Abstract
The "CTD code" links the combinatorial potential of the modifications found on the Rpb1 C-terminal domain (CTD) to the growing group of CTD binding effectors. The genetic dissection of serine 2 and serine 7 function within the CTD in both budding and fission yeast reveals distinct in vivo requirement.
Keywords: CTD, RNA polymerase II, cell differentiation, phosphorylation
Introduction
During transcription, many aspects of mRNA synthesis are intricately connected by the sequential modifications of the carboxyl-terminal domain (CTD) of the largest subunit of the RNA polymerase II, Rpb1. The structural plasticity of the CTD enables modifications-dependent interactions with various protein complexes required for maturation of the nascent RNA molecule or alterations of the template chromatin. Five of the seven residues found in the tandem heptapeptide repeats (Y1S2P3T4S5P6S7) forming the CTD can be phosphorylated or glycosylated, and the proline residues can be isomerized between the cis and trans conformations. Some modifications are stage specific during transcription and the phosphorylation status of serine 2 (S2), serine 5 (S5) and serine 7 (S7) residues specify the position of the polymerase along the transcription unit. A variety of reviews of that process are available.1-9
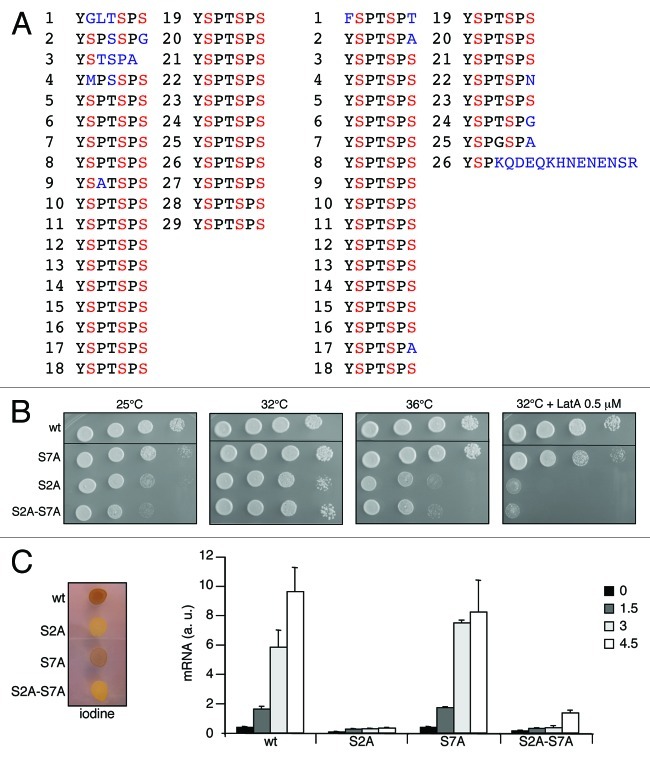
In all the species where this was analyzed, cell viability is contingent on a minimal number of repeats, usually about one third of the native number (native length is 29 repeats in fission yeast, 26 repeats in budding yeast, Figure 1A).
Figure 1. The specific requirements of fission yeast S2 are independent of S7. (A) The CTD sequence in fission yeast (left) and budding yeast (right). Red residues indicate S2, S5 and S7. Substituted residues or divergent regions are marked in blue. (B) Spot dilution assay of wt, rpb1 S7A, rpb1 S2A and rpb1 S2A-S7A strains grown on rich media (YES) at 25°C, 32°C and 36°C, or supplemented with 0.5 μM Latrunculin A (Enzo Life Sciences) at 32°C. (C) Left: Homothallic wt, rpb1 S7A, rpb1 S2A and rpb1 S2A-S7A strains were mixed and plated for 48h on mating medium before iodine staining to reveal sterility. Iodine vapors stain starch present in the S. pombe spores. Right: Relative quantification (RQ) of the ste11 mRNA determined by quantitative RT-PCR using the ΔΔct method in a wt, rpb1 S7A, rpb1 S2A and rpb1 S2A-S7A strains at indicated times (hours) during nitrogen starvation. a.u.: arbitrary units.
The fission yeast Schizosaccharomyces pombe is the only organism where a genetic dissection covering the entire heptad repeat is available,10,11 which led to three important conclusions. First, four residues out of seven cannot be replaced by alanine: Y1, P3, S5, P6, and the requirement of S5 is circumvented by the artificial tethering of mRNA capping enzymes to the CTD.11 Second, S2 is nearly completely dispensable for vegetative growth but plays a key role in sexual differentiation through transcriptional activation of ste11, which encodes the master regulator of the differentiation switch.10 Finally, it was reported that replacing S7 erases the gene-specific requirement of S2.11
In the budding yeast Saccharomyces cerevisiae, pioneer work from the Corden laboratory reported that the substitution of either S2 or S5 was lethal,12 which was confirmed by others.5
It is noteworthy that most of theses studies were based on partially truncated, often plasmid born, mutants.
Here we present and compare the analyses of full length, authentic CTD phosphorylation mutants integrated at the endogenous locus in both budding yeast and fission yeast. These experiments challenge previous conclusions about the universality of CTD function and requirement.
The Gene-Specific Requirement of Fission Yeast S2 for Sexual Differentiation is Independent of S7
We previously constructed and integrated at the rpb1 locus a full-length mutant with the exact sequence of the fission yeast CTD except that all S2 that were replaced by alanine (A). The resulting S2A mutant showed only very mild defects during vegetative growth but was nearly sterile due to its inability to induce the expression of ste11 in response to nitrogen starvation. The ste11 gene encodes the master transcription factor required to switch fission yeast cells deprived of nitrogen to sexual differentiation. The phenotypes were identical when the S2 kinase Lsk1 was inactivated.10 These results were later confirmed by three independent studies.11,13-15 Using a truncated version of the combined S2A and S7A mutants (14 repeats), Schwer and Shuman reported that replacing S7 by A7 could bypass the requirement of S2 phosphorylation for sexual differentiation. This conclusion was supported by an increased expression of ste11 at a late time point (5 h) during the induction by nitrogen starvation.11
We have constructed and integrated full length, authentic fission yeast S7A and S2A-S7A mutants and analyzed their phenotypes. As reported before, the S2A mutant is thermosensitive at 25°C and 36°C, is sensitive to LatA (Latrunculin A, a drug destabilizing actin), and exhibits very low mating and meiosis efficiency. As shown in Figure 1B-C, the S7A mutant was nearly indistinguishable from the wild type in all tested conditions. Aside from a very subtle sensitivity to LatA and cold temperature, the induction of ste11 during nitrogen starvation was normal, which resulted in efficient mating, as witnessed by positive iodine staining. In contrast, the double S2A-S7A mutant behaved similarly to the single S2A and no suppression of either the thermosensitive phenotype, the sensitivity to LatA, and the defect in ste11 induction were observed (Fig. 1B and C). Therefore, we conclude that S2 phosphorylation plays a specific role in the biological processes underlying these phenotypes, and that this role is independent of S7 phosphorylation. These data also confirm that neither S2 nor S7 phosphorylation play an essential role in vegetative conditions.
A Budding Yeast S2A Mutant is Viable but Synthetic Lethal With S7A
The very first genetic analysis of CTD function by cis mutations was reported in 1995.12 Plasmid shuffling experiments showed that a truncated version of the S2A mutant (harboring 18 repeats) was unable to sustain growth and it was concluded that S2 phosphorylation is essential in that species. This was later confirmed using a more severely truncated version,5 implying that S2 phosphorylation is essential, which fits well with the well described links between S2 phosphorylation and very important cellular processes.1,7 These data notwithstanding, it is unexpected that the gene encoding the main S2 kinase, Ctk1, is not essential in budding yeast, albeit its deletion impairs growth.16 This paradox is resolved by the fact that evidences exist that the Bur1 kinase also participates in S2 phosphorylation17 and that the absence of a particular CTD kinase could allow opportunistic kinases to fulfill its job.18 Nevertheless, the level of S2 phosphorylation is markedly reduced in the absence of CTK1.
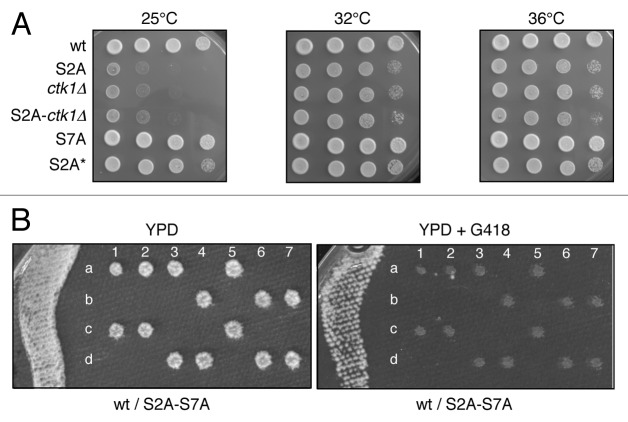
Building on the data obtained in fission yeast, we generated a full length, authentic S2A mutant and integrated it at the locus in a budding yeast diploid. Tetrad analysis revealed that the S2A mutant was viable but slowly growing. The growth rate was very similar to what is observed in the absence of CTK1 or in a double ctk1 S2A mutant (Fig. 2A). These data reinforce the conclusion that Ctk1 is the main S2 kinase in vivo. In contrast, the S7A mutant grew similarly to the wild type. While in the process of generating the S2A mutant, we obtained a S2A mutant allele that retained the first four wild type repeats, instead of being a complete S2A version. As shown in Figure 2A, this mutant (S2A*) is barely affected for growth, which confirms that besides phosphorylation, the overall length of the CTD is also important for the function.6
Figure 2. A budding yeast CTD S2A mutant is viable but synthetic lethal with S7A. (A) Spot dilution assay of wt, rpb1 S2A, ctk1::kanR, ctk1::kanR rpb1 S2A, rpb1 S7A strains and a rpb1 S2A mutant strain harboring the first four wild type copies of the CTD heptad (S2A*) grown on rich media (YPD) at 25°C, 32°C and 36°C. (B) Tetrad analysis of a sporulated diploid RPB1 / rpb1 S2A-S7A. Spores were germinated on YPD for two days and replicated overnight on YPD supplemented with G418. The four spores dissected from a tetrad are indicated a, b, c, d. The tetrads are numbered from 1 to 7.
In strong contrast to fission yeast, the double S2A-S7A mutant was lethal, as shown by tetrad analysis (Fig. 2B). The simplest explanation of this phenomenon is that an essential function of the CTD is redundantly shared between S2 and S7 phosphorylation in budding yeast.
Discussion
The comparison of fission yeast and budding yeast has often been a strong lever in the understanding of complex biological processes, the evolutionary distance between these species highlighting key principles from species-specific details. The genetic dissection of the requirement of phospho-acceptors within the CTD supports that only S5 phosphorylation is an essential feature of the transcription cycle. The elegant experiment reported by Schwer and Shuman strongly supports that the recruitment of capping enzymes to the CTD soon after transcription initiation is the main cause underlying the essential presence of S5. In contrast, neither S2 nor S7 phosphorylations are absolutely required for viability. While the phosphorylation of S7 is likely to be important only for specific processes in the two yeasts, a much stronger growth defect arises from the absence of S2 in S. cerevisiae, suggesting that S2 phosphorylation affects much more broadly transcription in budding yeast than in fission yeast. The discrepancy between the proposed requirement of S2 phosphorylation to recruit essential component of the transcription machinery and the fact that the S2A mutant is viable may be explained by the redundant use of S7 phosphorylation in some cases, as suggested by the co-lethality of the double S2A-S7A mutant, or by CTD independent recruitment mechanisms. This possibility was evoked recently for Spt6 and Pcf11.19
Another issue raised by the present study is the behavior of truncated mutants. Even though these mutants behave as wild type in vegetative growth conditions, they also showed defects in both fission yeast and budding yeast in challenging conditions. This is clearly exemplified by the viability of the full length S2A mutant in budding yeast, compared with truncated versions. Interestingly, a mutation of the mediator subunit Med13 (sca1::hisG) in budding yeast could efficiently suppress both the lethality of the truncated S2A mutant or CTD truncations.20 In conclusion, and as underlined previously,6 the CTD length is a key parameter of CTD function, together with CTD modifications.
Finally, our data reinforce the idea that fission yeast requires S2 phosphorylation in very specific context, which may indicate that it evolved a somewhat more flexible CTD code allowing tighter gene specific regulation during the expression of particular transcription programs.
The understanding of the biological roles of the CTD code calls for additional work and the strains described in this study are available to help with their implementation.
Material and Methods
General methods
General yeast methods were performed as described.21-23 Media used: YES (fission yeast rich medium), EMM (fission yeast minimal medium), EMM-N (fission yeast minimal medium lacking a nitrogen source), YPD (budding yeast rich medium), SPO (budding yeast sporulation medium).
Generation and integration of CTD mutants
In order to generate the exact fission yeast and budding yeast CTD sequence containing either serine 2, serine 7 or both mutated to alanine, long oligonucleotides of about 650 bp, flanked by restriction sites (BamHI-AscI), harboring the mutations were synthesized (Integrated DNA Technologies) and cloned in pIDT-smart. They were then moved to pFA6a-3HA-kanMX624 in order to replace the HA tag by the mutated CTD. The resulting CTD-kanMX6 cassettes were amplified with primers harboring homology to the rpb1 locus and transformed into a diploid strain to allow integration with the help of the KanR selection marker. In each case, five to ten tetrads were dissected and the proper integration and sequence checked. All sequences are available upon request.
Quantitative analysis of ste11 expression
RT-Q-PCR were performed as described.10
Acknowledgments
We thank the GEMO laboratory for discussions and Isabelle Georis for help with budding yeast tetrad dissection. This work was supported by the grants FRFC 2.4510.10, Credit aux chercheurs 1.5.013.09 and MIS F.4523.11 to D.H. D.H. is a FNRS Research Associate.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21066
References
- 1.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–6. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009;10:1213–9. doi: 10.1038/embor.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–6. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho EJ. RNA polymerase II carboxy-terminal domain with multiple connections. Exp Mol Med. 2007;39:247–54. doi: 10.1038/emm.2007.28. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Greenleaf AL, Stiller JW. The essential sequence elements required for RNAP II carboxyl-terminal domain function in yeast and their evolutionary conservation. Mol Biol Evol. 2008;25:719–27. doi: 10.1093/molbev/msn017. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol. 2010;27:2628–41. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–91. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 9.Drogat J, Hermand D. Gene-specific requirement of RNA polymerase II CTD phosphorylation. Mol Microbiol. 2012;84:995–1004. doi: 10.1111/j.1365-2958.2012.08071.x. [DOI] [PubMed] [Google Scholar]
- 10.Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, et al. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr Biol. 2010;20:1053–64. doi: 10.1016/j.cub.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 11.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–8. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–33. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karagiannis J, Bimbó A, Rajagopalan S, Liu J, Balasubramanian MK. The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell. 2005;16:358–71. doi: 10.1091/mbc.E04-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saberianfar R, Cunningham-Dunlop S, Karagiannis J. Global gene expression analysis of fission yeast mutants impaired in Ser-2 phosphorylation of the RNA pol II carboxy terminal domain. PLoS One. 2011;6:e24694. doi: 10.1371/journal.pone.0024694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sukegawa Y, Yamashita A, Yamamoto M. The fission yeast stress-responsive MAPK pathway promotes meiosis via the phosphorylation of Pol II CTD in response to environmental and feedback cues. PLoS Genet. 2011;7:e1002387. doi: 10.1371/journal.pgen.1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–67. [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–62. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bataille AR, Jeronimo C, Jacques PÉ, Laramée L, Fortin MÈ, Forest A, et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–70. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 20.Yuryev A, Corden JL. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–71. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bamps S, Westerling T, Pihlak A, Tafforeau L, Vandenhaute J, Mäkelä TP, et al. Mcs2 and a novel CAK subunit Pmh1 associate with Skp1 in fission yeast. Biochem Biophys Res Commun. 2004;325:1424–32. doi: 10.1016/j.bbrc.2004.10.190. [DOI] [PubMed] [Google Scholar]
- 22.Fersht N, Hermand D, Hayles J, Nurse P. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 2007;35:5323–37. doi: 10.1093/nar/gkm527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermand D, Nurse P. Cdc18 enforces the S-phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol Cell. 2007;6:553–63. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–51. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]