Abstract
Molecular imaging, the visualization, characterization and measurement of biological processes at the cellular, subcellular level, or even molecular level in living subjects, has rapidly gained importance in the dawning era of personalized medicine. Molecular imaging takes advantage of the traditional diagnostic imaging techniques and introduces molecular imaging probes to determine the expression of indicative molecular markers at different stages of diseases and disorders. As a key component of molecular imaging, molecular imaging probe must be able to specifically reach the target of interest in vivo while retaining long enough to be detected. A desirable molecular imaging probe with clinical translation potential is expected to have unique characteristics. Therefore, design and development of molecular imaging probe is frequently a challenging endeavor for medicinal chemists. This review summarizes the general principles of molecular imaging probe design and some fundamental strategies of molecular imaging probe development with a number of illustrative examples.
Keywords: Molecular imaging, imaging probe design, imaging probe development strategies
INTRODUCTION
Molecular imaging can be defined as the in vivo visualization, characterization and measurement of biological processes at the molecular and cellular levels [1], or more broadly as a technique to directly or indirectly monitor and record the spatiotemporal distribution of molecular or cellular processes for biochemical, biological, diagnostic, or therapeutic application [2]. During the last decades, the field of molecular imaging has grown exponentially. Molecular imaging can provide a whole-body readout in an intact system, which is much more relevant and reliable than in vitro/ex vivo studies; decrease the workload and accelerate the drug development process; provide more statistically accurate results through longitudinal studies which can be performed in the same living subject; facilitate lesion detection in cancer patients and patient stratification; and assess individualized anti-cancer treatment and dose accuracy [3, 4]. The great potential of molecular imaging offers many opportunities in the diagnosis and management of diseases such as cancer, neurological and cardiovascular diseases. With the accomplishment of human genome project, rapid advancements of molecular and cell biology have been achieved, and ever-increasing molecular targets and biomarkers have been identified, especially those aberrantly expressed in tumor malignancy, invasiveness, and metastasis [5, 6]. Non-invasive detection of molecular markers can allow for much earlier diagnosis, earlier treatment, better prognosis, and improved staging and management, which is expected to have a major impact in the personalized medicine era.
To date, a good number of imaging modalities for preclinical research and clinical settings has been developed. The predominant imaging modalities can be generally sorted out into two categories. On the one hand, some of imaging modalities, such as ultrasound and computed tomography (CT), can provide anatomical information which is depended on anatomical changes associated with the disease. On the other hand, some of imaging modalities can offer functional information of the disease and track biochemical processes in vivo. The imaging modalities belonging to the second category include positron emission tomography (PET), single photon emission computed tomography (SPECT), optical bioluminescence/fluorescence imaging, molecular magnetic resonance imaging (mMRI), and magnetic resonance spectroscopy (MRS) [4]. PET and SPECT in addition offer the possibility of quantification of diseases associated biochemical processes. Many hybrid systems that combine two or more modalities are also commercially available and others are currently under development [7–9]. Among the various imaging modalities, some of modalities, such as PET, SPECT, and optical imaging, require the injection of molecular probes in the tested subject in order to acquire the imaging signal, while the others, such as optical imaging and mMRI can follow the disease either through the exogenous molecular probes or through the endogenous molecules [4]. Because of the pivotal role of imaging probes in the realization of the power of molecular imaging, the design and development of the biologically active probe is becoming one of the major subjects of molecular imaging [4, 10–12].
MOLECULAR IMAGING PROBE
Molecular imaging probe is an agent used to visualize, characterize and quantify biological processes in living systems [1, 4]. It can be referred to many other terms, such as tracer, contrast agent, and molecular beacon. A molecular imaging probe typically comprises a signal agent, a targeting moiety, and a linker connecting the targeting moiety and the signal agent (Fig. 1). The signal agent usually produces signal for imaging purpose. The applicability of imaging probe to imaging modality can be determined in terms of the physical property of the label moiety. For example, a PET imaging probe requires a positron-emitting radionuclide as the label whereas a SPECT imaging probe employs a gamma-emitting radionuclide. The signal agent can be radionuclides (PET and SPECT), bioluminescence or fluorescent molecules (optical imaging), magnetic molecules (MRI), and microbubbles (ultrasound). The targeting moiety interacts with a target or a biomarker in a specific biological process. The targeting moiety can be widely considered as any targeting ligand including but not limited to small molecule, peptide, protein, antibody and its fragments, and nanoparticle. The linker used in a molecular imaging probe can couple the targeting moiety with the signal agent, minimize the interaction between the targeting moiety and the signal agent and, most importantly, modify the pharmacokinetics of the imaging probe. It is well-documented that the linker has a profound impact on the biodistribution of the imaging probe [1, 4, 13–15]. Various types of linkers can be used to modulate the pharmacokinetics of the imaging probe. The length, flexibility, hydrophilicity, and charges (cationic, anionic, and neutral) are the key factors to be considered to select an appropriate linker. Because many aspects need to be optimized to obtain the best imaging outcome, in some cases not all three components - the signal agent, the linker, and the targeting moiety are simultaneously present.
Fig. (1).
Schematic representation of molecular imaging probe.
Molecular imaging probes can be treated as a special class of pharmaceuticals. The conventional pharmaceuticals are designed and developed to intervene and alter the biological processes of diseases and lead to a positive outcome of treatment, and at the same time show minimal toxicity in the normal tissues. Therefore, the efficacy and safety are usually two of the most important considerations for a drug. In comparison, molecular imaging probes are designed to detect the diseases non-invasively and reflect the biochemical information of diseases in the format of images and, thus, provide critical means for scientists and clinicians to better identify and understand the diseases. The distinctive utilities of a molecular imaging probe differentiate itself from a conventional drug. Overall, a desirable molecular imaging probe with clinical translation potential is expected to have the following unique characteristics:
High binding affinity to target. It is a prerequisite to achieve ideal accumulation of the imaging probe in the targeted tissues. Molecular imaging generally favors the acquisition of the images at early time after administration of a molecular probe. To obtain high uptake of the imaging probe to the target within limited circulation time frame requires that the imaging probe has binding property of fast on-rate (Kon) and slow off-rate (Koff).
High specificity to target. Non-specific imaging probes do not have a well-defined set of molecular targets, and they are usually used to monitor the downstream and overall effects of diseases such as changes in blood flow and perfusion. In contrast, target-specific molecular imaging probes can interact with particular biomarkers, such as enzyme, receptor, and transporters, which are involved in various biological processes associated with particular cell populations and subcellular compartments. Because the target-specific imaging probes are able to provide information of distinct biological processes at molecular level, they are very useful for understanding the biology of specific disease. In addition, highly specific binding can reduce the non-specific uptake of the imaging probe to other tissues and, in turn, simplify the quantification analysis of imaging outcome.
High sensitivity. To detect the biochemical process of the disease, especially at an early stage, frequently requires spying on the aberrant of a very small amount of targets. The molecular imaging probe, therefore, must be highly sensitive, so that the minimal amount of probe is needed to obtain a good quality image. Additionally, in contrast to a therapeutic drug, a molecular imaging probe needs to reduce the perturbation as well as pharmacological effects to biological systems and processes at a minimal level. Thus, high sensitivity is crucially demanded for a good imaging probe.
High contrast ratio. The conclusions derived from low quality images can be sometimes misleading. High contrast images with high target-to-background or signal-to-noise ratio ensure appropriate interpretation of physiological and pathological conditions of the diseases. A molecular imaging probe with high uptake and slow wash-out in target tissue, and low uptake and fast clearance from normal organs can provide the improved imaging quality.
High stability in vivo. Although only trace amount of imaging probe is normally given to the living subjects, maintenance of the intact structure of an imaging probe is a big challenge because numerous enzymes or proteases present in serum or targeted tissue may degrade the imaging probe. The image information given from the metabolites of the imaging probe undoubtedly complexifies the imaging readout and usually makes the understanding of disease highly vague. The quality of the image as well as the validity of the quantitative analysis of the images heavily depends on the in vivo stability of the imaging probe.
Low immunogenicity and toxicity. The molecular imaging probes are generally administrated in low dose and their pharmacological effects can be negligible. As a special category of pharmaceuticals, however, the biological effects cause by an imaging probe still need to be closely monitored. A molecular imaging probe should have minimal or acceptable level of immunogenicity and toxicity before it can be safely employed in human.
Production and economical feasibility. The low cost and excellent availability of molecular imaging probes are advantageous for their wide distribution and clinical routine use. High cost and difficult production dramatically hinder the application of an imaging probe albeit it may fulfill all other characteristics described above.
As a result, it remains highly challenging for medicinal chemists to design and develop desirable molecular imaging probes with clinical applicability.
MOLECULAR IMAGING PROBE DESIGN
The basic understanding of molecular biology behind the identified disease for imaging is the prerequisites to start design of a molecular imaging probe. Appropriate target associated with the biological process of the disease needs to be identified, selected, and even validated in a testing system. The design and development of molecular imaging probe will unlikely be successful unless the molecular target is well defined and its biological role is well characterized. It is fortunate that the past two decades of biomedical research have yielded an enormous amount of information about the molecular events that take place during the development of a specific disease. With the explosion of biological data and information generated from various genomics, proteomics, and chemogenomics approaches, bioinformatics methods have become increasingly useful to extract or filter valuable targets by the combination of biological ideas with computer tools or statistical methods [16–20]. In recent years, a variety of databases, such as Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo), ware-housing high-throughput microarray data of tumor expression profiles and bioinformatics initiatives, such as Oncomine [16,17], have been developed to accelerate the discovery of diagnostic markers for human diseases. With the availability of these biological data-mining tools, not only the appropriate target linked to a specific disease can be more readily identified than before, but also the location and distribution (intercellular, cell membrane, extracellular matrix, etc.) of the molecular targets can be examined. According to the biological information, appropriate molecular platforms can be selected and designed to obtain probes with capability to reach the targeting sites. In addition, the molecular imaging strategies are largely relied on the nature of molecular targets, which in turn affects the molecular imaging probe design. For instance, imaging probes are generally designed for indirect imaging of relatively low amount of biomarkers, such as deoxyribonucleic acid (DNA) or messenger ribonucleic acid (mRNA), whereas for the molecular targets with abundant expression such as proteins, design of a probe to directly interact with the targets for imaging is a valid approach [21, 22].
It is quite often that the imaging probe can bind to the biological target beautifully in vitro; however, the in vivo imaging results are rather frustrated. It is understandable that the biological subject is a complex system and it has many sophisticated defense mechanisms to prevent exogenous molecule, such as a molecular imaging probe, from performing its intended function. A molecular probe has to possess reasonable features to overcome several biological or pharmacological barriers, so that it can ultimately reach the specific target at sufficient concentration and retain long enough time for imaging. First of all, the molecular imaging probe must remain intact in the circulation to evade the reticuloendothelial system (RES). In addition, the molecular imaging probe must be able to reach and accumulate into the targeted tissues such as tumor. Moreover, if the molecular imaging probe is designed to image the intracellular target, the imaging probe has to penetrate the cell membrane and stay inside of the cells. One of major goals of molecular imaging probe design is to maximize probe’s ability of crossing these biological barriers.
Because molecular imaging probe is a subtype of pharmaceuticals, the knowledge and experience gained from conventional drug design can facilitate molecular imaging probe design. Lipinski's Rule of Five is a rule of thumb to evaluate druglikeness, or determine if a chemical compound with a certain pharmacological or biological activity has properties that would make it a likely orally active drug in humans. It has been widely used as a guideline for drug design. Although such a guideline may not be exactly suitable for molecular imaging probe design, physicochemical properties of molecular imaging probes, such as ionization constant (pKa value), lipophilicity (LogP value), and stability, are generally considered to be key factors related to probes’ in vivo pharmacokinetics including adsorption, distribution, metabolism, and excretion. It is important to determine these properties in silico or experimentally at the beginning stage of design and development of an imaging probe [23, 24]. Ionization is a factor which is closely related to the solubility and membrane permeability of a molecule. Many molecular imaging probes contain ionizable groups and possess different charges within the physiological pH range. The overall charge or the distribution of the charge of a molecular imaging probe affects its solubility, permeability, and the binding to the active site. In addition, lipophilicity is a key parameter which describes a molecule to distribute between a lipid and a hydrophilic phase. It reflects the ability of an imaging probe to penetrate the lipid bilayer, as well as provides some ideas of the probe’s metabolism and elimination. For example, the clearance of an imaging probe with high lipophilicity is usually through the hepatobiliary system, whereas a hydrophilic imaging probe is more likely washed out through the renal system. Finally, the imaging probe must be stable enough within a period of imaging time to obtain the images with good quality. It is worthy to mention that advances in high performance liquid chromatography/mass spectrometry (HPLC/MS) instrumentation and procedures provide a powerful tool to identify the metabolites of the imaging probe [25]. High-sensitivity and high-resolution LC/MS instrumentation is capable of detecting the mass peak of an imaging probe and its metabolites at tracer dose level in animal or human tissues. The knowledge of metabolic transformations gained from HPLC/MS data can help in further modification of the imaging probe and increasing its in vivo stability.
The general approaches of molecular imaging probe design are depicted in Fig. 2. Coupling of a targeting moiety with a signal agent through a linker/delivery vehicle connection is the most common approach (Fig. 2A). The targeting moiety, such as a small molecule, a peptide, and a protein, can be conjugated with a signal agent to generate an imaging probe [26]. One targeting moiety can also be conjugated with several signal agents for multimodality molecular imaging [27]. Moreover, multiple targeting moieties binding to various targets can be constructed and labeled with one or multiple signal agents for multiple targets imaging [28]. In order to tune the pharmacokinetics of an imaging probe, a linker, such as polyethylene glycol chain, poly-amino acids, can be selected to connect the targeting moiety and the signal agent. Furthermore, a delivery vehicle can also be incorporated into an imaging probe for improving its pharmacokinetics. Many nanoparticles, both inorganic and organic, can serve as targeting moieties as well as the delivery vehicles [29–32]. The targeting moieties or signal agents can be coupled either on the surface of the nanoparticles [29–34] or inside of the nanoparticles [35, 36]. The second approach represents coupling of two signal agents through a targeting moiety (Fig. 2B). Activatable probes or smart probes belong to this type of design [37–39]. In most cases, the targeting moiety is a specific enzyme substrate. Upon the interaction of the substrate with the enzyme, the imaging signal can be generated, increased, or decreased. In the third approach, a signal agent is served as a core while various targeting moieties can be conjugated to the signal agent (Fig. 2C) [28]. Overall, selection of the design approach depends on many factors, including the specific target and imaging modality. In order to generate an imaging probe with ideal in vivo performance, each component of a molecular imaging probe must be carefully investigated.
Fig. (2).
Schematic representation of general approaches of molecular imaging probe design.
STRATEGIES FOR DEVELOPMENT OF MOLECULAR IMAGING PROBES
In general, the strategies of molecular imaging probe development can be categorized into two major classes: random approach and rational approach. Under some circumstances, two approaches can be combined together to develop molecular imaging probes.
Random Approach
In the past two decades, High-throughput screening (HTS) technique has become the paramount hit identification method and the major source of leads in medium and large pharmaceutical companies. Much earlier in the drug development process using HTS could save significant time and money. HTS technique is also the most commonly used random approach for identification of novel molecular imaging probes to new biological targets. A molecule probe library can be generated using chemical approaches. Combinatorial chemistry, such as one-bead-one-compound method [40–43], has been explored and demonstrated as a powerful approach for probe development. Hundreds and thousands of compounds could be prepared in a short period of time and used for bioactivity testing. The best hit will then be labeled with signal agent and tested in vivo as an imaging probe. Besides using chemistry approach, biotechniques such as HTS in vitro display technologies can also be employed to generate a library of imaging probes. In vitro phage, bacterial, yeast, ribosome and mRNA display technologies have been established. The rapid progress of these display technologies has led to the discovery of many novel small peptides and proteins with high affinity and specificity to a variety of molecular targets [44–46]. The relatively small size of these peptides leads to fast clearance rate and rapid tumor accumulation which are desirable properties of molecular imaging probes. Among all in vitro display techniques, phage display has demonstrated its great success in the imaging probe development [47–52]. A random phage-displayed peptide library comprises a vast population of phages, each of which expresses a unique peptide sequence on its surface. Once a binding peptide is fished out, signal agent can be tagged onto the peptide and further optimized for in vivo imaging. Display technologies in conjunction with rational protein engineering have also been used to construct non-immunoglobulin protein libraries with a defined protein scaffold. Several small protein scaffolds have been evaluated and demonstrated great potential for development of molecular probes for various targets [53–58]. Recently, in vivo HTS approach of molecular imaging probe development has been reported [59]. This work combines the advantages of combinatorial chemistry, site specific solid-phase radiolabeling, and in vivo imaging for the rapid screening of molecular imaging probes, whereby a decision regarding the potential of an imaging probe can be made rapidly.
Rational Approach
Rational design and development of molecular imaging probes is an inventive process of finding new molecular imaging probes. It usually takes advantages of the knowledge gained from the biological targets and requires extensive experience in the development of imaging probes. Several methods are practically used and have demonstrated a number of successes.
Generation from Naturally-Occurring Bioactive Molecules
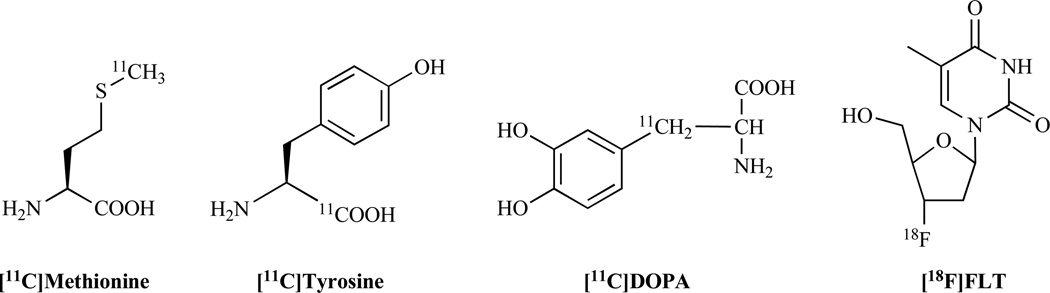
Numerous naturally-occurring bioactive molecules have been discovered over the past decades. Many of these molecules are involved in both normal and disease biological process. These molecules can interact with enzymes, receptors, transporters, and proteins, and provided great sources and foundations for molecular imaging probe development. For instance, many amino acids have been converted to PET probes for imaging tumor metabolism, such as L-[1-11C]tyrosine (11C-tyrosine) [60, 61] and L-[methyl-11C]methionine (11C-methionine) [62, 63] (Fig. 3). L-DOPA (L-3,4-dihydroxyphenylalanine) is the precursor to the neurotransmitters dopamine, norepinephrine (noradrenaline), and epinephrine (adrenaline). Radiolabeled L-DOPA has been designed to reveal the presynaptic dopaminergic function in healthy and pathologic states of humans [64]. 3'-18F-fluoro-3'-deoxy-L-thymidine (18F-FLT), a radiolabeled thymidine analog, has been widely used for imaging tumor proliferation [65] (Fig. 3). In addition, many molecular imaging probes are developed based on the naturally-occurring peptides, including radiolabeled somatostatin and octreotide [66, 67], bombesin analogs [68], neurotensin peptides [69, 70], and α-MSH peptides [71, 72].
Fig. (3).
Examples of molecular imaging probes generated from naturally-occurring bioactive molecules.
Generation from Drugs or Drug Candidates
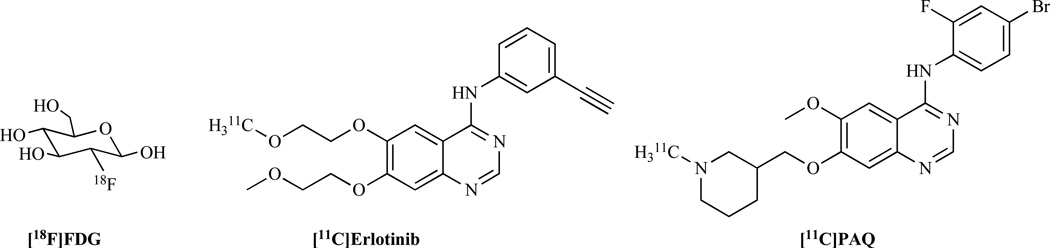
Routinely used drugs as well as drug candidates in clinical trial provide valuable sources for molecular imaging probe development. The most successful PET probe, 18F-fluoro-deoxy-D-glucose (18F-FDG) (Fig. 4), is a good example. 2-Deoxy-D-glucose (DG) was first developed in 1960 as a chemotherapeutic agent to inhibit glucose use by cancer cells [73, 74]. DG enters the cell similar to glucose and is converted to deoxyglucose 6-phosphate which, however, does not undergo further metabolism and is trapped in the cell [75]. Therefore, the design of the FDG molecule is based on labeling carbon-2 atom in DG with 18F. Incidentally the C–F bond, which is more stable than C–H bond, is chemically unrecognized by hexokinase. In 1976, Dr. Wolf and his colleagues developed the synthesis of 18F-FDG to study the cerebral glucose metabolism based on PET [76]. In the early 1990s, 18F-FDG PET started to be used in conjunction with whole-body imaging protocols. To date, 18F-FDG is a model PET radiopharmaceutical and is regarded as the “molecule of the century” in nuclear medicine.
Fig. (4).
Examples of molecular imaging probes generated from drugs or drug candidates.
Excluding FDG, many other molecular imaging probes based on clinical used drugs have been reported. For example, erlotinib is a drug used to treat non-small cell lung cancer, pancreatic cancer and several other types of cancer. It is a tyrosine kinase inhibitor, which acts on the epidermal growth factor receptor (EGFR). [11C]-erlotinib (Fig. 4) has been synthesized and evaluated in nude mice bearing lung cancer xenografts of A549, NCI358, and HCC827 [77]. The results demonstrate that [11C]-erlotinib can be used as a new PET probe for the identification of erlotinib-responding tumors. In addition, (R,S)-N-(4-bromo-2-fluorophenyl)-6-methoxy-7-((1-methyl-3-piperidinyl)methoxy)-4-quinazolinamine (PAQ) is a typosine kinase inhibitor with high affinity for the vascular endothelial growth factor receptor 2 (VEGF-2), which plays an important role in tumor angiogensis. 11C-labeled PAQ (Fig. 4) has been developed and tested in breast and melanoma cancer xenografts [78]. The results suggest that [11C]PAQ has potential as a PET tracer for in vivo imaging of VEGF-2 expression.
Overall, this approach is considered as a high risk and high return method. The addition of a labeling tag may affect the bioactivity of the drug or drug candidate. However, if the bioactivity of the drug or drug candidate remains the same after coupling with the labeling moiety, there is high likelihood of the resulting imaging probe being ultimately translated into the clinical applications.
Generation from Established Molecular Imaging Probes
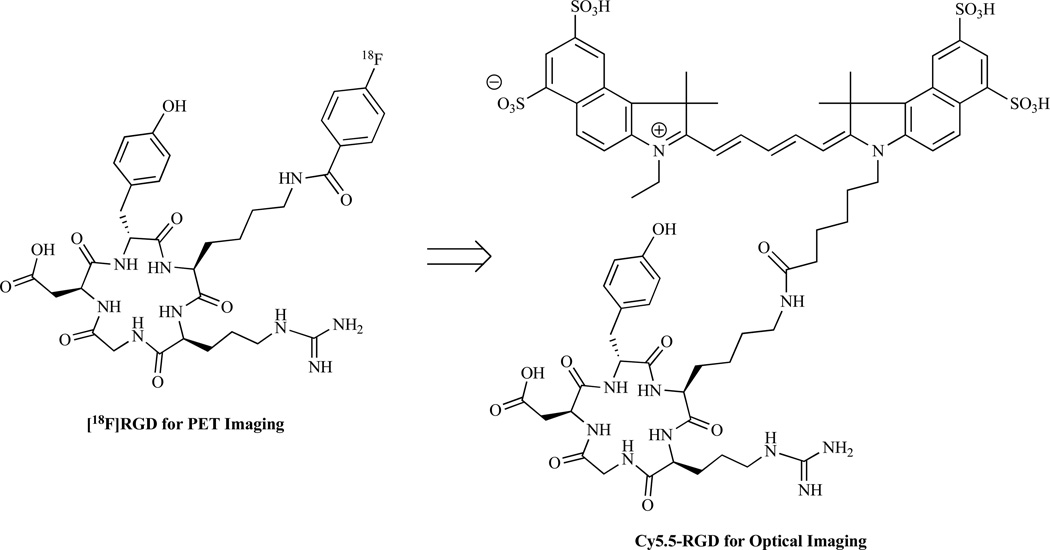
A good number of molecular imaging probes have been developed for various imaging modalities. To further extend their applications in molecular imaging, the established imaging agents could be converted into novel imaging probes by using different labeling moieties and labeling methodologies. For example, arginine-glycine-aspartic acid (RGD) containing peptides have been extensively studied as PET and SPECT agents for imaging integrin αvβ3. After coupling with near infrared (NIR) optical dye Cy5.5, the new generated peptide could also be used for optical imaging [79–81] (Fig. 5). Additionally, α-MSH analogs were initially developed as SPECT probes for melanocortin receptor 1 targeted melanoma imaging [82]. By using positron-emitter radionuclides, such as 18F and 64Cu, PET probes based on α-MSH analogs can be rapidly obtained [83,84]. Because the established imaging probe has already been well evaluated in vitro and in vivo, the generation from the established imaging agents has the potential in development of novel imaging probes with high success rate within relatively short time frame.
Fig. (5).
An example of a molecular imaging probe generated from an established molecular imaging probe.
Recently, molecular imaging and contrast agent database (MICAD) has been established by National Institutes of Health. This imaging agent database offers biomedical researchers easy access to a large number of molecular probes. It will help the researchers to quickly identify the probes for further optimization and modification, and thus facilitate the molecular imaging probe development.
Computer-Aided Molecular Imaging probe Development
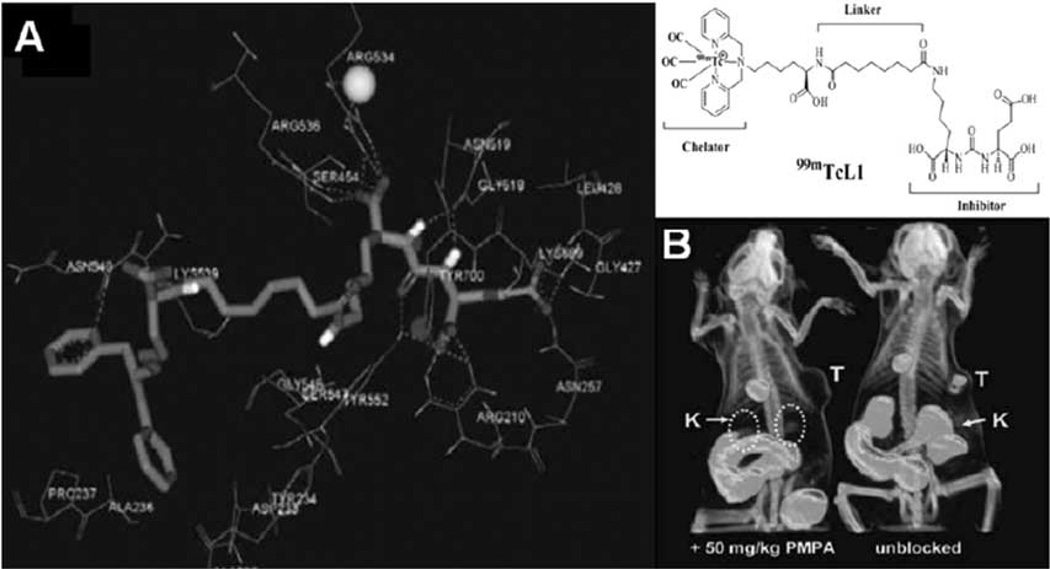
Computer-aided drug design (CADD) has been widely used in pharmaceutical development. It uses computational chemistry to discover, enhance, or study drugs and related biologically active molecules. The most fundamental goal is to predict whether a given molecule will bind to a target and if so how strongly. Ideally the computational method should be able to predict affinity before a compound is synthesized. In practice, it still takes several iterations of design, synthesis, and testing before an optimal molecule is discovered. Overall, CADD have accelerated discovery by reducing the number of iterations required and in addition have often provided more novel ligand structures. The application of CADD approach in imaging probe development has recently been reported. For example, molecular modeling technique has been used in development of SPECT imaging probe targeting prostate-specific membrane antigen (PSMA) [85] (Fig. 6). The X-ray complex structure of PSMA and its inhibitor was retrieved from protein data bank (PDB). The newly designed imaging probe was then docked onto the active binding site of PSMA. The binding model was analyzed and the affinity was then predicted. The results suggest that in silico modeling technique can hasten the development of PSMA-specific imaging probe. Another example is the development of a near-infrared fluorescent non-peptidic bivalent probe for integrin αvβ3 imaging [86]. Through molecular modeling approach, the binding affinities of a series of ligands to integrin αvβ3 were calculated. The ligand showing highest binding affinity in silico was then synthesized and coupled with Cy5.5 for optical imaging. The in vitro and in vivo results demonstrate molecular modeling is a valid approach to predict the binding affinity before the synthesis of imaging probe.
Fig. (6).
An example of computer-aided molecular imaging probe design. A) Binding mode of L1 to the active site of PSMA. B) SPECT-CT imaging of LNCaP (PSMA+) tumor bearing mice with [99mTc]L1 with (left) and without (right) blockade of PSMA using the potent, selective PSMA inhibitor, PMPA, as the blocking agent. Lack of radiopharmaceutical in both the tumor and kidneys (another PSMA+ site) upon cotreatment with PMPA provides a further check on PSMA-specific binding. Images were acquired from 30–60 min postinjection. T = tumor; K = kidney. Reproduced with permission from Ref. 85: Banerjee, S.R., et al. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA). J. Med. Chem., 2008, 51, 4504–4517.
PERSPECTIVE AND CONCLUSION
Molecular imaging as an emerging field has demonstrated importance to help further steps toward personalized medicine. As a key component of molecular imaging, design and development of novel imaging probes are far from trivial. A wide variety of targeting ligands (small molecules, peptides, peptidomimetics, and antibodies) have been conjugated with various labeling moieties for MRI, ultrasound, optical, PET, SPECT, and multimodality imaging. Non-invasive imaging has shown clinical applications in many aspects including lesion detection, patient stratification, drug development, treatment monitoring, and dose optimization.
It is expected that random and rational strategies will continue to play a central role in design and development of molecular imaging probes. Under many circumstances, these two basic approaches have to be combined together to discover the novel imaging probes for specific target. The advanced chemistry and biology techniques will likely pave the way of design and development of molecular imaging probe. For example, “click chemistry” [87, 88] offers chemists a platform for general, modular and high yielding synthetic transformations for constructing highly diverse molecules. Because click chemistry is a modular approach, it is possible to fine tune the new molecular imaging probes and enhance their pharmacokinetic and pharmacodynamic profiles [89, 90]. Click chemistry has proven itself to be superior in satisfying many criteria (e.g., biocompatibility, selectivity, yield, stereospecificity, and so forth); thus, one can expect it will consequently become a more routine strategy in the near future for a wide range of applications for imaging probe development [91]. In addition, various in vitro display techniques facilitate the findings of novel ligands with high binding affinity. These biological techniques will continue to be a useful tool in design and development of imaging probes. The complete sequencing of human genome has directed a new era of system biology. Genomic and proteomic molecular profiling technologies are transforming to various disease research [92, 93]. New technologies such as microarray analysis, genomics, proteomics, high-throughput screening, and computer-aided molecular design will accelerate the discovery process of novel imaging probes.
One major component of molecular medicine is nanomedicine, where the ultimate goal is that multifunctional nanoparticles containing both therapeutic components and multimodality imaging labels can allow for efficient, specific in vivo delivery of drugs and accurate, quantitative assessment of the therapeutic efficacy non-invasively over time [94]. The major application of multifunctional nanoparticles will likely not be disease diagnosis but rather for “theranostics”, the diagnostic therapy for individual patients [95]. Most of developed nanoparticles to date are far from optimal for clinical applications. In future efforts towards developing novel nanoparticle-based specific targeting probes, a number of factors need to be optimized in parallel including biocompatibility, pharmacokinetics, targeting efficacy, acute and chronic toxicity, ability to escape from the RES, and cost-effectiveness [96].
Finally, to foster the continued design and development of molecular imaging probes, cooperative efforts are needed from cellular/molecular biologist to identify and validate new imaging targets, medicinal chemists/radiochemists to design, synthesize, and characterize the imaging probes, and engineers/medical physicists to develop high sensitivity and high resolution imaging device and better imaging reconstruction algorithms. Close partnerships among academic researchers, clinicians, pharmaceutical industries, the NCI, and FDA are required to succeed in fast translation of molecular imaging probes from bench to bedside.
REFERENCES
- 1.Weissleder R, Mahmood U. Molecular imaging. Radiology. 2001;219(2):316–333. doi: 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- 2.Thakur M, Lentle BC. Report of a summit on molecular imaging. Radiology. 2005;236(3):753–755. doi: 10.1148/radiol.2363051160. [DOI] [PubMed] [Google Scholar]
- 3.Cai W, Rao J, Gambhir SS, Chen X. How molecular imaging is speeding up antiangiogenic drug development. Mol. Cancer Ther. 2006;5(11):2624–2633. doi: 10.1158/1535-7163.MCT-06-0395. [DOI] [PubMed] [Google Scholar]
- 4.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 5.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J. Proteome. Res. 2005;4(4):1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- 6.Hartwell L, Mankoff D, Paulovich A, Ramsey S, Swisher E. Cancer biomarkers: a systems approach. Nat. Biotechnol. 2006;24(8):905–908. doi: 10.1038/nbt0806-905. [DOI] [PubMed] [Google Scholar]
- 7.Beyer T, Townsend DW, Brun T, Kinahan PE, Charron M, Roddy R, Jerin J, Young J, Byars L, Nutt R. A combined PET/CT scanner for clinical oncology. J. Nucl. Med. 2000;41(8):1369–1379. [PubMed] [Google Scholar]
- 8.Catana C, Wu Y, Judenhofer MS, Qi J, Pichler BJ, Cherry SR. Simultaneous acquisition of multislice PET and MR images: initial results with a MR-compatible PET scanner. J. Nucl. Med. 2006;47(12):1968–1976. [PubMed] [Google Scholar]
- 9.Even-Sapir E, Lerman H, Lievshitz G, Khafif A, Fliss DM, Schwartz A, Gur E, Skornick Y, Schneebaum S. Lympho-scintigraphy for sentinel node mapping using a hybrid SPECT/CT system. J Nucl. Med. 2003;44(9):1413–1420. [PubMed] [Google Scholar]
- 10.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer. 2002;2(9):683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 11.Weissleder R. Scaling down imaging: molecular mapping of cancer in mice. Nat. Rev. Cancer. 2002;2(1):11–18. doi: 10.1038/nrc701. [DOI] [PubMed] [Google Scholar]
- 12.Weissleder R. Molecular imaging in cancer. Science. 2006;312(5777):1168–1171. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 13.Chen X. Multimodality imaging of tumor integrin alphavbeta3 expression. Mini Rev. Med. Chem. 2006;6(2):227–234. doi: 10.2174/138955706775475975. [DOI] [PubMed] [Google Scholar]
- 14.Haubner R, Wester HJ, Weber WA, Mang C, Ziegler SI, Goodman SL, Senekowitsch-Schmidtke R, Kessler H, Schwaiger M. Noninvasive imaging of alpha(v)beta3 integrin expression using 18F-labeled RGD-containing glycopeptide and positron emission tomography. Cancer Res. 2001;61(5):1781–1785. [PubMed] [Google Scholar]
- 15.Liu S, Edwards DS. Bifunctional chelators for therapeutic lanthanide radiopharmaceuticals. Bioconjug. Chem. 2001;12(1):7–34. doi: 10.1021/bc000070v. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes DR, Chinnaiyan AM. Bioinformatics strategies for translating genome-wide expression analyses into clinically useful cancer markers. Ann. N. Y. Acad. Sci. 2004;1020:32–40. doi: 10.1196/annals.1310.005. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YP, Chen F. Identifying targets for drug discovery using bioinformatics. Expert. Opin. Ther. Targets. 2008;12(4):383–389. doi: 10.1517/14728222.12.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Klee EW, Finlay JA, McDonald C, Attewell JR, Hebrink D, Dyer R, Love B, Vasmatzis G, Li TM, Beechem JM, Klee GG. Bioinformatics methods for prioritizing serum biomarker candidates. Clin. Chem. 2006;52(11):2162–2164. doi: 10.1373/clinchem.2006.072868. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov. Today. 2009;14(3–4):147–154. doi: 10.1016/j.drudis.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Liu S. Radiolabeled cyclic RGD peptides as integrin alpha (v)beta(3)-targeted radiotracers: maximizing binding affinity via bivalency. Bioconjug. Chem. 2009;20(12):2199–2213. doi: 10.1021/bc900167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shokeen M, Anderson CJ. Molecular imaging of cancer with copper-64 radiopharmaceuticals and positron emission tomography (PET) Acc. Chem. Res. 2009;42(7):832–841. doi: 10.1021/ar800255q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avdeef A. Physicochemical profiling (solubility, permeability and charge state) Curr. Top. Med. Chem. 2001;1(4):277–351. doi: 10.2174/1568026013395100. [DOI] [PubMed] [Google Scholar]
- 24.Wildman SA, Crippen GM. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999;39(5):868–873. [Google Scholar]
- 25.Ma Y, Lang L, Reyes L, Tokugawa J, Jagoda EM, Kiesewetter DO. Application of highly sensitive UPLC-MS to determine biodistribution at tracer doses: validation with the 5-HT1A ligand [(18)F]FPWAY. Nucl. Med. Biol. 2009;36(4):389–393. doi: 10.1016/j.nucmedbio.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Z, Chen J, Quinn TP, Jurisson SS. Radioiodination of rhenium cyclized alpha-melanocyte-stimulating hormone resulting in enhanced radioactivity localization and retention in melanoma. Cancer Res. 2004;64(4):1411–1418. doi: 10.1158/0008-5472.can-03-0193. [DOI] [PubMed] [Google Scholar]
- 27.Deroose CM, De A, Loening AM, Chow PL, Ray P, Chatziioannou AF, Gambhir SS. Multimodality imaging of tumor xenografts and metastases in mice with combined small-animal PET, small-animal CT, and bioluminescence imaging. J. Nucl. Med. 2007;48(2):295–303. [PMC free article] [PubMed] [Google Scholar]
- 28.Li ZB, Wu Z, Chen K, Ryu EK, Chen X. 18F-labeled BBN-RGD heterodimer for prostate cancer imaging. J. Nucl. Med. 2008;49(3):453–461. doi: 10.2967/jnumed.107.048009. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotechnol. 2005;16(1):63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Han M, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001;19(7):631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 31.Ye Y, Bloch S, Achilefu S. Polyvalent carbocyanine molecular beacons for molecular recognitions. J. Am. Chem. Soc. 2004;126(25):7740–7741. doi: 10.1021/ja049441z. [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Li WP, Anderson CJ, Kao J, Nikiforovich GV, Achilefu S. Synthesis and characterization of a macrocyclic near-infrared optical scaffold. J. Am. Chem. Soc. 2003;125(26):7766–7767. doi: 10.1021/ja034186o. [DOI] [PubMed] [Google Scholar]
- 33.Cai W, Chen K, Li ZB, Gambhir SS, Chen X. Dual-function probe for PET and near-infrared fluorescence imaging of tumor vasculature. J. Nucl. Med. 2007;48(11):1862–1870. doi: 10.2967/jnumed.107.043216. [DOI] [PubMed] [Google Scholar]
- 34.Diagaradjane P, Orenstein-Cardona JM, Colon-Casasnovas NE, Deorukhkar A, Shentu S, Kuno N, Schwartz DL, Gelovani JG, Krishnan S. Imaging epidermal growth factor receptor expression in vivo: pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clin. Cancer Res. 2008;14(3):731–741. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- 35.Yasuhara A, Katami T, Shibamoto T. Dioxin formation during combustion of nonchloride plastic, polystyrene and its product. Bull. Environ. Contam. Toxicol. 2005;74(5):899–903. doi: 10.1007/s00128-005-0666-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Zhou J. Improving the signal sensitivity and photostability of DNA hybridizations on microarrays by using dye-doped core-shell silica nanoparticles. Anal. Chem. 2004;76(18):5302–5312. doi: 10.1021/ac049472c. [DOI] [PubMed] [Google Scholar]
- 37.Asanuma D, Kobayashi H, Nagano T, Urano Y. Fluorescence Imaging of Tumors with "Smart" pH-Activatable Targeted Probes. Methods Mol. Biol. 2009;574:47–62. doi: 10.1007/978-1-60327-321-3_5. [DOI] [PubMed] [Google Scholar]
- 38.Elias DR, Thorek DL, Chen AK, Czupryna J, Tsourkas A. In vivo imaging of cancer biomarkers using activatable molecular probes. Cancer Biomark. 2008;4(6):287–305. doi: 10.3233/cbm-2008-4602. [DOI] [PubMed] [Google Scholar]
- 39.Lee S, Park K, Kim K, Choi K, Kwon IC. Activatable imaging probes with amplified fluorescent signals. Chem. Commun. (Camb.) 2008;(36):4250–4260. doi: 10.1039/b806854m. [DOI] [PubMed] [Google Scholar]
- 40.Lam KS, Lebl M, Krchnak V. The "one-bead-one-compound" combinatorial library method. Chem. Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 41.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 42.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nat. Chem. Biol. 2006;2(7):381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 43.Xiao W, Yao N, Peng L, Liu R, Lam KS. Near-infrared optical imaging in glioblastoma xenograft with ligand-targeting alpha 3 integrin. Eur. J. Nucl. Med. Mol. Imaging. 2009;36(1):94–103. doi: 10.1007/s00259-008-0920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosse RJ, Rothe A, Power BE. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 2006;15(1):14–27. doi: 10.1110/ps.051817606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothe A, Hosse RJ, Power BE. In vitro display technologies reveal novel biopharmaceutics. FASEB J. 2006;20(10):1599–1610. doi: 10.1096/fj.05-5650rev. [DOI] [PubMed] [Google Scholar]
- 46.Uchiyama F, Tanaka Y, Minari Y, Tokui N. Designing scaffolds of peptides for phage display libraries. J. Biosci. Bioeng. 2005;99(5):448–456. doi: 10.1263/jbb.99.448. [DOI] [PubMed] [Google Scholar]
- 47.Dennis MS, Eigenbrot C, Skelton NJ, Ultsch MH, Santell L, Dwyer MA, O'Connell MP, Lazarus RA. Peptide exosite inhibitors of factor VIIa as anticoagulants. Nature. 2000;404(6777):465–470. doi: 10.1038/35006574. [DOI] [PubMed] [Google Scholar]
- 48.Hyde-DeRuyscher R, Paige LA, Christensen DJ, Hyde-DeRuyscher N, Lim A, Fredericks ZL, Kranz J, Gallant P, Zhang J, Rocklage SM, Fowlkes DM, Wendler PA, Hamilton PT. Detection of small-molecule enzyme inhibitors with peptides isolated from phage-displayed combinatorial peptide libraries. Chem. Biol. 2000;7(1):17–25. doi: 10.1016/s1074-5521(00)00062-4. [DOI] [PubMed] [Google Scholar]
- 49.Kumar SR, Deutscher SL. 111In-labeled galectin-3-targeting peptide as a SPECT agent for imaging breast tumors. J. Nucl. Med. 2008;49(5):796–803. doi: 10.2967/jnumed.107.048751. [DOI] [PubMed] [Google Scholar]
- 50.Kumar SR, Quinn TP, Deutscher SL. Evaluation of an 111In-radiolabeled peptide as a targeting and imaging agent for ErbB-2 receptor expressing breast carcinomas. Clin. Cancer Res. 2007;13(20):6070–6079. doi: 10.1158/1078-0432.CCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 51.Meiring MS, Litthauer D, Harsfalvi J, van Wyk V, Badenhorst PN, Kotze HF. In vitro effect of a thrombin inhibition peptide selected by phage display technology. Thromb. Res. 2002;107(6):365–371. doi: 10.1016/s0049-3848(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 52.Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273(5274):458–464. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 53.Cheng Z, Kramer DJ, Padilla De Jeuus O, De A, Webster JM, Gheysens O, Levi J, Namavari M, Wang S, Paik JM, Zhang R, Lee B, Hoerner J, Grade H, Syud FA, Gambhir SS. Radiometals labeled synthetic affibody molecules for imaging of HER2 expression. Mol. Imaging Biol. 2010 doi: 10.1007/s11307-009-0256-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang L, Kimura RH, Miao Z, Ren G, Liu HG, Gambhir SS, Cochran JR, Cheng Z. Evaluation of a 64Cu-labeled cystine-knot peptide based on agouti related protein scaffold for tumor αvβ3 integrin PET imaging. J. Nucl. Med. 2010;51(2):251–258. doi: 10.2967/jnumed.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miao Z, Ren G, Liu H, Jiang L, Kimura RH, Cochran JR, Gambhir SS, Cheng Z. An engineered knottin peptide labeled with 18F for PET imaging of αvβ3 Integrin expression. Bioconjug. Chem. 2009;20(12):2342–2347. doi: 10.1021/bc900361g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webster JM, Zhang R, Gambhir SS, Cheng Z, Syud FA. Engineered two-helix small proteins for molecular recognition. Chembiochem. 2009;10(8):1293–1296. doi: 10.1002/cbic.200900062. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Z, De Jesus OP, Namavari M, De A, Levi J, Webster JM, Zhang R, Lee B, Syud FA, Gambhir SS. Small-animal PET imaging of human epidermal growth factor receptor type 2 expression with site-specific 18F-labeled protein scaffold molecules. J. Nucl. Med. 2008;49(5):804–813. doi: 10.2967/jnumed.107.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res. 2009;69(6):2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gagnon MK, Hausner SH, Marik J, Abbey CK, Marshall JF, Sutcliffe JL. High-throughput in vivo screening of targeted molecular imaging agents. Proc. Natl. Acad. Sci. USA. 2009;106(42):17904–17909. doi: 10.1073/pnas.0906925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Boer JR, Pruim J, van der Laan BF, Que TH, Willemsen AT, Albers FW, Vaalburg W. L-1-11C-tyrosine PET in patients with laryngeal carcinomas: comparison of standardized uptake value and protein synthesis rate. J. Nucl. Med. 2003;44(3):341–346. [PubMed] [Google Scholar]
- 61.Plaat B, Kole A, Mastik M, Hoekstra H, Molenaar W, Vaalburg W. Protein synthesis rate measured with L-[1-11C]tyrosine positron emission tomography correlates with mitotic activity and MIB-1 antibody-detected proliferation in human soft tissue sarcomas. Eur. J. Nucl. Med. 1999;26(4):328–332. doi: 10.1007/s002590050394. [DOI] [PubMed] [Google Scholar]
- 62.Davis J, Yano Y, Cahoon J, Budinger TF. Preparation of 11C-methyl iodide and L-[S-methyl-11C]methionine by an automated continuous flow process. Int. J. Appl. Radiat. Isot. 1982;33(5):363–369. doi: 10.1016/0020-708x(82)90150-8. [DOI] [PubMed] [Google Scholar]
- 63.Lindholm P, Leskinen S, Nagren K, Lehikoinen P, Ruotsalainen U, Teras M, Joensuu H. Carbon-11-methionine PET imaging of malignant melanoma. J. Nucl. Med. 1995;36(10):1806–1810. [PubMed] [Google Scholar]
- 64.Tedroff J, Aquilonius SM, Hartvig P, Bredberg E, Bjurling P, Langstrom B. Cerebral uptake and utilization of therapeutic [beta-11C]-L-DOPA in Parkinson's disease measured by positron emission tomography. Relations to motor response. Acta Neurol. Scand. 1992;85(2):95–102. doi: 10.1111/j.1600-0404.1992.tb04005.x. [DOI] [PubMed] [Google Scholar]
- 65.Cobben DC, Jager PL, Elsinga PH, Maas B, Suurmeijer AJ, Hoekstra HJ. 3'-18F-fluoro-3'-deoxy-L-thymidine: a new tracer for staging metastatic melanoma? J. Nucl. Med. 2003;44(12):1927–1932. [PubMed] [Google Scholar]
- 66.Cescato R, Erchegyi J, Waser B, Piccand V, Maecke HR, Rivier JE, Reubi JC. Design and in vitro characterization of highly sst2-selective somatostatin antagonists suitable for radiotargeting. J. Med. Chem. 2008;51(13):4030–4037. doi: 10.1021/jm701618q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwekkeboom DJ, de Herder WW, Kam BL, van Eijck CH, van Essen M, Kooij PP, Feelders RA, van Aken MO, Krenning EP. Treatment with the radiolabeled somatostatin analog [177 Lu-DOTA 0, Tyr3]octreotate: toxicity, efficacy, and survival. J. Clin. Oncol. 2008;26(13):2124–2130. doi: 10.1200/JCO.2007.15.2553. [DOI] [PubMed] [Google Scholar]
- 68.Schally AV, Nagy A. Cancer chemotherapy based on targeting of cytotoxic peptide conjugates to their receptors on tumors. Eur. J. Endocrinol. 1999;141(1):1–14. doi: 10.1530/eje.0.1410001. [DOI] [PubMed] [Google Scholar]
- 69.Garcia-Garayoa E, Blauenstein P, Blanc A, Maes V, Tourwe D, Schubiger PA. A stable neurotensin-based radiopharmaceutical for targeted imaging and therapy of neurotensin receptor-positive tumours. Eur. J. Nucl. Med. Mol. Imaging. 2009;36(1):37–47. doi: 10.1007/s00259-008-0894-y. [DOI] [PubMed] [Google Scholar]
- 70.Nock BA, Nikolopoulou A, Reubi JC, Maes V, Conrath P, Tourwe D, Maina T. Toward stable N4-modified neurotensins for NTS1-receptor-targeted tumor imaging with 99mTc. J. Med. Chem. 2006;49(15):4767–4776. doi: 10.1021/jm060415g. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, Cheng Z, Hoffman TJ, Jurisson SS, Quinn TP. Melanoma-targeting properties of (99m)technetium-labeled cyclic alpha-melanocyte-stimulating hormone peptide analogues. Cancer Res. 2000;60(20):5649–5658. [PubMed] [Google Scholar]
- 72.Chen J, Cheng Z, Owen NK, Hoffman TJ, Miao Y, Jurisson SS, Quinn TP. Evaluation of an (111)In-DOTA-rhenium cyclized alpha-MSH analog: a novel cyclic-peptide analog with improved tumor-targeting properties. J. Nucl. Med. 2001;42(12):1847–1855. [PubMed] [Google Scholar]
- 73.Laszlo J, Humphreys SR, Goldin A. Effects of glucose analogues (2-deoxy-D-glucose, 2-deoxy-D-galactose) on experimental tumors. J. Natl. Cancer Inst. 1960;24:267–281. [PubMed] [Google Scholar]
- 74.Pacak J, Cerny M. History of the first synthesis of 2-deoxy-2-fluoro-D-glucose the unlabeled forerunner of 2-deoxy-2-[18F] fluoro-D-glucose. Mol. Imaging Biol. 2002;4(5):352–354. doi: 10.1016/s1536-1632(02)00083-5. [DOI] [PubMed] [Google Scholar]
- 75.Sols A, Crane RK. Substrate specificity of brain hexokinase. J. Biol. Chem. 1954;210(2):581–595. [PubMed] [Google Scholar]
- 76.Ido TWC, Casella V, Fowler JS, Wolf AP, Reivich M, Kuhl DE. Labeled 2-deoxy-D-glucose analogs. 18F-labeled 2-deoxy-2-fluoro-D-glucose, 2-deoxy-2-fluoro-D-mannose and 14C-2-deoxy-2-fluoro-D-glucose. J. Label. Compd. Radiopharm. 1978;14(2):175–183. [Google Scholar]
- 77.Memon AA, Jakobsen S, Dagnaes-Hansen F, Sorensen BS, Keiding S, Nexo E. Positron emission tomography (PET) imaging with [11C]-labeled erlotinib: a micro-PET study on mice with lung tumor xenografts. Cancer Res. 2009;69(3):873–878. doi: 10.1158/0008-5472.CAN-08-3118. [DOI] [PubMed] [Google Scholar]
- 78.Samen E, Thorell JO, Lu L, Tegnebratt T, Holmgren L, Stone-Elander S. Synthesis and preclinical evaluation of [(11)C]PAQ as a PET imaging tracer for VEGFR-2. Eur. J. Nucl. Med. Mol. Imaging. 2009;36(8):1283–1295. doi: 10.1007/s00259-009-1111-3. [DOI] [PubMed] [Google Scholar]
- 79.Chen X, Conti PS, Moats R. A In vivo near-infrared fluorescence imaging of integrin alphavbeta3 in brain tumor xenografts. Cancer Res. 2004;64(21):8009–8014. doi: 10.1158/0008-5472.CAN-04-1956. [DOI] [PubMed] [Google Scholar]
- 80.Cheng Z, Levi J, Xiong Z, Gheysens O, Keren S, Chen X, Gambhir SS. Near-infrared fluorescent deoxyglucose analogue for tumor optical imaging in cell culture and living mice. Bioconjug. Chem. 2006;17(3):662–669. doi: 10.1021/bc050345c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng Z, Wu Y, Xiong Z, Gambhir SS, Chen X. Near-infrared fluorescent RGD peptides for optical imaging of integrin alphavbeta3 expression in living mice. Bioconjug. Chem. 2005;16(6):1433–1441. doi: 10.1021/bc0501698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Froidevaux S, Calame-Christe M, Tanner H, Sumanovski L, Eberle AN. A novel DOTA-alpha-melanocyte-stimulating hormone analog for metastatic melanoma diagnosis. J. Nucl. Med. 2002;43(12):1699–1706. [PubMed] [Google Scholar]
- 83.Cheng Z, Xiong Z, Subbarayan M, Chen X, Gambhir SS. 64Cu-labeled alpha-melanocyte-stimulating hormone analog for microPET imaging of melanocortin 1 receptor expression. Bioconjug. Chem. 2007;18(3):765–772. doi: 10.1021/bc060306g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng Z, Zhang L, Graves E, Xiong Z, Dandekar M, Chen X, Gambhir SS. Small-animal PET of melanocortin 1 receptor expression using a 18F-labeled alpha-melanocyte-stimulating hormone analog. J. Nucl. Med. 2007;48(6):987–994. doi: 10.2967/jnumed.107.039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee SR, Foss CA, Castanares M, Mease RC, Byun Y, Fox JJ, Hilton J, Lupold SE, Kozikowski AP, Pomper MG. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA) J. Med. Chem. 2008;51(15):4504–4517. doi: 10.1021/jm800111u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li F, Liu J, Jas GS, Zhang J, Qin G, Xing J, Cotes C, Zhao H, Wang X, Diaz LA, Shi ZZ, Lee DY, Li KC, Li Z. Synthesis and evaluation of a near-infrared fluorescent non-peptidic bivalent Integrin alpha(v)beta(3) antagonist for cancer imaging. Bioconjug. Chem. 2010;21(2):270–278. doi: 10.1021/bc900313d. [DOI] [PubMed] [Google Scholar]
- 87.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few Good reactions. Angew. Chem. Int. Ed. Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 88.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov. Today. 2003;8(24):1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 89.Hausner SH, Marik J, Gagnon MK, Sutcliffe JL. In vivo positron emission tomography (PET) imaging with an alphavbeta6 specific peptide radiolabeled using 18F-"click" chemistry: evaluation and comparison with the corresponding 4-[18F]fluorobenzoyl- and 2-[18F]fluoropropionyl-peptides. J. Med. Chem. 2008;51(19):5901–5904. doi: 10.1021/jm800608s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li ZB, Wu Z, Chen K, Chin FT, Chen X. Click chemistry for (18)F-labeling of RGD peptides and microPET imaging of tumor integrin alphavbeta3 expression. Bioconjug. Chem. 2007;18(6):1987–1994. doi: 10.1021/bc700226v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nwe K, Brechbiel MW. Growing applications of "click chemistry" for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009;24(3):289–302. doi: 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Workman P. The impact of genomic and proteomic technologies on the development of new cancer drugs. Ann. Oncol. 2002;13(Suppl. 4):115–124. doi: 10.1093/annonc/mdf648. [DOI] [PubMed] [Google Scholar]
- 93.Wulfkuhle J, Espina V, Liotta L, Petricoin E. Genomic and proteomic technologies for individualisation and improvement of cancer treatment. Eur. J. Cancer. 2004;40(17):2623–2632. doi: 10.1016/j.ejca.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 94.Cai W, Chen X. Nanoplatforms for targeted molecular imaging in living subjects. Small. 2007;3(11):1840–1854. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 95.Del Vecchio S, Zannetti A, Fonti R, Pace L, Salvatore M. Nuclear imaging in cancer theranostics. Q. J. Nucl. Med. Mol. Imaging. 2007;51(2):152–163. [PubMed] [Google Scholar]
- 96.Cai W, Niu G, Chen X. Imaging of integrins as biomarkers for tumor angiogenesis. Curr. Pharm. Des. 2008;14(28):2943–2973. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]