Abstract
The transcription factor MyoD is a master regulator of skeletal muscle differentiation. The finding that G9a, an enzyme principally involved in histone H3 lysine 9 di-methylation (H3K9me2), methylates MyoD, identifies previously unappreciated mechanisms by which chromatin modifiers regulate the transcriptional activity of non-histone substrates to control cellular differentiation programs.
Keywords: G9a, PRC2, chromatin modifiers, differentiation, methylation, skeletal muscle, transcription, transcription factors
Introduction
In a landmark study published over two decades ago, the transcription factor MyoD was found to be sufficient to convert fibroblasts into myoblasts.1 MyoD, which is expressed only in skeletal muscle due to silencing of its promoter by DNA methylation in non-muscle cells,2 is a member of the basic helix-loop-helix (bHLH) transcription factor family and the founding member of Myogenic Regulatory Factors (MRF) that include Myf5, MRF4 and Myogenin.3,4 All MRFs are competent in inducing muscle differentiation when expressed in non-muscle cells, although with varying efficiencies. However, in vivo, MRFs are expressed at different times and have distinct functions. In developing mouse embryos, muscle progenitor cells present in somites migrate and give rise to body and limb muscles. Wnt and Sonic Hedgehog signals from the dorsal neural tube, surface ectoderm and notochord induce expression of the paired-box transcription factors Pax3 and Pax7 in muscle progenitor cells resulting in myogenic cell specification. Subsequently Myf5 and MyoD expression is induced committing cells to the muscle lineage. Myf5 is the first MRF to be expressed in dorsal-medial somites that give rise to trunk and intercostal muscles, and MyoD is expressed in dorso-lateral somitic cells that give rise to developing body wall and limb muscles. MRF4 is transiently expressed in the myotome, and re-expressed later during differentiation, whereas Myogenin is expressed only in differentiating muscle cells.5,6 Consistent with their distinct expression patterns, genetic evidence has demonstrated overlapping but distinct roles for each MRF. Loss of MyoD in mice does not result in overt defects in muscle differentiation due to functional redundancy with Myf5. Consistently, mice lacking both MyoD and Myf5 exhibit a defect in formation of myoblasts and consequently reduced muscle mass.7 Myogenin null mice develop myoblasts but exhibit a defect in differentiation,8,9 whereas MRF4 plays a role in both early and late muscle development.10
Regulating the Regulators
MRFs contain a basic domain for DNA-binding, a HLH domain for dimerization, and transactivation domain(s) (Fig. 1). All MRFs heterodimerize with ubiquitously expressed E-proteins HEB, E2–2, and E2A proteins (E12/E47). MRF-E protein heterodimers bind E-box sites (CANNTG) present in promoters and enhancers of target genes with high affinity, whereas MRF homodimers bind poorly to DNA.3,4 The myocyte enhancer factor 2 (MEF2) family, that includes MEF2-A, -B, -C and -D cooperate with MRFs to activate the differentiation program. While MEF2 factors do not have myogenic activity on their own, they increase the efficiency of conversion of non-muscle cells to muscle when expressed together with MRFs.11
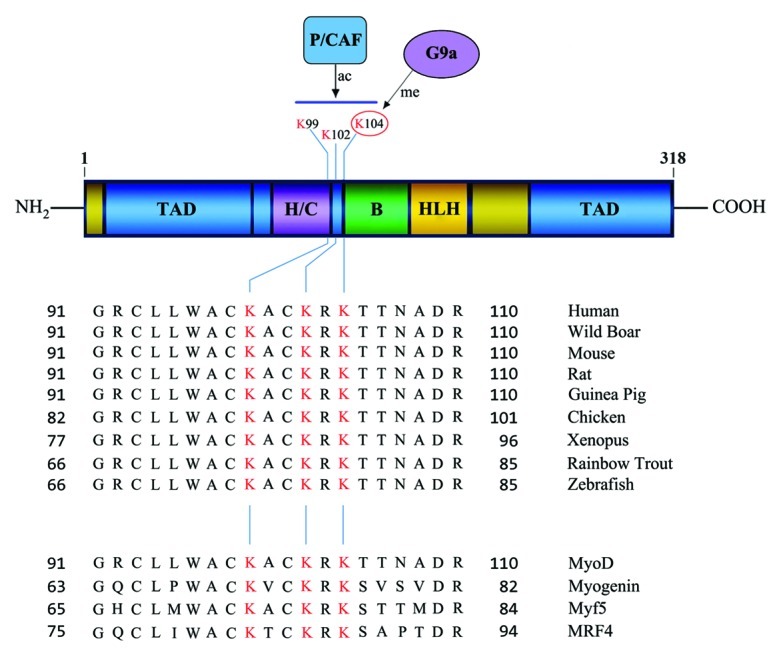
Figure 1. Schematic representation of the MyoD domain structure (upper panel). The basic (B) DNA-binding domain; helix-loop-helix (HLH) dimerization domain; transactivation domain(s) (TAD); and the cysteine-histidine rich region (H/C) are shown. Numbers indicate amino acid residues. Alignment of MyoD cDNA from various species (middle panel) show three highly conserved lysine (K) residues (highlighted in red) that are acetylated by P/CAF upon differentiation. K104 is methylated by G9a in undifferentiated myoblasts (numbering based on the human/mouse cDNA). These lysine residues are conserved in all MRFs (lower panel).
In cultured cells, as well as in vivo, MyoD is expressed in myoblasts prior to the onset of differentiation (Fig. 2). Upon differentiation cues, MyoD promotes an irreversible cell cycle exit via regulation of the p21Cip promoter, and also upregulates expression of early and late differentiation genes including myogenin, myosin heavy chain (MHC), muscle creatine kinase, and Troponin T.12 Given its ability to regulate the entire differentiation program, MyoD activity in skeletal myoblasts is kept in check by association with a number of proteins. For instance, Inhibitor of differentiation (Id) proteins are expressed at high levels in myoblasts, and efficiently heterodimerize with E-proteins. Thus E-proteins are sequestered away from MyoD, reducing its ability to activate transcription of downstream target genes. Twist, a bHLH protein, competes with MyoD-E protein heterodimers for binding to E-box sites, and also titrates out E12/E47 via dimerization preventing MyoD-dependent gene expression. Similarly, other bHLH transcription factors Sharp-1, MyoR and Mist form ‘inactive’ heterodimers with MRFs or E-proteins that are unable to activate transcription and thereby antagonize MyoD activity.13,14
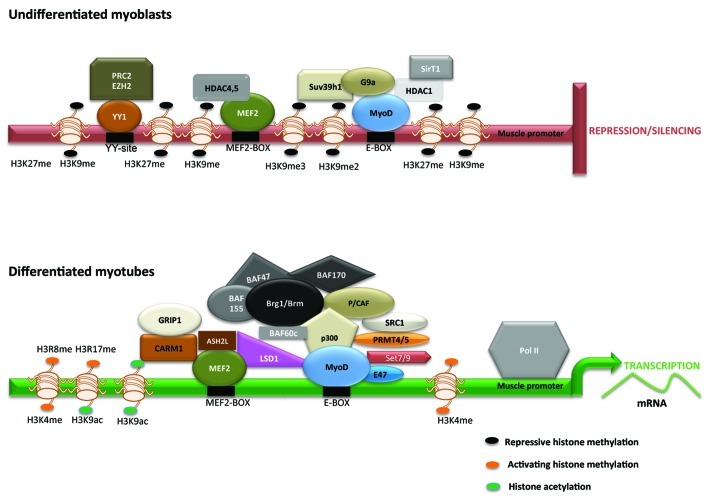
Figure 2. Model for epigenetic regulation of muscle promoters in undifferentiated and differentiated muscle cells. Transcriptional repression of muscle gene expression in undifferentiated cells is achieved through recruitment of HDACs (HDAC1, HDAC4/5, and SirT1) and HMTs (Suv39h1, G9a, Ezh2), that interact with MyoD, MEF2, and YY1 as indicated. HMTs mediate two signature repressive chromatin marks H3K9me2/me3 and H3K27me3 that restrict MyoD and MEF2 activity. In differentiated cells, recruitment of HATs (p300, P/CAF, SRC1, GRIP1), HMTs (Set7/9, PRMT4/5, CARM1, Ezh1; Ash2L), demethylases (JMJD2A, LSD1); and SWI/SNF Brg1/Brm chromatin remodeling complexes permit extensive reprograming of muscle promoters resulting H3K9ac and H3K14ac, and H3K4, H3R8, and H3R17 methylation that allow for an open chromatin configuration and activation of MyoD- and MEF2-dependent gene expression.
Epigenetic Regulation of Skeletal Myogenesis
Epigenetic modifications constitute an additional mechanism to control MyoD and MEF2 activity through modulation of chromatin structure.15-17 Such modifications have largely been attributed to recruitment of chromatin modifying and remodeling complexes to muscle promoters. Two widely studied modifications are histone deacetylation/acetylation and methylation. Acetylation of ε-amino groups of lysine residues in histones tails is dynamically regulated by the antagonistic activities of histone deacetylases (HDACs) and histone acetyltransferases (HATs). Deacetylation of lysine (K) residues results in chromatin compaction and transcriptional repression, and conversely, histone acetylation is correlated with transcriptional activation. The role of histone methylation is more complex, and governed by opposing functions of histone methyltransferases (HMTs) and demethylases. Methylation can occur on lysines resulting in mono- di- or tri-methylation, whereas methylation of arginines results in mono or di-methylation (symmetric or asymmetric). Such modifications are linked to both transcriptional repression as well as activation. Lysine residues that undergo methylation include H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20. Methylation of H3K9 (H3K9me), H3K27 and H4K20 is associated with transcriptional repression, whereas H3K4 and H3K36 are associated with active transcription. On the other hand methylation of arginine-8 and -17 (H3R17me and H3R8me) is associated with transcriptional activation.
In undifferentiated myoblasts, deacetylation and methylation of histones are apparent on muscle promoters (Fig. 2) that are mediated by HDACs and histone methyltransferases (HMTs). All three classes of HDACs repress differentiation. HDAC1, which belongs to class I HDAC subfamily, is recruited to muscle promoters through its association with MyoD in myoblasts, and results in deacetylation of histones on late muscle promoters MCK and MHC. HDAC4 and HDAC5 (Class II HDACs) inhibit MEF2 factors blocking both early and late muscle differentiation genes.18 SirT1, a class III HDAC, whose activity relies on the cofactor NAD+, also forms a complex with MyoD and p300/CBP associated factor (P/CAF) in myoblasts inhibiting MyoD activity.19
Two families of HMTs are involved in inhibition of muscle gene expression in myoblasts. The Su(var)3–9 family SET domain containing proteins G9a and Suv39h1 have been shown to regulate skeletal myogenesis.20,21 G9a is present in euchromatin and mediates mono- and di-methylation of H3K9, whereas Suv39h1 mediates H3K9 tri-methylation and is enriched in heterochromatin. Both G9a and Suv39h1 are expressed in myoblasts, and interact with MyoD to block its activity and muscle differentiation. H3K9me serves as a platform for recruitment of heterochromatin protein 1 (HP1) that leads to stable silencing via formation of a heterochromatic structure. Whether G9a and Suv39h1 serve to functionally maintain an undifferentiated state by impacting H3K9me2 and H3K9me3 marks on distinct promoters, or act in sequence to mediate gene repression and silencing remain to be investigated. The Polycomb repressor complex (PRC2) contains three core subunits - Enhancer of zeste homolog 2 (Ezh2); Embryonic ectoderm Development (EED), and Suppressor of Zeste 12 (SUZ12). Ezh2, the catalytic subunit of PRC2, transfers a methyl moiety from S-adenosyl methionine to H3K27 resulting in H3K27me3 and thereby represses gene expression. Ezh2 is recruited on late myogenic promoters through the transcription factor YY1 and is found in complexes with HDAC1.22
During differentiation repressive methylation marks are replaced by transcriptionally permissive H3K4me3 signature on the myogenin promoter. Several mechanisms are involved in removal of repressive deacetylation and inhibitory methylation marks from muscle promoters. The expression of many co-repressors including HDAC1, G9a, Suv39h1, and Ezh2 are downregulated, HDAC4/5 are shuttled out of the nucleus by a CaMK-dependent mechanism, and the NAD+/NADH+ ratio decreases, reducing the inhibitory impact of SirT1.18-22 Growing evidence indicates that histone demethylases also play an active role in removing methylation marks from muscle promoters. The Jumonji-domain 2 (JMJD2) histone demethylase family has been shown to be involved in myogenesis. An isoform of JMJD2A (∆N-JMJD2A) which is expressed during differentiation, is recruited to the myogenin promoter and catalyzes demethylation of H3K9me2/3.23 Intriguingly, ∆N-JMJD2A lacks a demethylation domain suggesting that it may function by forming complexes with additional demethylases that have catalytic activity. UTX mediates removal of H3K27me3 marks, and is recruited to myogenin and MCK promoters.24 The second family of demethylases is Lysine specific demethylase 1 (LSD1), which removes mono- and di-methylated residues from H3K9 and H3K4 that are associated with repression and activation respectively. LSD1 interacts with both MyoD and MEF2, and in its absence, H3K9me2 is retained on the myogenin promoter suggesting that its pro-myogenic function is largely due to de-methylation of H3K9.25
In addition to removal of co-repressors, several co-activators actively reprogram muscle promoters during differentiation. The HMT Set7/9 plays a pro-myogenic role.26 Set7/9 competes with Suv39h1 for association with MyoD, and antagonizes Suv39h1-dependent H3K9me3. Moreover, Set7/9 directly associates with MyoD and increases H3K4me on myogenic promoters. Arginine methyltransferases Carm1/Prmt4 and Prmt5 that mediate H3R17me2 and H3R8me2 respectively also enhance myogenesis. Prmt5 interacts with MyoD and is important for activation of the myogenin promoter, whereas Carm1 is recruited by MEF2 on the MCK promoter and facilitates binding of the Brg1 ATP-dependent chromatin-remodeling enzyme. Brg1, the ATPase subunit of SWI/SNF, is required for nucleosome remodeling and correlates with the presence of Pol II holoenzyme on muscle promoters. SWI/SNF recruitment is dependent on p38 activity, thus linking extracellular signals to nucleosome remodelling.16 Recruitment of the Ash2L methyltransferase complex is facilitated by p38 MAPK mediated phosphorylation of MEF2D and establishes H3K4me3 marks on muscle specific promoters.27 Interestingly, Ezh1, a paralog of Ezh2, binds actively transcribed genes with regions of elevated H3K4me3 and is required for RNA Polymerase II occupancy.28 Two distinct HATs are recruited at the onset of differentiation that extensively reprogram muscle promoters and are essential for MyoD activation. p300/CBP directly associates with MyoD and acetylates histones H3 and H4. P/CAF recruitment on the other hand is mediated by p300/CBP, and is essential for acetylation of MyoD that stimulates its transcriptional activity and myogenic potential.29
Old Dogs, New Tricks: Transcription Factor Methylation
Our current understanding of epigenetic control of skeletal myogenesis has largely focused on histones which serve as targets for a variety of reversible post-translational modifications that modulate nucleosome structure and gene transcription. There is growing evidence however, that similar to histones, many transcription factors and other non-histone proteins undergo post-translational modifications such as acetylation and methylation that are mediated by chromatin modifiers. Direct acetylation and deacetylation of transcription factors has been shown to have positive and negative consequences on transcription. P/CAF-dependent MyoD acetylation is one example where such evidence is apparent in skeletal muscle.29 During differentiation, P/CAF directly acetylates MyoD at K99, K102 and K104 (Fig. 1). Acetylation of MyoD at these three lysine residues is key to its activation and ability to induce the expression of target genes during differentiation. Interestingly, mutation of these sites impairs muscle regeneration in mice demonstrating the importance of non-histone protein acetylation in vivo.30 Despite its significance, it unclear why acetylation of MyoD occurs primarily upon induction of differentiation, as P/CAF is expressed even in myoblasts.29,31 Several possibilities may account for it. In myoblasts, MyoD interacts with HDAC1, and may exist in a ‘deacetylated’ state. Consistent with this notion, HDAC1 can deacetylate MyoD in vitro. However, whether HDAC1 prevents P/CAF from associating with MyoD in myoblasts and thereby blocks MyoD acetylation, or whether MyoD undergoes additional post-translational modifications that prevent it from being a substrate for P/CAF has not been clarified. Our recent study, which provides evidence that MyoD is methylated in myoblasts provides some insights.22 The lysine methyltransferase G9a is expressed in myoblasts and in addition to mediating H3K9me2 on the myogenin promoter, also methylates MyoD at K104. Methylation of MyoD represses its transcriptional activity and ability to activate the myogenin promoter. Conversely, mutation of K104 to arginine (K104R) enhances MyoD-dependent activation of myogenin and muscle differentiation. While loss of function and gain of function experiments show that G9a modulates H3K9me2 mark on muscle promoters, MyoD methylation is critical in G9a-dependent inhibition of differentiation. Since the same lysine residue can be either acetylated or methylated, it is possible that methylation of MyoD prevents its P/CAF-dependent acetylation. K104 is conserved through evolution suggesting that it may be functionally relevant. Moreover, this site is also conserved through all MRFs (Fig. 1). Whether all MRFs are indeed methylated by G9a, and the consequences of such methylation in the function of MRFs remain to be investigated.
Similar to these findings, the transcription factor GATA4 was also recently reported to be methylated by the chromatin modifier PRC2 in fetal heart.32 Methylation at K299 blocks GATA4 transcriptional activity by preventing its association with and acetylation by p300. Interestingly, repression of myosin heavy chain α (Myh6) by PRC2 in fetal heart is dependent on GATA4 methylation, and independent of H3K27me3 which is the predominant activity of PRC2. MyoD and GATA4 methylation by G9a and Ezh2 respectively demonstrate an unconventional mechanism by which chromatin modifiers regulate differentiation programs by methylation of transcription factors as opposed to histones. More importantly, they demonstrate that such post-translational modifications are functionally relevant in repression of transcription factor activity and consequently differentiation of skeletal and cardiac muscles.
While largely understudied, it is likely that modulation of transcription factor activity by chromatin modifiers is a widespread phenomenon. Previous studies have shown that class II HDACs exhibit limited activity toward acetylated histones.33 Thus it remains to be clarified whether deacetylation of histones central to regulation of gene expression and differentiation by HDAC4/5, or alternatively whether non-histone proteins are the key targets. Similarly, the impact of HDAC1 in myogenesis may occur not only through deacetylation of histones, but perhaps MyoD, and other non-histone targets as well.
Cooperativity between different chromatin modifiers is likely the consequence of mutually exclusive modifications. Thus mechanisms must exist to ensure regulated access of chromatin modifiers to their substrates. While several examples of such regulated interactions and their functional consequences exist for histones, comparatively little is known on how mutually exclusive post-translational modifications of transcription factors are regulated. Future studies will shed light on how specific modifications are erased to establish others, and whether a methylation is a widespread regulatory switch to control transcription factor activity in various cellular differentiation programs.
Acknowledgments
Work in the authors’ laboratory is supported by the National Medical Research Council and the A*STAR Singapore Stem Cell Consortium.
Glossary
Abbreviations:
- CaMK
Ca2+/calmodulin-dependent protein kinases
- EED
Embryonic ectoderm Development
- Ezh1
Enhancer of zeste homolog 1
- Ezh2
Enhancer of zeste homolog 2
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMT
histone methyltransferase
- Id
Inhibitor of Differentiation
- JMJD2
Jumonji-domain 2
- LSD1
Lysine specific demethylase 1
- MCK
Muscle creatine kinase
- MEF2
myocyte enhancer factor 2
- MHC
Myosin heavy chain
- MRF
Muscle regulatory factors
- MyoR
Myogenic Repressor
- P/CAF
p300/CBP associated factor
- Pol II
RNA polymerase II
- PRC2
Polycomb repressor complex 2
- PRMT4/5
protein arginine methyltransferase 4/5
- SIRT1
silent mating type information regulation 2 homolog 1
- SRC
steroid receptor coactivator-1
- Su(var)3-9
Suppressor of variegation 3-9
- SUZ12
Suppressor of Zeste 12
- SWI/SNF
SWItch/Sucrose NonFermentable
- UTX
Ubiquitously transcribed tetratricopeptide repeat, X chromosome
- H3K9Ac
acetylated histone H3 at lysine 9
- H3K9me2/3
di- or tri-methylated histone H3 at lysine 9
- H3K27me3
tri-methylated histone H3 at lysine 27
- H3R8me2
di-methylated histone H3 at arginine 8
- H3R17me2
di-methylated histone H3 at arginine 17
- H3K4me
methylated histone H3 at lysine 4
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/20914
References
- 1.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 2.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–56. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 3.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 4.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–8. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 6.Tajbakhsh S, Cossu G. Establishing myogenic identity during somitogenesis. Curr Opin Genet Dev. 1997;7:634–41. doi: 10.1016/S0959-437X(97)80011-1. [DOI] [PubMed] [Google Scholar]
- 7.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–9. doi: 10.1016/0092-8674(93)90621-V. [DOI] [PubMed] [Google Scholar]
- 8.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–6. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 9.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, et al. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–5. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 10.Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–71. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 11.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 12.Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 2004;18:2348–53. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–73. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem. 2004;279:52643–52. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- 15.Yahi H, Philipot O, Guasconi V, Fritsch L, Ait-Si-Ali S. Chromatin modification and muscle differentiation. Expert Opin Ther Targets. 2006;10:923–34. doi: 10.1517/14728222.10.6.923. [DOI] [PubMed] [Google Scholar]
- 16.Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it’s time to exchange! Exp Cell Res. 2010;316:3073–80. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aziz A, Liu QC, Dilworth FJ. Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics. 2010;5:691–5. doi: 10.4161/epi.5.8.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/S0959-437X(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 19.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/S1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 20.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–34. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling BM, Bharathy N, Chung TK, Kok WK, Li S, Tan YH, et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2012;109:841–6. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–38. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verrier L, Escaffit F, Chailleux C, Trouche D, Vandromme M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011;7:e1001390. doi: 10.1371/journal.pgen.1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun. 2010;401:327–32. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, et al. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194:551–65. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–6. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–62. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–34. doi: 10.1016/S1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 30.Duquet A, Polesskaya A, Cuvellier S, Ait-Si-Ali S, Héry P, Pritchard LL, et al. Acetylation is important for MyoD function in adult mice. EMBO Rep. 2006;7:1140–6. doi: 10.1038/sj.embor.7400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–53. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–40. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]