Abstract
The Hedgehog signaling pathway regulates normal cell growth and differentiation. When deregulated, the Hedgehog pathway leads to tumorigenesis and supports more aggressive phenotypes of human cancers, such as progression, metastasis, and therapeutic resistance. The glioma-associated oncogene homolog 1 (GLI1) family of zinc finger transcription factors is the nuclear mediator of the Hedgehog pathway that regulates genes essential for various stages of tumor development and progression. Consequently, several components of the Hedgehog pathway are major targets of cancer therapy, including GLI1 and smoothened. Although the GLI1 gene was initially identified as an amplified gene in glioblastoma, its amplification was found to be relatively rare. No somatic mutations have been reported in the GLI1 gene. Notably, two decades after the discovery of the GLI1 gene, the GLI1 transcript was recently found to undergo alternative splicing forming two shorter isoforms, an N-terminal deletion variant (GLI1ΔN) and a truncated GLI1 (tGLI1). These variants appear to have different patterns of tissue expression and functions. Most notably, the tGLI1 isoform behaves as a gain-of-function GLI1 that can induce expression of genes not regulated by GLI1 and promotes more aggressive cancer phenotypes. Therefore, this review will focus on the structural and functional differences between these isoforms, and also on their contributions to important cancer cell characteristics, including proliferation, motility, invasion, and angiogenesis.
Introduction
The Hedgehog signaling pathway is critical to advanced forms of life as it is conserved in both vertebrates and invertebrates and involved in many biological processes (Dahmane and Ruiz I Altaba, 1999; Dahmane et al., 2001; Echelard et al., 1993; Roelink et al., 1994). The mammalian Hedgehog pathway is initiated by Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh) with Shh being the most potent of the three (Pathi et al., 2001) and more widely expressed (Ingham and McMahon, 2001). The Shh pathway plays a significant role in human cancer and is implicated in cancers that account for up to 25% of all human cancer deaths (Lum and Beachy, 2004). The main effectors of mammalian Shh signaling are the GLI transcription factors GLI1, GLI2, and GLI3 (Zhu and Lo, 2010). Shh addition to cells induces GLI protein translocation to the nucleus to affect gene transcription. Due to the importance of this pathway to human cancer, attempts to develop inhibitors have been extensive with a notable new small molecule inhibitor that acts upstream of the GLI1 proteins, Vismodegib/GDC-0449, and it is currently being evaluated in clinical trials (LoRusso et al., 2008; Rudin et al., 2009; Von Hoff et al., 2009).
The GLI1 gene is a unique member of the Shh pathway as it has no reported somatic mutations whereas several other Shh members do have mutations, including Shh, Patched (PTCH), Smoothened (SMO), Suppressor of Fused (SUFU), GLI2, and GLI3 (Kang et al., 1997; Radhakrishna et al., 1997; Raffel et al., 1997; Roessler et al., 1996; Xie et al., 1998). However, recent evidence indicates that the GLI1 transcript can undergo alternative splicing leading to the synthesis of an N-terminal deletion variant (GLI1ΔN) (Shimokawa et al., 2008) and truncated GLI1 variant (tGLI1) discovered in our laboratory (Lo et al., 2009). Efforts are ongoing to determine the relative importance of these splice variants to the full-length GLI1. To date, the GLI1ΔN variant appears to act on similar genes as GLI1, although with weaker activation, and is expressed in normal and cancerous tissues similar to GLI1 (Shimokawa et al., 2008). The tGLI1 variant, however, appears to only be expressed in tumor cells and tissues, but undetectable in normal tissues (Cao et al., 2012; Lo et al., 2009). Also, we have found that tGLI1 had gained the ability to regulate genes that are targeted by GLI1. tGLI1 but not GLI1 directly binds to and activates transcription of the CD24 gene in glioblastoma cells (Lo et al., 2009) and breast cancer cells (Cao et al., 2012), leading to increased cell motility and invasiveness. Most recently, we further reported that tGLI1 associates with the VEGF-A gene promoter leading to its activation (Cao et al., 2012). Consistent with the ability to enhance VEGF-A gene expression, tGLI1 expression stimulates the proliferation of vascular endothelial cells (Cao et al., 2012). These data strongly suggest that tGLI1 could be a more potent transcriptional regulator than GLI1 or GLI1ΔN. In light of these new insights, this review will summarize the structures and properties of the three GLI1 isoforms, their regulation by canonical and non-canonical cell signaling, and their differential impacts on tumor behaviors.
Structures and Properties of the GLI1 Isoforms
The human GLI1 gene was discovered in 1987 upon investigation into gene amplification in a human glioblastoma cell line (Kinzler et al., 1987). Investigators found a region of chromosome 12 to be amplified; however, this region did not correspond to any known oncogenes. The gene was termed GLI1 for the glioma tumor in which it was found (Kinzler et al., 1987) and was later mapped to a specific region of chromosome 12 at 12q13.3-14.1 (Arheden et al., 1989). This newly discovered GLI1 gene contained 3,318 base pairs giving rise to a 1,106-residue protein (Kinzler et al., 1988) that separates on a polyacrylamide gel to 150-kDa (Kinzler and Vogelstein, 1990). As shown in Figure 1, the GLI1 protein was found to contain five successive zinc finger DNA-binding motifs and belonged in the Kruppel family of zinc finger proteins (Kinzler et al., 1988). GLI1 was later found to bind to the 9-bp DNA sequence 5′-GACCACCCA-3′ (Kinzler and Vogelstein, 1990), in which only zinc fingers 2–5 were found to bind to the major groove of DNA (Pavletich and Pabo, 1993).
Figure 1.
Structures of the human GLI1 gene and the encoded full-length GLI1, GLI1ΔN, and tGLI1 isoforms. The full-length GLI1 gene is comprised of 12 exons, including the 5′-untranslated exon 1. The GLI1 coding region spans nt +79 to +3399 with the initiating methionine codon, ATG, at +79 in exon 2 (arrows). Exons are indicated as gray boxes while introns are shown by lines. The known functional domains of full-length GLI1 include the degron degradation signals (Dn and Dc; aa 77–116; 464–469), SUFU-binding domains (SU; aa 111–125 and C-terminus) (Dunaeva et al., 2003), zinc finger domains (ZF; aa 235–387), the nuclear localization signal (NLS; aa 380–420), and the transactivation domain (aa 1020–1091). Alternative splicing of GLI1 RNA can lead to the deletion of exons 1–3 totaling 128 amino acids in the N-terminus, forming the GLI1ΔN variant. The deletion of the entire exon 3 and part of exon 4 totaling 41 amino acids yields the tGLI1 isoform. Notably, tGLI1 retains all the known functional domains of the full-length GLI1.
In 2008, the first splice variant of GLI1 was discovered in which 128 amino acids were deleted from the N-terminus, and thus was termed GLI1ΔN (Shimokawa et al., 2008). As shown in Figure 1, the deletion of this region primarily affects the N-terminal SUFU-binding site and the N-terminal degron degradation signal of GLI1. The SUFU protein interacts with GLI1, and consequently sequesters GLI1 in the cytoplasm and prevents GLI1 from regulating gene transcription (Kogerman et al., 1999). However, GLI1ΔN variant displays a weakened ability to translocate to the nucleus and induce transcription of GLI1 target genes (Shimokawa et al., 2008). The GLI1ΔN variant can be detected in both normal and cancer cell lines (Shimokawa et al., 2008). However, we were unable to detect this variant in primary glioblastoma specimens (Lo et al., 2009).
In 2009, our laboratory discovered the novel tGLI1 isoform in which 41 amino acids are deleted, corresponding to alternative splicing of the entire exon 3 and part of exon 4 of the GLI1 gene (Lo et al., 2009). As shown in Figure 1, we found that tGLI1 retained all functional domains known to be present in GLI1 and preserved the ability to translocate to the nucleus, activate GLI1 target genes (e.g., PTCH1), and respond to Shh stimulation (Lo et al., 2009). Expression of tGLI1 was found to be tumor-specific as its presence was only detected in cell lines and primary specimens derived from human glioblastoma (Lo et al., 2009) and breast cancer (Cao et al., 2012), but not in normal brain tissues and other normal tissues. This distinctive pattern of tGLI1 differs from GLI1 and GLI1ΔN as both of these isoforms are expressed in both normal and cancer cells (Cao et al., 2012; Lo et al., 2009; Shimokawa et al., 2008). Detection of tGLI1 in the presence of GLI1 has proven difficult because of the following reasons. (1) There is a difference of only about 4.5 kD between the two proteins of large molecular weights (145-kD for tGLI1 versus 150-kD for GLI1), causing them to co-migrate in SDS-PAGE. To address this, our recent study has optimized an electrophoresis condition to resolve the two proteins (Cao et al., 2012). (2) Commercially available antibodies recognize both proteins (Cao et al., 2012; Lo et al., 2009) while antibodies specific to tGLI1 or GLI1 are not yet available. These technical difficulties could possibly have contributed to tGLI1 being discovered greater than 20 years after the discovery of GLI1. This also raises the question whether many functions attributed to GLI1 are, in fact, due to tGLI1. Therefore, the remainder of this review will be dedicated to describing the function of GLI1 and tGLI1 and their potential roles in the development of malignant cancer cell phenotypes.
Regulation of GLI1 Isoforms by the Hedgehog Signals
Canonical activation of GLI1 by Hedgehog signaling is initiated by Shh ligand binding to the receptor PTCH (Figure 2). Shh binding to PTCH releases PTCH-mediated constitutive suppression of the receptor SMO. Shh-mediated activation of SMO ultimately releases GLI1 from cytoplasmic sequestration by SUFU, allowing GLI1 to move to the nucleus and induce gene transcription (Kogerman et al., 1999). Mutations in some of the genes involved in the Shh-GLI1 pathway, such as SMO, PTCH, and SUFU, have been reported and these mutations are associated with the formation of cancer (Taylor et al., 2002; Villavicencio et al., 2000). No mutations have been reported for the GLI1 gene in normal or tumor tissue. Both splice variants tGLI1 and GLI1ΔN have been shown to be induced by Shh administration (Lo et al., 2009; Shimokawa et al., 2008). Thus, these alternatively spliced proteins are also controlled by the canonical Shh-GLI signaling pathway. It has yet to be investigated whether tGLI1 or GLI1ΔN interacts with components of the Shh pathway (e.g., direct SUFU cytoplasmic sequestration) similar to GLI1.
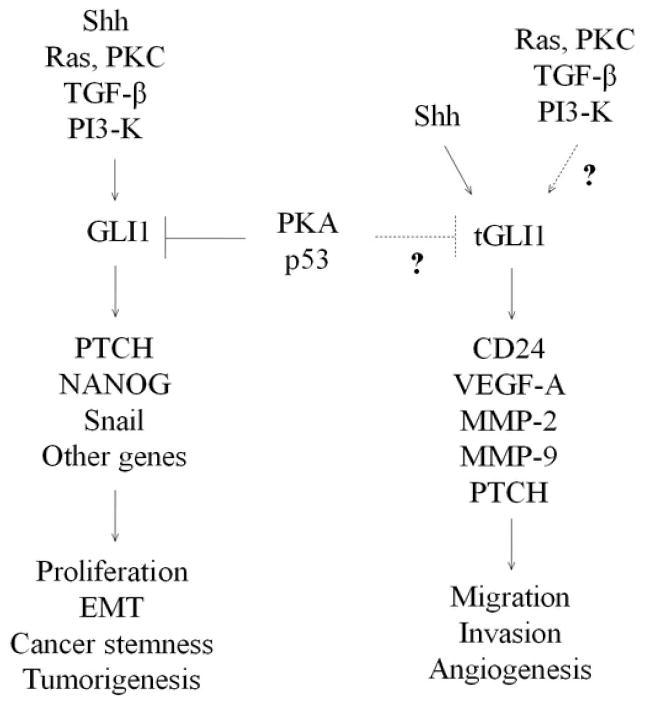
Figure 2.
GLI1 Isoforms and cancer characteristics. GLI1 is induced by Shh, Ras, TGF-β, and PI3-K, leading to induction of PTCH, NANOG, Snail, and other genes. Conversely, GLI1 is negatively regulated by PKA and p53. GLI1 expression is linked to proliferation, EMT, cancer stemness, and tumorigenesis. Shh can induce tGLI1 activity; however, it is unknown whether non-shh pathways can regulate tGLI1. Like GLI1, tGLI1 induces PTCH expression. Importantly, tGLI1 has gained the ability to transcriptionally upregulate expression of CD24, VEGF-A, MMP-2, and MMP-9 genes, and thereby promotes migration, invasion, and angiogenesis.
Regulation of GLI1 Isoforms by Non-hedgehog Signals
In addition to regulation by the canonical Shh signaling, recent evidence indicates that GLI1 can be regulated by several non-hedgehog signaling pathways (Figure 2). The Ras signaling pathway appears to regulate GLI1 as K-Ras expression induces GLI1-induced transcription (Seto et al., 2009) and suppression of GLI1 reduces the ability of K-Ras to induce cell transformation (Ji et al., 2007). This interaction appears to remain intact in vivo as mice overexpressing K-Ras form pancreatic adenocarcinoma that displays enhanced GLI1 expression despite deletion of the SMO gene (Nolan-Stevaux et al., 2009). In addition to Ras, GLI1 interacts with the tumor suppressor p53. Induction of p53 with oxaliplatin decreased cell proliferation, which was rescued by expression of GLI1 (Stecca and Ruiz I Altaba, 2009). To complement these results, expression of a dominant negative form of p53 induced tumor formation in frog embryos, which was reversed upon suppression of GLI1 (Stecca and Ruiz I Altaba, 2009). These and supporting experiments ultimately showed that p53 can inhibit the activity, localization to the nucleus, and protein levels of GLI1 while GLI1 was capable of repressing p53 protein expression (Stecca and Ruiz I Altaba, 2009). Lastly, GLI1 can interact with the TGF-β signaling pathway. TGF-β can induce expression of GLI1 despite inhibition of SMO (Dennler et al., 2007). This study also found that Smad3 or Smad4 knockdown prevented TGF-β induction of GLI1 (Dennler et al., 2007). Consistent with this, pancreatic adenocarcinoma cells that express GLI1 but are resistant to the SMO inhibitor cyclopamine are sensitive to TGF-β inhibition (Dennler et al., 2007). There is also some evidence that GLI1 can interact with other signaling pathways such as PI3K-AKT signaling (Stecca et al., 2007), protein kinase A (PKA) (Sheng et al., 2006), protein kinase C (PKC) (Cai et al., 2009), Notch (Schreck et al., 2010), and estrogen receptor-α (Xu et al., 2010).
While abundant evidence exists for non-canonical signaling pathway regulation of GLI1, investigations to determine the role of non-canonical signaling on levels and function of tGLI1 and GLI1ΔN have yet to be done. Regulation of these splice variants by signaling pathways may be due to regulation of the GLI1 parental gene. Conversely, tGLI1 and GLI1ΔN could have another layer of regulation by induction of splicing machinery. For example, inhibition of PKCι, a mediator of K-Ras-induced cell transformation suppresses RNA processing machinery (Guo et al., 2009). This possibly indicates that K-Ras-induced PKCι expression promotes activation of RNA processing machinery that could enhance the likelihood of splicing. In addition, loss of p53 is associated with increased presence of aberrant splicing (Moyret-Lalle et al., 2001; Turpin et al., 1999). Thus, the regulation of tGLI1 expression, which is only present in tumor tissue, is likely to be more complex than GLI1 with additional regulation of splicing machinery. The precise splicing molecules and conditions required for production of tGLI1 versus GLI1 have yet to be identified but will likely provide insight into the tumor-specific expression of tGLI1.
Expression Pattern of the GLI1 Isoforms and Their Effects on Proliferation
GLI1 has been reported to be overexpressed in normal tissues and several cancer tissues including glioblastoma (Cui et al., 2010; Lo et al., 2009), breast cancer (Cao et al., 2012; ten Haaf et al., 2009), and pancreatic adenocarcinoma (Nolan-Stevaux et al., 2009) among several others. GLI1ΔN has expression in both normal and cancer tissues (Shimokawa et al., 2008) whereas tGLI1 expression has only been detected in cancer tissues, glioblastoma and breast cancer, to date (Cao et al., 2012; Lo et al., 2009). Thus, these GLI1 forms may play a role in cancer development and progression.
Cancer can be defined as a disease of uncontrolled cellular proliferation. The GLI1 isoforms have been shown to be involved in proliferation of cells. For example, ectopic GLI1 expression in embryonic mouse brains leads to neural tissue outgrowth (Hynes et al., 1997; Stecca and Ruiz I Altaba, 2009). Inhibition of Shh-GLI1 signaling by cyclopamine or siRNA targeting of GLI1 reduces cell proliferation and tumor size (Clement et al., 2007; Wang et al., 2010). Epithelial expression of GLI1 leads to increases in proliferation as well as anchorage-independent proliferation (Kimura et al., 2005; Li et al., 2006; Yoon et al., 2002). GLI1 expression in mammary glands leads to hyperplasia and tumor formation (Fiaschi et al., 2009) whereas knockdown of GLI1 decreases proliferation and enhances apoptosis in inflammatory metastatic breast cancer cells (Thomas et al., 2011). However, the effects of tGLI1, relative to GLI1, on tumor proliferation remain largely unknown. Our recent study indicated that breast cancer cells expressing tGLI1 had a higher rate of proliferation compared to cells with GLI1 (Cao et al., 2012). Our unpublished data in glioblastoma xenografts are in support of the notion that tGLI1 promotes tumor growth to a greater extent than GLI1.
GLI1 Isoforms and Tumor Migration and Invasion
The progression of cancer is characterized by cells acquiring an enhanced ability to migrate to and invade adjacent tissues, both of which can precede metastasis. The SMO inhibitor cyclopamine can decrease melanoma cells’ ability to form lung metastases (Stecca et al., 2007) and reduce invasiveness of prostate cancer cells (Sheng et al., 2004). GLI1 expression is positively correlated with tumor grade and lymph node status (Stein et al., 1999; ten Haaf et al., 2009; Wang et al., 2010), indicating a role for GLI1 in metastasis. Our laboratory showed that tGLI1 had a stronger propensity than GLI1 to promote glioblastoma and breast cancer cell migration and invasion in vitro (Cao et al., 2012; Lo et al., 2009). Using glioblastoma xenograft mouse model, we further showed that tumor cells expressing tGLI1 were significantly more infiltrative than those with GLI1 (Lo et al., 2009). Subsequently, DNA microarray analysis and biochemical validations have led us to discover CD24 as a novel target of tGLI1 but not GLI1 and that CD24 is required for the enhanced migration and invasion of tGLI1-expressing cells (Lo et al., 2009). Thus, the evidence to date suggests tGLI1 to be a stronger promoter of tumor migration and invasion compared to GLI1 in two tumor types examined to date, glioblastoma and breast cancer. It remains unknown whether these observations can be extended to other types of human cancers.
GLI1 Isoforms and Epithelial-Mesenchymal Transition (EMT)
Another cell characteristic of epithelial-derived cancer cells is EMT, a de-differentiation program characterized by the loss of E-cadherin (epithelial marker) and the gain of vimentin and fibronectin (mesenchymal markers) (Yang et al., 2004). E-cadherin expression downregulation is typically mediated by transcriptional repressors, Slug, Snail, and TWIST (Lo et al., 2007). Different studies reported different effects of GLI1 on E-cadherin as some indicated that GLI1 represses E-cadherin by induction of Snail (Li et al., 2006; Louro et al., 2002) while others showed that GLI1 promotes E-cadherin expression and redistribution toward the cell membrane (Liao et al., 2009; Neill et al., 2008). However, reports are consistent with GLI1 induction of other EMT markers such as Snail (Li et al., 2006; Liao et al., 2009; Louro et al., 2002). There has been no investigation on whether tGLI1 can affect EMT. However, the discrepancies between studies on the role of GLI1 in EMT may be due to unknown tGLI1 expression in some studies leading to an apparent difference in results concerning the role of GLI1 in EMT.
GLI1 Isoforms and Tumor Angiogenesis
Progression of cancer turns on the angiogenic switch whereby tumor cells tip the balance toward pro-angiogenic environment, via production of pro-angiogenic factors (e.g., VEGF), which enhances tumor growth as angiogenesis is required for tumor growth beyond 2 mm in diameter (Bergers and Benjamin, 2003). The GLI1 pathways also appear to play a role in tumor angiogenesis as cyclopamine can reduce angiogenesis and production of pro-angiogenic factors from pancreatic adenocarcinoma (Nakamura et al., 2010). Specific knockdown of GLI1 can reduce VEGF production by glioma stem cells reducing their ability to promote angiogenesis in vitro (Hsieh et al., 2011). Notably, tGLI1 appears to be a stronger inducer of angiogenesis as medium from tGLI1-expressing cells promotes a stronger in vitro angiogenesis response compared to GLI1-expressing cells (Cao et al., 2012). Further analysis revealed VEGF-A as a novel and direct target of tGLI1 but not GLI1 (Cao et al., 2012). Thus, data showing that GLI1 promotes angiogenesis by VEGF (Hsieh et al., 2011; Nakamura et al., 2010) may actually be due to unknown tGLI1 expression. As mentioned above, the difference in molecular weight between GLI1 and tGLI1 is approximately 4.5 kD and very difficult to detect. Studies assessing the role of GLI1 have not assessed any concurrent tGLI1 expression and, thus, possibly attributing the role of tGLI1 to GLI1. Overall, these data reinforce our initial conclusions that tGLI1 promotes an advanced and aggressive cancer cell phenotype characterized by enhanced proliferation, migration, invasion, and angiogenesis.
GLI1 Isoforms and Cancer Stem Cells
The role of tGLI1 in cancer stemness is still unknown. GLI1 has been shown to be involved in maintenance of cancer stem cells by promoting the expression of stem cell markers (e.g., NANOG) in gliomaspheres and have enriched expression in mammospheres (Liu et al., 2006; Stecca and Ruiz I Altaba, 2009; Zbinden et al., 2010). The role of tGLI1 in the cancer stem cell phenotype has yet to be elucidated but tGLI1 does directly induce CD24 expression, which has been shown to be necessary for both pancreatic cancer stem cells (Li et al., 2006) and gastric cancer stem cells (Song et al., 2011) possibly indicating further roles of tGLI1 that may have been attributed to GLI1.
GLI1 Isoforms and Therapeutic Resistance
GLI1 may be involved in promoting cancer cell resistance to chemotherapy, which is a characteristic of aggressive cancer cells and often leads to poor patient prognosis. Glioma patients have a positive correlation between recurrence of tumors and GLI1 expression (Cui et al., 2010). Furthermore, addition of SMO-GLI1 pathway antagonist cyclopamine to chemotherapy enhanced cell death and apoptosis of cancer cells compared to chemotherapy alone (Cui et al., 2010). Clearly, the GLI1 pathway can enhance therapeutic resistance in this setting. The effect of tGLI1 on cancer cell resistance to therapy has not been initiated. However, considering that tGLI1 tends to promote an aggressive cancer cell phenotype, it would be worthwhile to investigate the role of tGLI1 in cellular resistance to therapy to fully understand the breadth of effects tGLI1 has on cancer cell behavior.
Conclusions and Future Directions
The need for reassessment of the GLI1 gene is necessary considering that recent data has altered the paradigm for this gene. Most notably, two alternatively spliced variants have been discovered in the GLI1ΔN and tGLI1 forms. The GLI1ΔN variant has 128 amino acids deleted at the N-terminus (Shimokawa et al., 2008) while the tGLI1 form has only 41 amino acids deleted near the N-terminus (Lo et al., 2009). While the GLI1ΔN isoform does not yet appear to have drastically different expression pattern or gene targets compared to GLI1 (Shimokawa et al., 2008), the tGLI1 isoform is exclusively expressed in tumor tissue and has multiple direct transcriptional targets different from GLI1 (Cao et al., 2012; Lo et al., 2009). These novel direct targets of tGLI1 include CD24, VEGF-A, MMP-2, and MMP-9. All of these genes can contribute to an aggressive cell phenotype that leads to increased tumor size, increased cell motility, increased tumor invasiveness, and increased tumor angiogenesis with tGLI1 expression (Cao et al., 2012; Lo et al., 2009). Thus, future directions to study the GLI1 gene and its different splice variant isoforms will require investigation into the relative importance of these isoforms for different functions of cancer cells and their potential clinical impact.
The precise function of tGLI1 compared to GLI1 will be an important avenue of investigation. The difference between tGLI1 and GLI1 is 41 amino acids, which translates to approximately 4.5 kD on a resolving gel. This minute difference in size, combined with the fact that most commercially available antibodies detect both isoforms, indicates the possibility that some previous functions attributed to GLI1 may have been due to tGLI1. For example, some studies indicate that GLI1 promotes the loss of E-cadherin (Louro et al., 2002) in epithelial cells promoting EMT whereas other studies indicate GLI1 may induce E-cadherin expression (Li et al., 2006). This discrepancy could be due to unknown tGLI1 presence in one of these studies. Therefore, determining levels of tGLI1 and GLI1 is important to fully understand the implications of similar experiments. We have optimized both RT-PCR and SDS-PAGE conditions to effectively determine the levels of RNA transcripts and protein, respectively. While these methods are effective, an antibody to specifically detect tGLI1 without GLI1 detection, and vice-versa, will further be important to identifying levels of these two proteins in different tissues. Thus, more specific detection strategies will be necessary to advance the understanding the role of tGLI1 and GLI1 in cancer.
These findings are indeed exciting and suggest several other potential avenues of investigation. First, it will be important to determine what regulates tGLI1 expression levels and whether other members of the Shh pathway are required for its expression. Second, it will be important to determine if any of the non-canonical pathways that lead to GLI1 expression also regulate tGLI1 levels, potentially linking several signaling pathways to a novel mechanism for tumor progression. Third, to further understand what regulates tGLI1 levels, the precise splicing mechanisms will be important to understand as the expression of this splicing machinery will likely regulate the expression of tGLI1 levels. Fourth, the mechanism by which tGLI1 activates target genes will be important in identifying novel targets of tGLI1 that potentially contribute to the aggressive phenotype of tGLI1-expressing cells. Lastly, it will be important to measure the relative expression of GLI1/tGLI1 to determine if patients have altered prognosis with different expression profiles and whether these profiles have a correlation with effectiveness of broad chemotherapies as well as Shh pathway-targeted therapies. Together this information could potentially lead to effective patient stratification ultimately leading to more effective treatments and enhanced longevity.
Acknowledgments
This study was supported by the NIH grant K01-CA118423, DOD grants W81XWH-07-1-0390 and W81XWH-11-1-0600, the Beez Foundation, and Intramural Division of Surgical Sciences Dani P. Bolognesi, Ph.D., Award (to H.W. L.).
Footnotes
Disclosure
The authors report no conflicts of interest.
References
- Arheden K, Ronne M, Mandahl N, Heim S, Kinzler KW, Vogelstein B, Mitelman F. In situ hybridization localizes the human putative oncogene GLI to chromosome subbands 12q13.3-14.1. Hum Genet. 1989;82(1):1–2. doi: 10.1007/BF00288260. [DOI] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Cai Q, Li J, Gao T, Xie J, Evers BM. Protein kinase Cδ negatively regulates Hedgehog signaling by inhibition of Gli1 activity. J Biol Chem. 2009;284(4):2150–2158. doi: 10.1074/jbc.M803235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Geradts J, Dewhirst MW, Lo HW. Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene. 2012;31(1):104–115. doi: 10.1038/onc.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, De Tribolet N, Radovanovic I, Ruiz I, Altaba A. Hedgehog-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288(1–2):156–166. doi: 10.1016/j.jns.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz I, Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126(14):3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz I, Altaba A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128(24):5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- Dennler S, Andre J, Alexaki I, Li A, Magnaldo T, Ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67(14):6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Fiaschi M, Rozell B, Bergstrom A, Toftgard R. Development of mammary tumors by conditional expression of GLI1. Cancer Res. 2009;69(11):4810–4817. doi: 10.1158/0008-5472.CAN-08-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Wu S, Wang L, Wang RY, Wei X, Liu J, Fang B. Interruption of RNA processing machinery by a small compound, 1-[(4-chlorophenyl)methyl]-1H-indole-3-carboxaldehyde (oncrasin-1) Mol Cancer Ther. 2009;8(2):441–448. doi: 10.1158/1535-7163.MCT-08-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol. 2011;226(4):1118–1127. doi: 10.1002/jcp.22433. [DOI] [PubMed] [Google Scholar]
- Hynes M, Stone DM, Dowd M, Pitts-Meek S, Goddard A, Gurney A, Rosenthal A. Control of cell pattern in the neural tube by the zinc finger transcription factor and oncogene Gli-1. Neuron. 1997;19(1):15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates Hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282(19):14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15(3):266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24(25):4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236(4797):70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332(6162):371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10(2):634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1(5):312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Li X, Deng W, Nail CD, Bailey SK, Kraus MH, Ruppert JM, Lobo-Ruppert SM. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25(4):609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Siu MK, Au CW, Wong ES, Chan HY, Ip PP, Ngan HY, Cheung AN. Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30(1):131–140. doi: 10.1093/carcin/bgn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H-W, Zhu H, Cao X, Aldrich A, Ali-Osman F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69(17):6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67(19):9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso PM, Rudin CM, Borad MJ, Vernillet L, Darbonne WC, Mackey H, Dimartino JF, De Sauvage F, Low JA, Von Hoff DD. A first-in-human, first-in-class, phase (ph) I study of systemic Hedgehog (Hh) pathway antagonist, GDC-0449, in patients (pts) with advanced solid tumors. J Clin Oncol (Meeting Abstracts) 2008;26(15_suppl):3516. [Google Scholar]
- Louro ID, Bailey EC, Li X, South LS, Mckie-Bell PR, Yoder BK, Huang CC, Johnson MR, Hill AE, Johnson RL, Ruppert JM. Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res. 2002;62(20):5867–5873. [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304(5678):1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Moyret-Lalle C, Duriez C, Van Kerckhove J, Gilbert C, Wang Q, Puisieux A. p53 induction prevents accumulation of aberrant transcripts in cancer cells. Cancer Res. 2001;61(2):486–488. [PubMed] [Google Scholar]
- Nakamura K, Sasajima J, Mizukami Y, Sugiyama Y, Yamazaki M, Fujii R, Kawamoto T, Koizumi K, Sato K, Fujiya M, Sasaki K, Tanno S, Okumura T, Shimizu N, Kawabe J, Karasaki H, Kono T, Ii M, Bardeesy N, Chung DC, et al. Hedgehog promotes neovascularization in pancreatic cancers by regulating Ang-1 and IGF-1 expression in bone-marrow derived pro-angiogenic cells. PLoS One. 2010;5(1):e8824. doi: 10.1371/journal.pone.0008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill GW, Harrison WJ, Ikram MS, Williams TD, Bianchi LS, Nadendla SK, Green JL, Ghali L, Frischauf AM, O’Toole EA, Aberger F, Philpott MP. GLI1 repression of ERK activity correlates with colony formation and impaired migration in human epidermal keratinocytes. Carcinogenesis. 2008;29(4):738–746. doi: 10.1093/carcin/bgn037. [DOI] [PubMed] [Google Scholar]
- Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23(1):24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001;106(1–2):107–117. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Wild A, Grzeschik KH, Antonarakis SE. Mutation in GLI3 in postaxial polydactyly type A. Nat Genet. 1997;17(3):269–271. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- Raffel C, Jenkins RB, Frederick L, Hebrink D, Alderete B, Fults DW, James CD. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57(5):842–845. [PubMed] [Google Scholar]
- Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz I, Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell TM, et al. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell. 1994;76(4):761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14(3):357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, Lorusso PM, Von Hoff DD, De Sauvage FJ, Low JA. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, Eberhart CG. The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: a potential mechanism of therapeutic resistance. Clin Cancer Res. 2010;16(24):6060–6070. doi: 10.1158/1078-0432.CCR-10-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K, Mohri D, Tanaka Y, Ijichi H, Tateishi K, Kanai F, Kawabe T, Omata M. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol Carcinog. 2009;48(8):703–712. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- Sheng T, Chi S, Zhang X, Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. J Biol Chem. 2006;281(1):9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- Sheng T, Li C, Zhang X, Chi S, He N, Chen K, Mccormick F, Gatalica Z, Xie J. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3(1):29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgard R, Zaphiropoulos PG. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J Biol Chem. 2008;283(21):14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Yue W, Wei B, Wang N, Li T, Guan L, Shi S, Zeng Q, Pei X, Chen L. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PLoS One. 2011;6(3):e17687. doi: 10.1371/journal.pone.0017687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz I, Altaba A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104(14):5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz I, Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28(6):663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein U, Eder C, Karsten U, Haensch W, Walther W, Schlag PM. GLI gene expression in bone and soft tissue sarcomas of adult patients correlates with tumor grade. Cancer Res. 1999;59(8):1890–1895. [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Stavrou T, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- Ten Haaf A, Bektas N, Von Serenyi S, Losen I, Arweiler E, Hartmann A, Knuchel R, Dahl E. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer. 2009;9(1):298. doi: 10.1186/1471-2407-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ZI, Gibson W, Sexton JZ, Aird KM, Ingram SM, Aldrich A, Lyerly HK, Devi GR, Williams KP. Targeting GLI1 expression in human inflammatory breast cancer cells enhances apoptosis and attenuates migration. Br J Cancer. 2011;104(10):1575–1586. doi: 10.1038/bjc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin E, Dalle B, De Roquancourt A, Plassa LF, Marty M, Janin A, Beuzard Y, De The H. Stress-induced aberrant splicing of TSG101: association to high tumor grade and p53 status in breast cancers. Oncogene. 1999;18(54):7834–7837. doi: 10.1038/sj.onc.1203196. [DOI] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67(5):1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Lorusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, Darbonne WC, Marsters JC, Jr, De Sauvage FJ, Low JA. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang Z, Xu Z, Yin H, Bai L, Ma Z, Decoster MA, Qian G, Wu G. Activation of the sonic hedgehog signaling controls human pulmonary arterial smooth muscle cell proliferation in response to hypoxia. Biochim Biophys Acta. 2010;1803(12):1359–1367. doi: 10.1016/j.bbamcr.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, De Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Xu L, Kwon Y-J, Frolova N, Steg A, Yuan K, Johnson M, Grizzle W, Desmond R, Frost A. Gli1 promotes cell survival and is predictive of a poor outcome in ERα-negative breast cancer. Breast Cancer Res Treat. 2010;123(1):59–71. doi: 10.1007/s10549-009-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA, Jacob H, Walterhouse D, Iannaccone P. Gene Expression Profiling Leads to Identification of GLI1-binding Elements in Target Genes and a Role for Multiple Downstream Pathways in GLI1-induced Cell Transformation. J Biol Chem. 2002;277(7):5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz I, Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29(15):2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Lo H-W. The Human Glioma-associated Oncogene Homolog 1 (GLI1) Family of transcription factors in gene regulation and diseases. Curr Genomics. 2010;11:238–245. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]