Abstract
The ubiquitin proteasome system plays an important role in transcription. Monoubiquitination of activators is believed to aid their function, while the 26S proteasomal degradation of repressors is believed to restrict their function. What remains controversial is the question of whether the degradation of activators aids or restricts their function.
Keywords: Gal4, conditional protein degradation, transcription, ubiquitin
Introduction
Ubiquitin is a protein found ubiquitously in all eukaryotic cells. A 76 amino acid protein, it was first characterized as a proteolytic signal. Classically, a polyubiquitin chain of four or more molecules is best recognized as a signal for the degradation of proteins by the 26S proteosome.1 While this is mainly a “housekeeping” method for the cell to regulate the stability of proteins, it is becoming increasingly clear that the 26S proteasome is highly abundant in the nucleus and that the degradation of transcription factors is a means by which the cell regulates transcription.2 While there is no dispute that transcription factors are degraded and can display markedly different stability profiles during transcriptional activation compared with their basal states, the question remains of whether proteosomal degradation of transcription factors is a necessary event for the proper activation of transcription.3,4 Further investigation revealed that the ubiquitin moiety itself can be a signaling molecule outside of the context of the ubiquitin proteasome system, and monoubiquitination has been associated with various cellular processes such as endocytosis and DNA repair.2 Components of the 26S proteasome itself have also been implicated in transcriptional regulation such as the 19S ATPases.5
The Role of Monoubiquitination Signaling in Transcriptional Regulation
Aside from the various other roles monoubiquitination plays in the cell, it also has a significant part to play in the regulation of transcription. This was first demonstrated in a series of experiments performed in yeast where the deletion of an ubiquitin ligase, Met30, could severely diminish the activity of the viral protein 16 (VP16) transactivation domain.6 Consistently, while a fusion of the DNA-binding domain LexA and VP16 is capable of recruiting to the promoter positive transcription elongation factors, this recruitment can be greatly enhanced by the monoubiquitination of LexA-VP16.7 In addition, the monoubiquitination of the transcriptional activator Gal4 was suggested to aid its function by protecting it from the promoter-stripping activity of the proteasomal 19S ATPases.8 For the repressor MATα2, on the other hand, monoubiquitination was reported to lead to its removal from the promoter by the ATPase Cdc48,9 indicating that the monoubiquitination of a transcription factor can have opposite effects on its promoter occupancy.
Outside of yeast, a positive effect of the monoubiquitination of transcriptional activators can also be seen for transcription factors involved with cellular stress responses, where the transcriptional activity of p53 (ref. 10) and Foxhead box O4 (FOXO4) is increased upon monoubiquitination. Furthermore, monoubiquitination is known to enhance the stability of p53 and is important in the nuclear translocation of FOXO4, which implies that the ubiquitin moiety is crucial for various aspects of transcription factor function.11 Upon stimulation by hormones to induce positive cell-cycle progression, apoptosis antagonizing transcription factor (AATF) interacts with the E2 ubiquitin conjugating enzyme tumor susceptibility gene protein (TSG101), which results in its monoubiquitination. The monoubiquitination of this cell cycle regulator is known to increase both its transcriptional activity and stability much in the same manner as p53.12 The most famous and often cited example of how monoubiquitination regulates the activity of a transcription factor is found in the immune system, NF-κB, where a series of phosphorylation and ubiquitination events ultimately result in the degradation of its repressor IκBα and subsequent activation of NF-κB.13
The Proteolytic Role of the Proteasome in the Repression of Transcription
The first transcription factor shown to be degraded by the UPS, MATα2, is a repressor.14 MATα2, which represses the a-specific genes in α cells, has a short half-life of only 5 min. The protein degradation of MATα2 is not conditional but permanent, and it is believed to be necessary in order to restrict repressor function and to allow α cells to switch their mating type. MATα2 is stable in α cells lacking the ubiquitin-conjugating enzymes Ubc4/6, and ubc4/6 mutant α cells are deficient for mating type switching,15 which is consistent with this hypothesis. It could have been argued that a Ubc4/6 target other than MATα2 is the true cause for the switching defect; however, overexpression of MATα2 from a strong promoter also interferes with mating type switching,15 confirming the hypothesis that mating type switching requires UPS-mediated MATα2 restriction. The first example for the conditional degradation of a transcriptional repressor was provided by Cup9, whose protein degradation is stimulated by the presence of peptides in the culture media.16 Cup9 represses the PTR2 gene, and Ptr2 imports peptides, which bind and stimulate Ubr1, the E3 ubiquitin ligase for Cup9. This means that peptides accelerate their own uptake by stimulating the degradation of the transcriptional repressor of the gene encoding their importer. In the absence of Cup9, PTR2 is expressed constitutively, while in the absence of Ubr1, Cup9 remains stable and PTR2 remains repressed in the presence of peptides, confirming the hypothesis that PTR2 activation requires UPS-mediated Cup9 restriction.
Another example for the conditional degradation of a transcriptional repressor is Mig2, which is stable in cells grown with glucose and rapidly degraded upon galactose induction.17 Contrary to Mig1, which is not controlled by the UPS but by subcellular localization,18 expression of Mig2 from a multi-copy vector does not inhibit transcriptional activation of the GAL genes.17 Furthermore, the deletion of the MIG2 gene does not result in the derepression of the GAL genes in glucose-grown cells, which lead to the suggestion that Mig2 has no role in the glucose repression of the GAL genes.19 However, Mig2 expressed even from a multi-copy vector is still completely degraded in galactose-induced cells,17 indicating that Mig2 does not display the MIG (Multi-copy Inhibition of GAL gene expression) phenotype because the Mig2 protein is never actually overexpressed in galactose-induced cells. Furthermore, the simultaneous deletion of two PEST sequences from Mig2 leads to a galactose-stable Mig2 deletion derivative, and the overexpression of this galactose-stable Mig2 deletion derivative inhibits GAL gene activation just like the overexpression of Mig1.17 These results indicate that the role of Mig2 in the glucose repression of the GAL genes is redundant and fully compensated for by other repressors of the GAL genes like Mig1 and Gal80. Galactose induction of the GAL genes, on the other hand, still requires the UPS-mediated restriction of Mig2.
The Proteolytic Role of the Proteasome in the Activation of Transcription
Logically restricting the activation of transcription by simply removing the transcriptional activators themselves through degradation by the 26S proteasome is a good proposition for the cell. Such a system would require far less resources than synthesizing several other proteins to sequester transcriptional activators or to transcribe wholesale transcriptional repressors each time the cell needs to reduce the transcription of a given gene. The caveat is that several transcription factors do exactly that: regulate transcription through interfering with another transcription factor, but generally it is more economical to use the existing 26S proteasome for such purposes. This principle is demonstrated in the function of the cell cycle regulators. Cyclins, cyclin-dependent kinases and other cell cycle regulators are targeted for degradation when they are not needed and the ubiquitin proteasome system no longer degrades these proteins when there is a need for an increase in their cellular levels.20 Another piece of evidence for this mode of control is the regulation of β-catenin by Wnt. Glycogen synthase kinase-3β (GSK-3β) phosphorylates β-catenin which is subsequently ubiquitinated and then degraded by the 26S proteasome in the absence of Wnt. In the presence of Wnt, GSK-3β is rendered inactive by the binding of Wnt to its cell surface receptors. Under these conditions β-catenin remains stable and fully capable of activating downstream genes. Thus the cell is able to maintain a low basal level of transcription for genes controlled by β-catenin but can potentially induce a rapid spike in expression if the need arises.21
The Active Turnover of Transcription Factors
All experimental evidence shows that transcriptional repressors are restricted by the UPS and that the failure to degrade repressors results in repression under inappropriate conditions. Transcriptional activators are restricted by the UPS as well. The F-box protein Grr1, for example, targets Gal4, the transcriptional activator of the GAL genes, for UPS-mediated protein degradation in S. cerevisiae cells grown under non-activating conditions (with raffinose). In raffinose-grown wild-type cells, Gal4 is degraded and the GAL genes are not expressed, while in raffinose-grown cells lacking Grr1, Gal4 is stable and activates transcription of the GAL genes, confirming that the UPS restricts Gal4 and that the failure to degrade the activator Gal4 results in activation of the GAL genes under inappropriate conditions.22 In the same article, however, the authors (Tansey group) also proposed that the UPS-mediated degradation of Gal4 under activating conditions is actually required for its function. They drew this surprising conclusion from another correlation. The F-box protein Mdm30/Dsg1 targets Gal4 for UPS-mediated protein degradation in cells grown under activating conditions (with galactose). In galactose-grown wild-type cells, Gal4 is degraded and the GAL genes are expressed, while in galactose-grown cells lacking Mdm30/Dsg1, Gal4 is stable and fails to activate transcription of the GAL genes.22 The daring hypothesis that Gal4 requires UPS-mediated degradation in order to function was supported by the Deshaies group, which reported that the proteasome inhibitor MG132 prevents galactose induction of the GAL genes.23 One possible explanation is that promoter-bound activators misfold over time and that the periodic clearance of activators from a promoter is essential for high-level gene expression. An in vivo competition assay performed by the Johnston and Kodadek groups, on the other hand, showed that Gal4 remains stably bound to the GAL1 promoter in galactose-induced cells.3 The authors thus concluded that proteolytic turnover of the Gal4 transcription factor cannot be required for its function in vivo. The in vivo competition assay was subsequently challenged by the Tansey and Deshaies groups, who claimed that the assay was unsuitable, and that promoter-bound, active Gal4 was indeed susceptible to competition in vivo.4 However, the real question of whether the proteolytic degradation of Gal4 was indeed required for its activator function remained unanswered. The most obvious possibility that an Mdm30/Dsg1 target other than Gal4 was the true cause for the galactose utilization defect of cells lacking Mdm30 and of cells treated with MG132 had not been pursued. By now, two groups (including our own) have shown that the additional deletion of the gene encoding the inhibitor Gal80 restores transcriptional activation of the GAL genes in cells lacking Mdm30/Dsg1.24,25 This demonstrates that any effect Mdm30/Dsg1 has on Gal4 is not required for its activation function once Gal80 is removed. We went on to show that Mdm30/Dsg1 indeed targets Gal80, which is stable in cells grown with glucose, for galactose-induced protein degradation. Furthermore, galactose-stable N-terminal deletion derivatives of Gal80 prevent galactose induction of the GAL1 gene, confirming that galactose induction requires UPS-mediated restriction of the repressor Gal80 rather than UPS-mediated stimulation of the activator Gal4.25
The Ubiquitin Clock
There is, however, a clear overlap between activation domains and degradation signals, and most transcriptional activators are unstable even under activating conditions.26 Many models have been proposed to explain why transcriptionally competent activators are degraded.27,28 The same question could have been asked for MATα2, which is a transcriptionally competent repressor that is constantly degraded. The most likely explanation is that the cell simply restricts the life span of its transcription factors in order to be able to quickly respond to changes in the environment or in developmental programs. The ubiquitin clock and timer models take further into account that monoubiquitination aids transcriptional activators like Gal4 or Src3,8,29 which means that those activators will occupy their respective target promoters and activate transcription only for as long as it takes to extend the ubiquitin chain from one to four, following which the activator is degraded by the 26S proteasome (Fig. 1). In both cases, kinases have been implicated in the ubiquitination: the Mediator component Srb10/Cdk8 for Gal4 and GSK3 for Src3.22,29
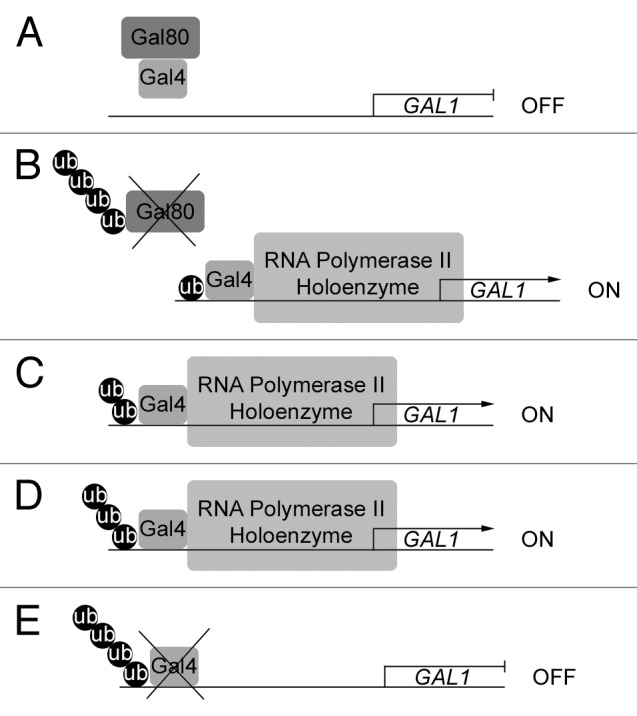
Figure 1. The ubiquitin timer model for Gal4. (A) In cells grown under non-inducing conditions, Gal80 binds Gal4 and blocks its activation domain. The GAL genes are off. (B) Upon galactose induction, SCFMdm30 monoubiquitinates Gal4, which stabilizes its binding to the GAL gene promoter, as ubiquitinated Gal4 is protected from the promoter-stripping activity of the 19S subunit of the proteasome. SCFMdm30 further polyubiquitinates Gal80, leading to its degradation by the 26S proteasome. This frees Gal4’s activation domain and allows it to recruit the RNA Polymerase II holoenzyme. The GAL genes are switched on. (C) SCFMdm30 adds ubiquitin to the ubiquitin moiety of monoubiquitinated Gal4, which continues to activate transcription of the GAL genes. (D) SCFMdm30 extends the ubiquitin chain to three, while Gal4 continues to activate transcription of the GAL genes. (E) Once SCFMdm30 has added a fourth ubiquitin moiety, polyubiquitinated Gal4 is degraded by the 26S proteasome and the GAL genes are switched off.
Another key component is that the ubiquitin clock model also accounts for the relative activities of the ubiquitin ligases, ubiquitin elongases and deubiquitinating enzymes (DUBs) unique to each promoter complex as a factor in the stability of a transcriptional activator. For example, DUBs can significantly impact the stability of a transcription factor through removal of ubiquitin moieties from the polyubiquitin chain. Thus, a long-lived complex simply possesses a highly active DUB under this model. This model removes the 26S proteasome as the sole determinant of activator stability accounting for cases where proteosomal turnover of activators and transcriptional activity can be easily divorced such as ER-α.30
One protein whose stability is regulated by a DUB is Tbp1, the TATA-binding protein. It was observed that in yeast, transcriptional activation requires protection of Tbp1 by the DUB Ubp3.31 The notion that a DUB exists and specifically targets Tbp1, a crucial component of the transcriptional machinery, has various implications. One of these is that the stability of these proteins is under very precise control by the cell, implying a more deliberate process than simple “recycling.”
Conclusion
As noted, transcription factors are degraded just like all other proteins. The open question remains of whether the degradation of transcriptional activators simply restricts their action, or if it is also necessary for their activation function. A transcriptional defect after the application of a proteasome inhibitor can also be interpreted as the result of a negative feedback loop or misfolded and inactive transcriptional machinery. The loss of activation after removing an ubiquitin ligase or a mutation in a transcription factor that renders it unable to be ubiquitinated could also be a function of a monoubiquitination signaling defect. Although great progress has been made toward understanding how the ubiquitin proteasome system is involved in the regulation of transcription, a great deal of research is needed before a definitive picture emerges.
Glossary
Abbreviations:
- MIG
multi-copy inhibitor of GAL gene expression
- UPS
ubiquitin proteasome system
- VP16
viral protein 16
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21249
References
- 1.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- 2.Varshavsky A. Three decades of studies to understand the functions of the ubiquitin family. Methods Mol Biol. 2012;832:1–11. doi: 10.1007/978-1-61779-474-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–7. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- 4.Collins GA, Lipford JR, Deshaies RJ, Tansey WP. Gal4 turnover and transcription activation. Nature. 2009;461:E7–, discussion E8. doi: 10.1038/nature08406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, Johnston SA. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 2007;21:112–23. doi: 10.1101/gad.1493207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–3. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 7.Kurosu T, Peterlin BM. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr Biol. 2004;14:1112–6. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Archer CT, Kodadek T. The hydrophobic patch of ubiquitin is required to protect transactivator-promoter complexes from destabilization by the proteasomal ATPases. Nucleic Acids Res. 2010;38:789–96. doi: 10.1093/nar/gkp1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–6. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.van der Horst A, de Vries-Smits AMM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 12.Burgdorf S, Leister P, Scheidtmann KH. TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J Biol Chem. 2004;279:17524–34. doi: 10.1074/jbc.M313703200. [DOI] [PubMed] [Google Scholar]
- 13.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[κ]B activity. Annu Rev Immunol. 2000;18:621–63. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 14.Richter-Ruoff B, Wolf DH, Hochstrasser M. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–2. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 15.Laney JD, Hochstrasser M. Ubiquitin-dependent degradation of the yeast Mat(alpha)2 repressor enables a switch in developmental state. Genes Dev. 2003;17:2259–70. doi: 10.1101/gad.1115703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–83. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 17.Lim MK, Siew WL, Zhao J, Tay YC, Ang E, Lehming N. Galactose induction of the GAL1 gene requires conditional degradation of the Mig2 repressor. Biochem J. 2011;435:641–9. doi: 10.1042/BJ20102034. [DOI] [PubMed] [Google Scholar]
- 18.De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–18. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–91. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- 22.Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–99. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–6. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Chen G, Liu W. Alterations in the interaction between GAL4 and GAL80 affect regulation of the yeast GAL regulon mediated by the F box protein Dsg1. Curr Microbiol. 2010;61:210–6. doi: 10.1007/s00284-010-9598-1. [DOI] [PubMed] [Google Scholar]
- 25.Ang K, Ee G, Ang E, Koh E, Siew WL, Chan YM, et al. Mediator acts upstream of the transcriptional activator Gal4. PLoS Biol. 2012;10:e1001290. doi: 10.1371/journal.pbio.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molinari E, Gilman M, Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 1999;18:6439–47. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodadek T, Sikder D, Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–4. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–40. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 30.Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/S1097-2765(03)00090-X. [DOI] [PubMed] [Google Scholar]
- 31.Chew BS, Siew WL, Xiao B, Lehming N. Transcriptional activation requires protection of the TATA-binding protein Tbp1 by the ubiquitin-specific protease Ubp3. Biochem J. 2010;431:391–9. doi: 10.1042/BJ20101152. [DOI] [PubMed] [Google Scholar]


