activity, albeit much weaker than in CI66 (Figure 3E). The apparent difference in neocentromere activity may be a result of the fact that K10L2 was heterozygous in PI 620831, whereas it is homozygous in CI66.
K10L2 is genetically linked to a TR-1 neocentromere-activating factor
Neocentromere activity on Ab10 is conferred by trans-acting factor(s) that are linked to the knob repeats they activate (Hiatt et al. 2002). To test whether K10L2 contains a similar linked transacting factor, we crossed a heterozygous K10L2 plant (r1_K10L2/R1_N10) to a normal (r1_N10/r1_N10) plant and scored progeny for neocentromere activity. Nine plants with the K10L2 knob (K10L2/N10) and seven cytologically normal siblings (N10_N10) were assayed at meiosis II. All K10L2-containing plants showed neocentromeres at TR-1 repeats (and not at 180-bp repeats), whereas there were no neocentromeres in any of the N10/N10 plants. These data demonstrate that a factor activating TR-1 activity is genetically linked to K10L2 (by a distance of ∼6 cM or less).
K10L2 occurs on an N10-like chromosome
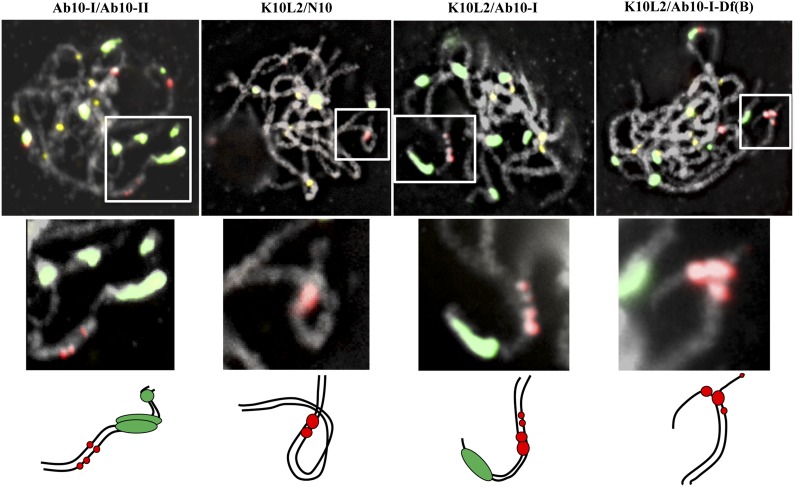
Prior data have shown that Ab10 haplotypes do not pair with N10 during the pachytene substage of meiosis (Rhoades and Dempsey 1966; Rhoades and Dempsey 1988b). To make similar tests, we examined plants from four chromosome 10 pairings, where the K10L2 chromosome was derived from CI66: Ab10-I/Ab10-II (which have a similar structure) (Kanizay et al. 2012), K10L2/Ab10-I, K10L2/Ab10-I-Df(B), and K10L2/N10. These assays revealed that the K10L2 chromosome does not show consistent pairing behavior with any chromosome except N10 (Figure 4).
Figure 4.
K10L2 pairs more consistently with N10 than with Ab10. Plants heterozygous for different forms of chromosome 10 were assayed at the pachytene substage of meiosis I. Ab10-I paired well with Ab10-II (in 6 of 10 cells assayed, as depicted), and K10L2 paired well with N10 (in 10 of 10 cells, as depicted). In contrast, K10L2 did not pair in an orderly way with either Ab10-I or Ab10-I-Df(B) (each pairing looked different; the ones depicted are examples). TR-1 repeats are shown in red, 180-bp repeats in green, and centromeres in yellow. Boxed areas are enlarged below, and illustrated with cartoons to show the inferred pairing arrangements.
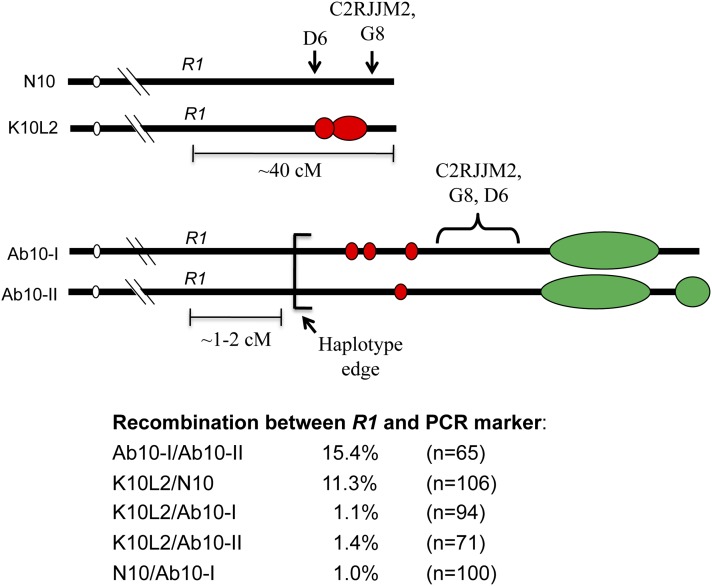
In addition, we measured recombination between the R1 gene and a set of three dominant PCR markers derived from Ab10-I (Figure 5). Prior data indicate that the R1 locus is ∼1–2 cM away from the edge of the Ab10 haplotype (Rhoades and Dempsey 1985) and that the three molecular markers map within the haplotype (Kanizay et al. 2012). As shown in Figure 5, we observed ∼15% recombination between Ab10-I and Ab10-II using these markers, consistent with the fact that Ab10-I and Ab10-II have a similar overall structure (Kanizay et al. 2012). Recombination between K10L2 and N10 was measured at 11%, while recombination between K10L2 and either Ab10-I or Ab10-II was <2%, strongly suggesting that the K10L2 knob lies on a chromosome that is more similar to N10 than Ab10.
Figure 5.
Recombination between different chromosome 10 variants. Chromosomes with differing R1 alleles were made heterozygous and then crossed as females to tester lines. Recombination was measured between R1 and a second PCR marker that maps to within the Ab10 haplotype. The inferred locations of the PCR markers are shown relative to the B73 genome assembly (although the markers are dominant to either K10L2 or Ab10 and do not amplify from N10). In the pairing of N10 and Ab10, the genetic distance between R1 and any marker within the haplotype is <2 cM, due to the large inversion that defines the haplotype. Measured recombination values are shown, where n is the number of progeny assayed.
K10L2 confers weak or no meiotic drive
The observation that K10L2 produces TR-1 neocentromere activity led us to address whether it causes meiotic drive. To this end, we created lines heterozygous for Ab10-I/N10, Ab10-II/N10, and K10L2/N10 and testcrossed them as females to tester lines such that we could track the segregation of chromosomes using linked R1 alleles. Consistent with prior data, we found that both Ab10-I and Ab10-II averaged >70% transmission by this assay (Table 1).
Table 1.
Meiotic drive of chromosome 10 variants in different pairings
| Female genotype | Male genotype | Driving haplotype | Average % transmission, season 1 (GA) | Average % transmission, season 2 (HA) | Average % transmission, season 3 (HA) |
| r1_Ab10-I/ R1-nj_N10 | R1-st_N10/ R1-st_N10 | Ab10-I | 71%** [n = 8578 (24)] | 79%** [n = 3665 (16)] | 79%**[n = 783 (11)] |
| r1_Ab10-II/ R1-nj_N10 | R1-st_N10/ R1-st_N10 | Ab10-II | 70%**[ n = 7390 (25)] | 77%**[ n = 2860 (12)] | 79%**[ n = 441 (6)] |
| r1_K10L2/ R1-nj_N10 | R1-st_N10/ R1-st_N10 | K10L2 | 51%**[ n = 10,970 (27)] | 51%[ n = 5316 (15)] | 51%[ n = 1705 (9)] |
| r1_K10L2/ R1-st_N10 | r1_N10/ r_N10 | K10L2 | N/A | 52%**[ n = 4650 (16)] | 53%**[ n = 3498 (15)] |
| r1_K10L2/ R1_N10 | r1_N10/ r1_N10 | K10L2 | N/A | 50%[ n = 5322 (21)] | 51%*[ n = 5250 (15)] |
| Total K10L2/N10 × N10/N10 | 51%**[ n = 10,970 (27)] | 51%*[ n = 15,288 (52)] | 52%**[ n = 10,453 (39)] | ||
| R1_Ab10-I/r1_K10L2 | r1_N10/r1_N10 | Ab10-I | N/A | 54%**[ n = 3958 (13)] | 52%**[ n = 6134 (17)] |
| R1_Ab10-II/r1_K10L2 | r1_N10/r1_N10 | Ab10-II | N/A | 60%**[ n = 3592 (21)] | 56%**[ n = 5828 (23)] |
| R1_Ab10-I/r1_Ab10-II | r1_N10/r1_N10 | — | N/A | 52%[ n = 845 (18)] | N/A |
n, total number of seeds counted, with the number of ears in parentheses. Significant deviations from Mendelian expectation are indicated by *P < 0.05 or **P < 0.01. GA, crosses were performed in Georgia; HA, crosses were performed in Hawaii; N/A, cross was not performed in that year.
The data for K10L2 derived from three seasons suggest that it displays weak meiotic drive when paired with N10 (Table 1). In seasons one and two, K10L2 showed an average of 51% segregation over N10, and in season three, 52% segregation. When the results from all three seasons were combined, a chi-squared test indicated that the overall 51% segregation exceeded the Mendelian expectation of 50%. It is important to note that these levels of meiotic drive are on the edge of detectability and that we did not observe K10L2 drive in all experiments (for instance, seasons 2 and 3 for the cross R1-nj_N10/ r1_K10L2 (Table 1)). It is clear, however, that K10L2 segregates at levels that exceed what is observed for any known Ab10 deficiency or meiotic drive mutant, all of which are transmitted at 45% or less through the female (Hiatt and Dawe 2003b).
K10L2 reduces Ab10-mediated meiotic drive
Prior data suggest that knobs on other chromosomes compete with each other and that larger knobs tend to be favored (Kikudome 1959; Buckler et al. 1999). We wondered how K10L2 would perform in head-to-head pairings with Ab10. Positive controls involving Ab10-I/N10 and Ab10-II/N10 for these seasons showed very strong meiotic drive—nearing 79% (Table 1). However, meiotic drive was dramatically reduced in crosses involving Ab10-I/K10L2 and Ab10-II/K10L2. Ab10-I was transmitted to <54% of progeny, and Ab10-II to <60% progeny when paired with K10L2. We also testcrossed individuals heterozygous for Ab10-I and Ab10-II and found that segregation of the two Ab10s was not statistically different from Mendelian expectations.
A TR-1-only knob on 6L displays meiotic drive in the presence of Ab10-I
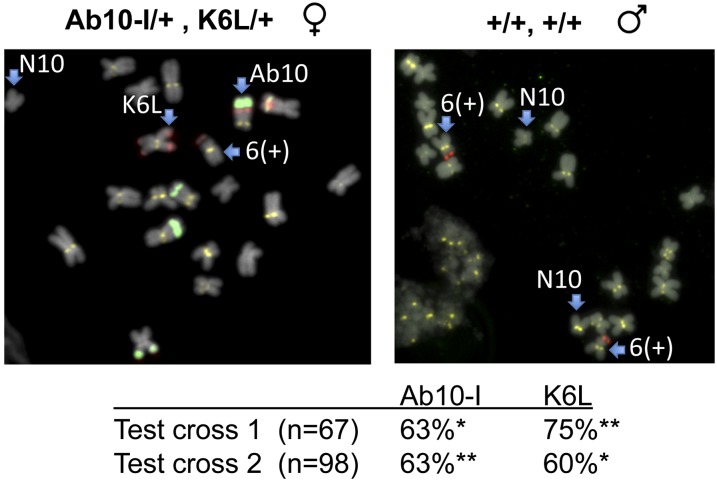
Although K10L2 does not encode a complete meiotic drive haplotype, the competition experiments suggest that it may respond to the genes that confer meiotic drive on Ab10-I. This cannot be tested directly because the pairing of K10L2 with Ab10 results in suppression. However, we can address the more general question of whether Ab10 has the capacity to drive a TR-1 knob in trans. To this end we testcrossed a line that was heterozygous for both Ab10-I and a TR-1-only knob on the long arm of chromosome 6 (K6L; see Figure 6). The presence or absence of the K6L knob (and Ab10) was then assayed in a total of 165 progeny. The results demonstrate that the TR-1-rich K6L knob is preferentially transmitted when Ab10 is present (Figure 6).
Figure 6.
The TR-1-rich K6L knob shows meiotic drive in the presence of Ab10-I. Individuals of the genotype Ab10-I/+; K6L/+ were crossed to knobless line and 165 progeny scored. Chromosomes 6 and 10 from the parental lines are shown, where TR-1 repeats are highlighted in red, 180-bp repeats in green, and centromeres in yellow. Note that the K6L knob is on the long arm where the arrow is pointing (there is also a red signal on the short arm, but this is not a knob). The genetic transmission of Ab10-I and K6L is indicated in the table. Chi-squared tests were performed to verify deviation from Mendelian expectations (*P < 0.05, **P < 0.01).
Populations with Ab10 have statistically more mixed knobs than those with N10
Under a model where Ab10 drives TR-1 repeats, we would expect populations containing Ab10 to contain more knobs with TR-1. We examined 302 individuals from 37 maize landraces, scoring the total number of knobs composed exclusively of TR-1 repeats, the total number of knobs composed exclusively 180-bp repeats, and the total number of “mixed” knobs containing both TR-1 and 180-bp repeats (Figure 7, Table S1). Knobs are variable structures that may differ with respect to size and repeat content at homologous positions (Albert et al. 2010), and there was extensive knob heterozygosity and polymorphism in these diverse lines. The overall average was 17.6 knobs per individual, not counting chromosome 10.
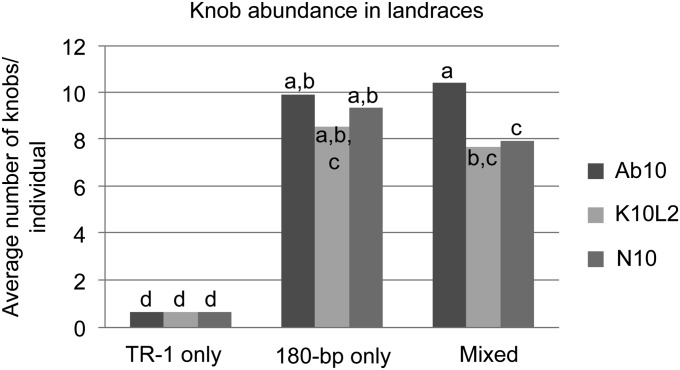
Figure 7.
Landraces with Ab10 have significantly more mixed knobs. The number of knobs composed exclusively of TR-1 repeats, exclusively of 180-bp repeats, or a mixture of both was counted in 302 individuals from 37 landraces. See Table S1 for complete data set. The individuals were separated into three groups (those with Ab10, those with N10, and those with K10L2) and knob content within each category compared by a one-way analysis of variance. The results supported four statistical categories indicated by letters (P < 0.01). Groups that have the same letter designation are not significantly different. The major result from this analysis is that mixed knobs are significantly more abundant when Ab10 is present, as compared to either N10 or K10L2.
We found that knobs composed entirely of TR-1 repeats are very rare (Figure 7). Of the remaining knobs, nearly half are mixed knobs. Mixed knobs were statistically more abundant in lines with Ab10 than lines containing either N10 or K10L2, while, surprisingly the number of 180-bp knobs was not higher in lines with Ab10 (Figure 7). These data suggest that in landraces with large numbers of knobs, mixed knobs are more competitive than knobs composed of either repeat alone.
Discussion
As originally described, the Ab10 drive system was thought to rely entirely on the dominant 180-bp neocentromere system, which includes both the large 180-bp knob and the linked neocentromere activating gene (Rhoades and Dempsey 1985). A second neocentromere system based on TR-1 was later discovered on Ab10 (Hiatt et al. 2002), but it was unclear how TR-1 repeats participated in the process—whether as an enhancer to facilitate drive by the 180-bp system or as a potential competitor. Here we provide new data suggesting that TR-1 repeats can act as competitors with 180-bp repeats when paired in opposition, but may also function to facilitate meiotic drive when the two forms of repeat are linked in coupling. The primary evidence comes from detailed studies of an unusual chromosome 10 variant carrying the TR-1-rich knob K10L2. Our results and interpretations can be summarized as follows:
K10L2 is a prevalent TR-1-rich knob. Although K10L2 is rare in modern inbred maize (1/103 tested—only the CI66 inbred; Albert et al. 2010), it is surprisingly abundant in landraces and teosintes. McClintock and coworkers observed K10L2 in 8% (110/1246) of landrace populations and 42% (23/54) of teosinte populations, which is comparable to the frequency of Ab10 (18 and 35%, respectively; McClintock et al. 1981). We found K10L2 in 12 landraces and observed in all cases that it was composed entirely of TR-1 repeats (Table S1). The prevalence and size of the K10L2 knob suggests it is under positive selection, either for its inherent capacity to confer weak meiotic drive or for its role as a suppressor of the 180-bp neocentromere system.
Lines with K10L2 show neocentromere activity at TR-1 repeats but not 180-bp repeats. In a small mapping population, we showed that a TR-1 neocentromere-activating gene is linked to the K10L2 knob. Since K10L2 resembles Ab10 in key ways, it could in principle be a deletion derivative of a complete Ab10 haplotype. However, both our pachytene pairing analyses and genetic mapping data suggest that K10L2 resides on a chromosome that is more similar to N10 than Ab10 (Figures 4 and 5). These data indicate that K10L2 and Ab10 have separate evolutionary origins or, at the least, are linked to structural rearrangements that hinder genetic exchange between the chromosomes.
TR-1 repeats can participate in meiotic drive. The fact that K10L2 is transmitted only slightly over Mendelian levels (Table 1) raises the question of whether TR-1 repeats have the capacity to display meiotic drive. Our assays of the TR-1-rich K6L knob establish that TR-1 repeats can indeed be strongly driven in the presence of Ab10 (Figure 6). We have previously postulated that Ab10 encodes an as yet unknown factor that helps to maintain the orientation of knobs between meiosis I and meiosis II, perhaps through an interaction with the nuclear envelope (Hiatt and Dawe 2003a). It appears that the chromosome carrying K10L2 lacks this factor; however, it can presumably make use of it in trans.
K10L2 competes with Ab10. We have previously assumed that Ab10 has no natural competitors except other variants of Ab10 (Kanizay et al. 2012). However, the data presented here demonstrate that K10L2 can function as a strong competitor even though it is a poor driver. We can see evidence of this conflict in the pairing of Ab10-II (lacking TR-1) and K10L2 (lacking knob 180) (Table 1). In this pairing, Ab10-II suffers by losing much of its segregation advantage (we found a reduction from 77 to 60% in season 2 and from 79 to 56% in season 3), and the chromosome carrying K10L2 presumably gains by outcompeting other knobless N10 chromosomes in the population. These data clearly establish that TR-1 repeats compete with 180-bp repeats for segregation to progeny.
Mixed knobs composed of both TR-1 repeats and 180-bp repeats are commonly observed and statistically more common when Ab10 is present (Figure 7). These results indicate that mixed knobs are very successful when Ab10 is present and suggest that they may be particularly effective in diverse populations where multiple knobs are in direct competition with each other. The prevalence of mixed knobs may also reflect the fact that TR-1 repeats evolved more recently: 180-bp repeats are abundant in the maize relative Tripsacum (Dennis and Peacock 1984), whereas TR-1 repeats are not (Hsu et al. 2003). Previous authors have argued that knobs form in locations that optimize their chances of being preferentially transmitted by Ab10 (Buckler et al. 1999). Therefore it is possible that the abundant 180-bp repeat had already occupied the best locations before TR-1 arose, forcing TR-1 to invade loci occupied by 180-bp repeats to take advantage of Ab10-mediated meiotic drive.
In summary, our results suggest that TR-1 repeats have the capacity to function as a counterbalance to the dominant 180-bp system. In particular, the TR-1-rich K10L2 chromosome functions as an Ab10 suppressor, and this presumably acts to reduce the overall abundance of the 180-bp repeat that relies on Ab10. Similarly, other TR-1-based knobs (such as K6L) may have evolved to suppress the effects of powerful 180-bp knobs on 6L (chromosome 6L has four different knob sites; McClintock et al. 1981). In the larger perspective, the presence of two competitive knob repeats might help to limit the potential for runaway escalation in knob size, which might be the natural outcome if there were only one form of knob repeat and larger knobs are more fit in the context of meiotic drive. In fact, published data suggest that Zea species with fewer TR-1 repeats (e.g., Zea huehuetenangensis and Z. luxurians) have far larger 180-bp-based knobs when compared with cultivated maize or its direct ancestors Z. parviglumis and Z. mexicana, which have substantial amounts of TR-1 (Albert et al. 2010). However, in many instances, and even within the Ab10 haplotype (e.g., Ab10-I), TR-1 can function in conjunction with the 180-bp repeat to facilitate preferential transmission. TR-1 repeats can be either a friend or foe to 180-bp repeats: when they face off on opposite chromosomes they naturally compete, and presumably both suffer as an outcome, but when they work together, they often benefit.
The idea of an intragenomic conflict between repeat arrays has been heavily discussed in recent years relative to the centromere drive hypothesis, which posits that rapid centromere evolution is the outcome of an arms race between repeats and their binding proteins (Henikoff et al. 2001). Neocentromeres are similar to centromeres in the fundamental way that repeats and binding proteins interact to move chromatin on the spindle apparatus. However, outside of this superficial similarity, the parallels are few: neocentromeres lie on chromosome arms, they cannot align on the metaphase plate unless they are linked to a true centromere (Yu et al. 1997), and none of the major kinetochore proteins localize to active neocentromeres (Dawe and Hiatt 2004). Nevertheless, our data clearly support the central tenant of the centromere drive hypothesis, which is that by mediating chromosome motility, tandem repeats can take on selfish qualities that result in unexpected patterns of evolution.
Supplementary Material
Acknowledgments
We thank Takeo Angel Kato for his kind correspondence, thoughts, and the K10L2 image used in Figure 2C. We also thank Rashin Ghaffari and Margaret Glover for help with FISH screening and Josiah Zachary and Andrew Donovan for help with DNA extractions and PCR. This work was supported by a grant from the National Science Foundation (NSF-0951091).
Footnotes
Communicating editor: A. Houben
Literature Cited
- Albert P. S., Gao Z., Danilova T. V., Birchler J. A., 2010. Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet. Genome Res. 129: 6–16. [DOI] [PubMed] [Google Scholar]
- Ananiev E. V., Phillips R. L., Rines H. W., 1998. A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: Are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95: 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., Phelps-Durr T. L., Buckler C. S. K., Dawe R. K., Doebley J. F., et al. , 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W., 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Clarke J. D., 2009. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009(3) doi: 10.1101/pdb.prot5177 [DOI] [PubMed] [Google Scholar]
- Dawe R. K., Cande W. Z., 1996. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc. Natl. Acad. Sci. USA 93: 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R. K., Hiatt E. N., 2004. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 12: 655–669. [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Peacock W. J., 1984. Knob heterochromatin homology in maize and its relatives. J. Mol. Evol. 20: 341–350. [DOI] [PubMed] [Google Scholar]
- Gemayel R., Vinces M. D., Legendre M., Verstrepen K. J., 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44: 445–477. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hiatt E. N., Dawe R. K., 2003a Four loci on abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt E. N., Dawe R. K., 2003b The meiotic drive system on maize abnormal chromosome 10 contains few essential genes. Genetica 117: 67–76. [DOI] [PubMed] [Google Scholar]
- Hiatt E. N., Kentner E. K., Dawe R. K., 2002. Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell 14: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F. C., Wang C. J., Chen C. M., Hu H. Y., Chen C. C., 2003. Molecular characterization of a family of tandemly repeated DNA sequences, TR-1, in heterochromatic knobs of maize and its relatives. Genetics 164: 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay L. B., Pyhäjärvi T., Lowry E. G., Hufford M. B., Peterson D. G., et al. , 2013. Diversity and abundance of the abnormal chromosome 10 meiotic drive complex in Zea mays. Heredity doi: 10.1038/hdy.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikudome G., 1959. Studies on the phenomenon of preferential segregation in maize. Genetics 44: 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley A. E., 1945. Abnormal segregation during megasporogenesis in maize. Genetics 30: 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., Yamakake T., Blumenschein A., 1981. Chromosome Constitution of Races of Maize: Its Significance in the Interpretation of Relationships Between Races and Varieties in the Americas. Colegio de Postgraduados, Chapingo, Mexico. [Google Scholar]
- Mroczek R. J., Melo J. R., Luce A. C., Hiatt E. N., Dawe R. K., 2006. The maize Ab 10 meiotic drive system maps to supernumerary sequences in a large complex haplotype. Genetics 174: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Rhoades M. M., Pryor A. J., 1981. Highly repeated DNA-sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78: 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M., 1952 Preferential segregation in maize, pp. 66–80 in Heterosis, edited by J. W. Gowen. Iowa State College Press, Ames, IA. [Google Scholar]
- Rhoades M. M., Dempsey E., 1966. Effect of abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics 53: 989–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M., Dempsey E., 1985. Structural heterogeneity of chromosome 10 in races of maize and teosinte, pp. 1–18 in Plant Genetics, edited by Freeling M.Alan R. Liss, NY. [Google Scholar]
- Rhoades M., Dempsey E., 1988a Effect of K10-II on preferential segregation of chromosome 9. Maize Genet. Coop. News Lett. 62: 32–33. [Google Scholar]
- Rhoades M., Dempsey E., 1988b Structure of K10-II chromosome and comparison with K10-I. Maize Genet. Coop. News Lett. 62: 33–34. [Google Scholar]
- Shi J., Dawe R. K., 2006. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. G., Hiatt E. N., Chan A., Sweeney M., Dawe R. K., 1997. Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.