Abstract
Glioma-associated oncogene homolog 1 (GLI1) is the nuclear mediator of Hedgehog signaling that activates gene transcription via its zinc finger DNA-binding and transactivation domains. GLI1 plays a critical role in several cellular processes, including embryonic development, tumorigenesis, and tumor growth and progression. The human GLI1 gene was identified in 1987 as an amplified gene in glioblastoma. Somatic mutations have never been reported in the GLI1 gene in any cell or tumor type. Very recently in 2008–2009, the full-length GLI1 transcript was discovered to undergo alternative splicing to form two shorter isoforms, namely N-terminal deletion variant (GLI1ΔN) and truncated GLI1 (tGLI1). Emerging evidence suggests that the three structurally different GLI1 isoforms are distinctly different in their expression patterns and functions in the context of human cancers. The tGLI1 isoform, in particular, has been shown to gain the ability to modulate expression of the genes that are not regulated by GLI1 and to support the biology of more aggressive cancer. Consequently, a key focus of this chapter is to summarize and compare the properties of the three GLI1 isoforms and their relations to malignant biology of human cancers.
I. Introduction
The hedgehog signaling pathway is conserved in both vertebrates and invertebrates and is linked to a broad array of biological processes most prominently embryonic development (Ingham et al., 2011), tumorigenesis, and cancer progression (Yang et al., 2010; Zhu and Lo, 2010). Mammalian hedgehog signaling is initiated by three ligands: Sonic Hedgehog (Shh), Indian Hedghog (Ihh), and Desert Hedgehog (Dhh). Shh is the most potent of the three ligands (Pathi et al., 2001) and is expressed most widely in embryos and adult tissues (Ingham and McMahon, 2001). Aberrant Shh signaling is implicated in endodermally derived human cancers that account for up to 25% of human cancer deaths (Lum and Beachy, 2004). The main effectors of hedgehog signaling in Drosophila and mammals are Cubitus interruptus (Ci) and the GLI (glioma-associated oncogene homolog) proteins (GLI1, GLI2, GLI3), respectively. Hedgehog induces GLI protein translocation to the nucleus where transcription can be induced primarily by GLI1 and/or GLI2 or repressed primarily by GLI2 and/or GLI3. Via GLI-mediated gene regulation, Hedgehog plays an essential role in many important cellular processes in normal and cancerous cells. Consequent to the pivotal role that the Hedgehog pathway plays in tumor growth and progression, extensive efforts have been invested in the development of Hedgehog-targeted therapy (Ng and Curran, 2011; Yang et al., 2010). Notably, a small molecule inhibitor of smoothened (SMO), Vismodegib/GDC-0449, is being evaluated in several clinical trials for cancers (LoRusso et al., 2008; Rudin et al., 2009; Von Hoff et al., 2009).
The human GLI1 gene was discovered in 1987 by Vogelstein and colleagues, as an amplified gene in human glioblastoma multiforme, GBM (Kinzler et al., 1987). It was later found to encode for a zinc finger transcription factor belonging to the Kruppel family of zinc finger proteins (Kinzler et al., 1988). The clinical importance of GLI1 gene amplification is still unclear as early studies found low rates of GLI1 amplification ranging from 1.6% to 3.3% occurrence in gliomas (Bigner et al., 1987; Forus et al., 1993; Wong et al., 1987) whereas a more recent study reported a 22.6% occurrence in gliomas (Rao et al., 2010). Other cancers were observed to have varying ranges of occurrences for GLI1 amplification with the highest occurrence of 28% in rhabdomyosarcomas (Gordon et al., 2000). Amplification of the GLI1 gene has been shown to result in increased GLI1 gene expression (Reifenberger et al., 1994).
Somatic mutations have never been reported in the GLI1 gene in any cell or tumor type, unlike other components of the Hedgehog pathway that contain somatic mutations such as Shh, patched (PTCH), SMO, suppressor of fused (SUFU), GLI2, and GLI3 (Kang et al., 1997; Radhakrishna et al., 1997; Raffel et al., 1997; Roessler et al., 1996; Xie et al., 1998). Very recently in 2008 and 2009, the full-length GLI1 transcript was found to undergo alternative splicing to form two shorter isoforms, namely N-terminal deletion variant (GLI1ΔN) identified by Shimokawa et al. (2008) and truncated GLI1 (tGLI1) identified in our laboratory (Lo et al., 2009). While both GLI1 variants are under intensive investigations for their properties and functions, emerging evidence indicates that the three GLI1 isoforms differ in their expression patterns and their ability to regulate gene activation. In comparing tGLI1 to GLI1, our results indicated that although they display some overlapping properties, the two isoforms demonstrate distinctly different functions by regulating different sets of target genes (Cao et al., 2012; Lo et al., 2009). These differences may lead to their unique differences in mediating cellular processes important to tumor behaviors such as proliferation, migration, invasion, angiogenesis, and metastasis. To provide a comprehensive review of the GLI1 isoforms in relation to human cancers, we will summarize the structures and biochemical properties of the three GLI1 isoforms (Section II), provide an overview of the regulation of the GLI1 isoforms by the canonical and noncanonical signaling pathways (Section III), and compare the malignant phenotypes of human cancers that can be modulated by the GLI1 isoforms (Section IV).
II. Structures and Properties of GLI1 Isoforms
A. GLI1
The human GLI1 gene was identified in 1987 by Vogelstein and colleagues (Kinzler et al., 1987). In this study, the investigators found evidence of gene amplification in a human GBM cell line and mapped the gene to chromosome 12 (12q13-14.3). The gene locus did not correspond to already well-known oncogenes such as MYC or RAS, or other oncogenes known to be located on chromosome 12. Subsequently, the amplified gene was named as GLI for the glioma tumor in which it was discovered. The GLI gene was later mapped, more specifically, to 12q13.3-14.1 (Arheden et al., 1989). The GLI gene was found to contain 3318 bp as part of the open reading frame that predicted a 1106-residue protein with a calculated molecular mass of 118kDa (Kinzler et al., 1988). The encoded protein was detected by immunoblotting soon afterward and observed to migrate as a 150kDa protein (Kinzler and Vogelstein, 1990). It was later concluded that the discrepancy between calculated and apparent molecular mass was likely due to the intrinsic amino acid sequence as the GLI1 protein synthesized from in vitro translation migrated similarly as the 150kDa GLI1 protein isolated from the cells.
The GLI1 protein contains five successive repeats of a zinc finger DNA-binding consensus sequence (Fig. 6.1A). It was determined that the GLI1 protein belongs to the Kruppel family of zinc finger proteins as it contained the H–C link between zinc fingers common to this family of transcription factors. This finding defined GLI1 as the first human member of the Kruppel family of proteins (Kinzler et al., 1988). GLI1 was detected in the nuclei of the cells of cancer cell lines, xenografts, and primary tumor tissue, which is consistent with its structure being that of a zinc finger transcription factor (Kinzler and Vogelstein, 1990). In the nucleus, the GLI protein binds to a 9-bp DNA sequence of 5′-GACCACCCA-3′. Later analysis of GLI-DNA binding found that only fingers 2–5 bind to the major groove of DNA wrapping around for a full helical turn with fingers 4–5 having the most contact (Pavletich and Pabo, 1993). It was also reported that finger one does not even contact DNA but rather has protein–protein interaction with finger two. These early studies characterized the GLI gene to encode for a zinc finger transcription factor and upon discovery of GLI2 and GLI3, this original GLI gene was termed GLI1.
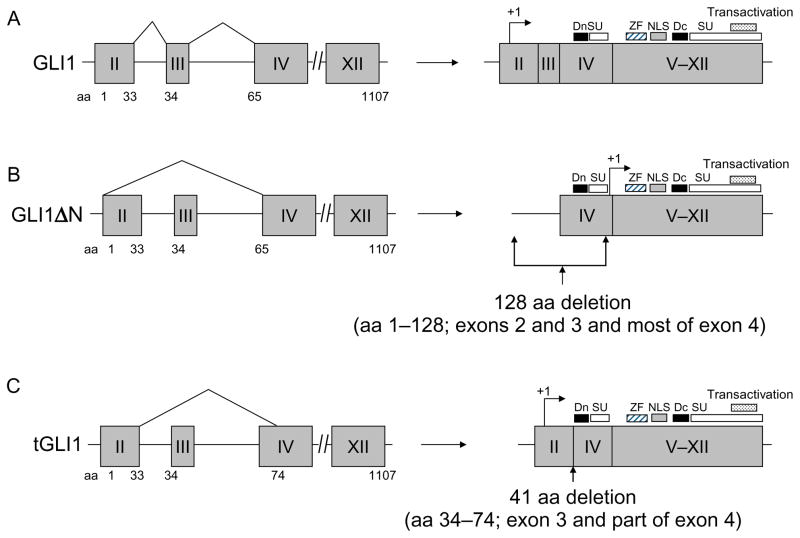
Figure 6.1.
Structures of the human GLI1 gene and the encoded full-length GLI1, GLI1ΔN and tGLI1 isoforms. (A) The full-length GLI1 gene comprises 12 exons, including the 5′-untranslated exon 1. The GLI1 coding region spans nt +79 to +3399 with the initiating methionine codon, ATG, at +79 in exon 2 (arrows). Exons are indicated as gray boxes while introns are shown by lines. The known functional domains of full-length GLI1 include the degron degradation signals (Dn and Dc; aa 77–116; 464–469), SUFU-binding domains (SU; aa 111–125 and C-terminus) (Dunaeva et al., 2003), zinc finger domains (ZF; aa 235–387), the nuclear localization signal (NLS; aa 380–420), and the transactivation domain (aa 1020–1091). (B) Alternative splicing of GLI1 RNA can lead to the deletion of exons 1–3 totaling 128 amino acids in the N-terminus, forming the GLI1ΔN variant. (C) The deletion of the entire exon 3 and part of exon 4 totaling 41 amino acids yields the tGLI1 isoform. Notably, tGLI1 retains all the known functional domains of the full-length GLI1.
B. GLI1ΔN
In 2008, Shimokawa et al. reported the existence of a splice variant of GLI1 that has 128 amino acids deleted from the N-terminus as a result of splicing exon 1 directly to exon 4 (Shimokawa et al., 2008). Due to the deletion of this region from the N-terminus, this variant was termed GLI1ΔN (Fig. 6.1B). Primary functional domains remained intact in the GLI1ΔN variant include the zinc finger domains, the transactivation domain, the nuclear localization signal (NLS), and the nuclear export signal. The SUFU-binding site was lost in GLI1ΔN. Notably, the SUFU-binding domain in the GLI1 protein interacts with cytoplasmic SUFU complex and consequently, is sequestered in the cytoplasm (Kogerman et al., 1999). In contrast to the prediction that GLI1ΔN would be enriched in the nucleus because of the loss of SUFU-binding domain, GLI1ΔN showed a weakened ability to translocate into the nucleus, which leads to weaker activation of GLI1 target genes such as GLI1 and PTCH. Further, GLI1ΔN was detected in several cancer cell lines derived from human tumors as well as in human embryonic kidney cells indicating it may have relevance in human tissues (Shimokawa et al., 2008). Our study showed that GLI1ΔN is undetectable in GBMs (Lo et al., 2009). The significance of GLI1ΔN in normal physiology and cancer biology is still largely unknown and remains to be defined.
C. tGLI1
In 2009, our laboratory reported the discovery of tGLI1 when analyzing cell lines and tumor tissue of GBM (Lo et al., 2009). As summarized in Fig. 6.1C, tGLI1 is a product of alternative splicing that lacks 41 amino acids corresponding to the entire exon 3 and part of exon 4 of the GLI1 gene. Analysis of the GLI1 gene in GBM cell lines and normal peripheral lymphocytes found no somatic abnormality in the GLI1 gene. tGLI1 retains all the known functional domains present in full-length GLI1, including the zinc finger domains, transactivation domain, NLS, degrons signals, and the SUFU-binding domains. In line with the preservation of these functional domains, tGLI1 retains its ability to translocalize into the nucleus, transactivate GLI1 binding sites and activates expression of PTCH1, a GLI1-regulated gene. Like GLI1, tGLI1 responds to Shh stimulation (Lo et al., 2009).
The expression pattern of tGLI1 is significant in the context of human cancers because currently available evidence suggests that tGLI1 is expressed in a tumor-specific fashion (Cao et al., 2012; Lo et al., 2009). For example, tGLI1 is highly expressed in cell lines and primary specimens of GBM but not in normal brain tissues and other normal tissues (Lo et al., 2009). More recently, we further reported that tGLI1 is frequently expressed in breast cancer cell lines and primary tumors but rarely in normal mammary tissues (Cao et al., 2012). This pattern of expression for tGLI1 is distinctly different than those of GLI1 and GLI1ΔN, which are expressed in both normal and cancer cells (Cao et al., 2012; Lo et al., 2009; Shimokawa et al., 2008).
Notably, the molecular weight difference between the tGLI1 and GLI1 proteins is only about 4.5kDa, which makes it difficult to distinguish the two isoforms using immunoblotting (146kDa for tGLI1 and 150kDa for GLI1). Commercially available antibodies appear to recognize both isoforms (Cao et al., 2012; Lo et al., 2009). These technical limitations may have contributed to the inability to uncover the existence of tGLI1 for more than 20years after the 1987 discovery of the GLI1 gene. These notions also suggest that some of the known GLI1 functions may be attributed to tGLI1 but not GLI1. Evidence supporting this speculation will be discussed in detail in Section IV. Importantly, we have recently optimized a gel electrophoresis condition that can effectively resolve the GLI1 and tGLI1 proteins (Cao et al., 2012). Although the combination of transcript analysis (by RT-PCR and DNA sequencing of the PCR product) with protein analysis (by gel electrophoresis followed by immunoblotting) adequately and accurately distinguish tGLI1 from GLI1, it is recognized that a tGLI1-specific antibody is needed to selectively detect the tGLI1 protein but not GLI1, in order to help further decipher the functions of the two isoforms and to gain a greater understanding of the long overlooked tGLI1 isoform. In Section IV, we will discuss in detail the transcriptional functions of tGLI1 and its ability to modulate several important malignant phenotypes of human cancers, including growth, migration, invasion, and angiogenesis.
III. Regulation of GLI1 Isoforms by Canonical and Noncanonical Pathways
A. Canonical pathway
Canonical Hedgehog signaling is initiated by binding of the ligand Shh to the 12-transmembrane receptors PTCH 1 and 2 (Fig. 6.2A). Shh binding to PTCHproteins relieves constitutive PTCHrepression of the 7-transmembrane protein receptor SMO. Activation of SMO leads to the release of GLI1 from SUFU-mediated cytoplasmic sequestration and to GLI1 nuclear translocalization (Kogerman et al., 1999). SMO also increases levels of full-length GLI2/3, which proceeds to induce transcription of GLI1. GLI1 acts on many target genes to regulate several physiological processes and its aberrant activity causes several pathologies including tumor progression.
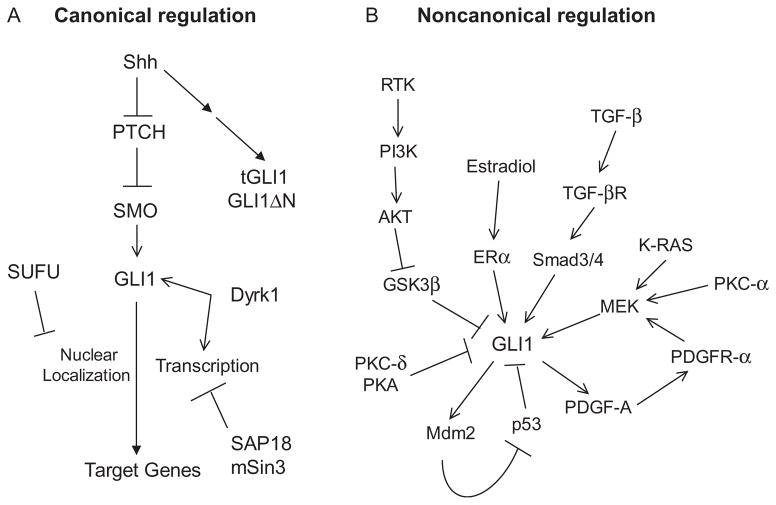
Figure 6.2.
Regulation of GLI1 the isoforms by the canonical and noncanonical pathways. (A) Canonical activation of GLI1 is initiated by Shh, which binds and inhibits PTCH1 relieving PTCH1-mediated suppression of SMO. Derepressed SMO releases GLI1 from SUFU-mediated cytoplasmic sequestration. In turn, GLI1 translocalizes to the nucleus to induce transcription of target genes. Dyrk1 enhances GLI1 activity. Both GLI1ΔN and tGLI1 respond to Shh; however, it is unknown whether they are subjected to the regulation by other components of the Hedgehog pathway. (B) GLI1 can be affected by several noncanonical pathways. Activators of GLI1 include TGF-β, K-Ras, PKC-α, PDGF-A, PI3K-AKT, and estradiol. PKC-δ, PKA, and p53 suppress GLI1. The response of tGLI1 or GLI1ΔN to noncanonical pathway activation is currently unknown.
GLI1 nuclear translocation and induction of transcription is further controlled by Shh pathway proteins. SUFU was cloned in 1999 as a 484 residue protein (Kogerman et al., 1999). SUFU physically interacts with GLI1 and the interaction traps GLI1 in the cytoplasm and prevents it from translocating into the nucleus. Expression of a GLI1 mutant that is constitutively localized to the nucleus also showed SUFU directly interacted with GLI1 while in the nucleus and reduced GLI1-mediated transcriptional activity (Kogerman et al., 1999). It was later discovered that SUFU forms a complex with SAP18 and mSin3, a transcription corepressor, and this complex directly binds GLI1 on DNA to repress GLI1-mediated transcription (Chen et al., 2002). While SUFU negatively regulates GLI1, the dual specificity Yak1-related kinase Dyrk1 appears to potentiate GLI1 activity. Dyrk1 phosphorylates GLI1 at multiple serine/threonine sites and induce nuclear accumulation of GLI1 (Mao et al., 2002). This study also showed that Dyrk1 enhances transcription by a mutant GLI1, which is constitutively localized to the nucleus, indicating Dyrk1 can enhance the process of GLI1-mediated transcription and not only nuclear localization.
Canonical hedgehog signaling is critical for Shh-induced patterning during embryonic development. For example, Shh-GLI1 signaling mediates the induction of floor plate cells in the posterial neural tube (Lee et al., 1997). Shh-GLI1 signaling also directly induced transcription of HNF-3β, a winged-helix transcription factor that mediated expression of floor plate differentiation in mice (Sasaki et al., 1997). Shh-GLI1 signaling also regulates pathological processes, most notably several cancer types (Kasper et al., 2006). Mutations in SMO, PTCH1, and SUFU have been reported and were associated with formation of cancer (Taylor et al., 2002; Villavicencio et al., 2000). The role of GLI1 in hedgehog-associated tumorigenesis is still not well understood with mixed results being reported (Kimura et al., 2005; Villavicencio et al., 2000).
Limited information is available in the regulation of GLI1ΔN and tGLI1. What is known is that Shh upregulates GLI1ΔN to a similar level as GLI1 (Shimokawa et al., 2008). It is also known that tGLI1 responds to Shh stimulation to a similar extent as GLI1, as reported in our study (Lo et al., 2009). However, the role of SUFU in regulating the cytoplasmic-nuclear shuttling of GLI1ΔN and tGLI1 is still not well understood. GLI1ΔN does not retain the N-terminal SUFU-binding domain, suggesting that its nuclear import may be regulated by the C-terminal SUFU-binding domain or by other means (Shimokawa et al., 2008). It is yet to be investigated whether tGLI1 interacts with SUFU, although it is expected that tGLI1 will associate with SUFU since tGLI1 retains the SUFU-binding domains. Also uninvestigated are the stability of GLI1ΔN and tGLI1 and their relationships to other components of the Hedgehog pathway such as SMO, GLI2, and GLI3. Taken together, future efforts are required to elucidate the regulatory role that the Hedgehog-PTCH-SMO signaling axis plays in GLI1ΔN and tGLI1.
B. Noncanonical pathway
Canonical activation of the Shh–PTCH–SMO pathway had been thought to be the primary mechanism for control of GLI1. However, evidence continues to accumulate that GLI1 can also be regulated by other signaling pathways and proteins outside of the canonical Shh–PTCH–SMO pathway (Fig. 6.2B). Elucidation of these pathways could potentially be important for treatment strategies as redundant regulation of GLI1 could lead to cancers that develop resistance to Shh-targeted therapies.
Gastric cancer patients showed a positive correlation between activation of ERK and PTCH expression, a GLI1 gene target (Seto et al., 2009). Expression of K-Ras increased GLI-induced transcription, which was suppressed by a MEK inhibitor. The effect of K-Ras-MEK signaling on GLI1 is independent of SUFU as knockdown of SUFU did not alter K-Ras induction of GLI1 (Seto et al., 2009). Further, suppression of GLI1 inhibits cell transformation induced by K-Ras overexpression ( Ji et al., 2007). Perhaps the strongest evidence that Ras can control GLI1 is that mice overexpressing K-Ras, which induces pancreatic adenocarcinoma, strongly expressed GLI1 and PTCH despite the fact that these mice lack the SMO gene (Nolan-Stevaux et al., 2009). Suppression of GLI1 in cancer cells isolated from these mice showed reduced indices of cell transformation and decreased K-Ras indicating positive feedback between GLI1 and K-Ras. Thus, the Ras–MEK–ERK pathway has the ability to regulate GLI1. Despite clear evidence for this regulation, the mechanism by which Ras–MEK regulates GLI1 levels has still not been discovered.
Another pathway that has recently gained attention for GLI1 regulation is the TGF-β-Smad pathway. Mice overexpressing TGF-β in the epidermis also overexpress GLI1 (Dennler et al., 2007). TGF-β treatment induces cellular expression of GLI1 that was independent of the SMO inhibitor cyclopamine (Dennler et al., 2007). Further, knockdown of Smad4 or Smad3 in these cells prevented GLI protein induction by TGF-β. The mechanism of GLI1 induction by TGF-β was further elucidated when loss of GLI2 prevented induction of GLI1 by TGF-β (Dennler et al., 2007). Adding to this, the GLI2 gene was found to have a TGF-β-responsive region (Dennler et al., 2009). Thus, it appears that TGF-β directly induces GLI2, which then promotes GLI1 production. Results also show pancreatic adenocarcinoma cells with enhanced GLI expression, but are resistant to cyclopamine, are sensitive to TGF-β inhibitors or GLI2 inhibition (Dennler et al., 2007). Since GLI1 expression can be controlled by GLI2 in response to TGF-β, it stands to reason that the tGLI1 and GLI1ΔN variants should be inducible by GLI2 in order to be responsive to TGF-β. However, exactly what drives expression of these variants is still unknown.
There is recent evidence to suggest the p53 tumor suppressor also can regulate GLI1. Intracranial tumor development in mice was seen to increase with p53 knockdown or with increased GLI1 expression but tumors growth was synergistic when both p53 was suppressed and GLI1 expression increased (Stecca and Ruiz i Altaba, 2009). Increasing p53 levels in these tumor cells by oxaliplatin treatment decreased cell proliferation, which was rescued by enhancement of GLI1 levels. Similarly, expression of a p53 dominant negative in frog embryos induced tumor formation that could be rescued with GLI1 suppression (Stecca and Ruiz i Altaba, 2009). Further, p53 knockdown decreased GLI1-induced transcription while p53 overexpression reduced GLI1-induced transcription. These data clearly indicate an inhibitory loop between GLI1 and p53. Another study found that GLI1 enhanced MDM2 interaction with p53, which led to p53 degradation indicating an indirect regulation by GLI1 (Abe et al., 2008). The mechanism of GLI1 regulation by p53 was not assessed and could be direct or indirect. Since GLI1 regulates p53 via its physical association with MDM2, it is difficult to predict whether tGLI1 and/or GLI1ΔN have influence on MDM2-p53 because it is not known whether the deletions within the two isoforms have an impact on their ability to interact with MDM2. In the case of tGLI1, we speculate that it may interact with proteins and promoter sequences in a different fashion compared to GLI1. This speculation is built on our findings showing that tGLI1, but not GLI1, binds to the promoters of CD24 and VEGF-A genes (Cao et al., 2012; Lo et al., 2009).
Signaling through the PI3K–AKT pathway appears to participate in GLI1 regulation, but perhaps not independent of Shh signaling. GLI1-induced transcription can be enhanced by IGF-1, which activates PI3K–AKT signaling, and Shh-induced GLI1 can be inhibited with PI3K inhibitors (Riobo et al., 2006). However, activation of PI3K signaling by IGF-1 could not induce GLI-mediated transcription independent of Shh. AKT knockdown reduced Shh-induced transcription by GLI1. Another study found that inhibition of PI3K signaling reduced cell proliferation induced by ectopic GLI1 expression (Stecca et al., 2007). This study showed PTEN, an endogenous inhibitor to PI3K signaling, could inhibit GLI1 nuclear localization and transcription. Evidence indicates a clear role for PI3K-AKT signaling in Shh-induced GLI1 but the precise mechanism for this regulation has not been made clear. Regulation of tGLI1 or GLI1ΔN by PI3K-AKT signaling is unknown.
Protein kinase A (PKA) negatively regulates GLI1. The first evidence of PKA-mediated negative regulation of GLI1 came from expression of dominant-negative PKA in mouse embryos leading to effects that mimic GLI1 hyperactivation (Epstein et al., 1996). More direct evidence showed that activation of PKA by forskolin resulted in an alteration of GLI1 localization from the nucleus to the cytoplasm (Sheng et al., 2006). PKA phosphorylates GLI1 at Thr374, which when mutated to valine prevented PKA-induced cytoplasmic localization (Sheng et al., 2006). Threonine 374 is adjacent to the NLS on GLI1. The phosphorylation-mimicking Thr374Asp mutant is sequestered in the cytoplasm. Consistent with these findings, activation of PKA decreased GLI1-mediated transcription indicative of GLI1 cytoplasmic localization. PKA negative regulation of GLI1 has also been shown previously in the context of embryonic development in which PKA inhibition led to hyperproliferation in several brain areas resulting in malformations (Epstein et al., 1996). Although PKA likely retains the ability to phosphorylate both tGLI1 and GLI1ΔN as Thr374 is present in both variants, this has not been established by experimental evidence.
PKC-α was found to enhance GLI1-mediated transcription in a MEK-dependent manner whereas PKC-δ inhibited GLI1-mediated transcription (Cai et al., 2009). GBM neurospheres that are resistant to Notch inhibitors were found to have increased GLI1 levels as the Notch target gene Hes1 directly interacted with the GLI1 promoter suggesting these cells become resistant by Notch-induced activation of GLI1 (Schreck et al., 2010). GLI1 expression can also induce phosphorylation of ERK2 that is dependent on PDGFR-α, which is a direct GLI1 target (Xie et al., 2001). In these cells, inhibition of MEK-ERK2 signaling or neutralizing antibodies to PDGF-A reduced enhanced proliferation rates due to GLI1 expression (Xie et al., 2001). Breast cancer cells exposed to 17β-estradiol displayed increased GLI1 levels that could be prevented with an estrogen receptor-α (ER-α) inhibitor (Xu et al., 2010). The potential for these pathways to regulate tGLI1 or GLI1ΔN levels, nuclear localization, or transcriptional activity is unclear as there is not currently a mechanism for how many of these noncanonical pathways influence GLI1 expression or activity.
GLI1 protein stability can be regulated by the noncanonical pathways. The GLI1 protein has two degron degradation signals with one on the N-terminus and the other on the C-terminus. The C-terminal degron signal within GLI1 had the motif of DSGVEM (Huntzicker et al., 2006), which is a consensus sequence recognized by the E3 ubiquitin ligase βTrCP ( Jiang and Struhl, 1998). Increased βTrCP expression resulted in decreased GLI1 protein levels (Huntzicker et al., 2006). GLI1 was seen to immunoprecipitate with βTrCP in a manner dependent on the presence of the DSGVEM motif. In addition, ubiquitinylated GLI1 was immunoprecipitated with βTrCP whereas mutation of the DSGVEM motif resulted in no detectable ubiqutinylated GLI1 (Huntzicker et al., 2006). Further deletion of residues 51–116 on the N-terminus of GLI1 resulted in further stabilization beyond that provided by deletion of the C-terminal degrons signal indicating an N-terminal degron was likely present. However, the N-terminal degrons signals were not affected by βTrCP or by SUFU suggesting an alternative method of degradation by this degron signal (Huntzicker et al., 2006). The importance of GLI1 protein stability controlled by these degron signals was illustrated in this same study by mice expressing GLI1 mutants. Mice expressing double mutants where both degrons were inactivated displayed a more severe phenotype of basal cell carcinoma compared to single mutants. The tGLI1 variant, although lacking several residues, is not missing residues in either degron signal and would not be expected to have altered degradation compared to GLI1. However, GLI1ΔN has lost the N-terminal degron signal but protein stability was not assessed in the one study examining this variant (Shimokawa et al., 2008).
IV. GLI1 Isoforms and Malignant Phenotypes of Cancer
GLI1 overexpression has been reported in normal tissues and also cancerous tissues, including those of GBM (Cui et al., 2010; Lo et al., 2009), hepatocellular carcinoma (Huang et al., 2006), gastric adenocarcinoma (Ma et al., 2005), esophageal cancer (Ma et al., 2006), breast cancer (Cao et al., 2012; ten Haaf et al., 2009), melanoma (Das et al., 2009), rhabdomyosarcoma (Roberts et al., 1989), and pancreatic adenocarcinoma (Nolan-Stevaux et al., 2009). Similar to GLI1, GLI1ΔN appears to be expressed in both normal and cancerous cells (Shimokawa et al., 2008). Currently available evidence suggests that tGLI1 is expressed in a tumor-specific fashion, at least in the two tumor types (GBM and breast carcinoma) examined to date (Cao et al., 2012; Lo et al., 2009). In line with the difference in expression patterns, the GLI1 and tGLI1 isoforms have been found to display overlapping and also distinctly different tissue- and tumor-specific functions (Fig. 6.3). In some instances, they differ in their ability to regulate expression of the genes that are important modulators of malignant phenotypes of human cancers, including proliferation, migration and invasiveness, progression, angiogenesis, cancer stem cell renewal, and drug resistance.
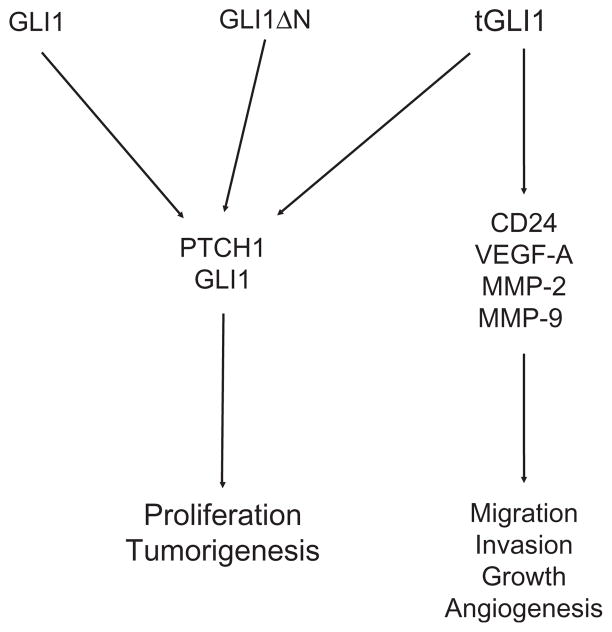
Figure 6.3.
Regulation of malignant phenotypes of cancer by GLI1 and tGLI1. Expression of all three GLI1 variants results in increased expression of PTCH1, an established GLI1 target gene. Expression of GLI1 leads to increased cell proliferation rates and other cellular processes. Expression of tGLI1 activates additional genes, namely VEGF-A, CD24, MMP-2, and MMP-9, leading to its ability to promote cancer cell growth, migration, invasion, and angiogenesis.
A. Proliferation
A major characteristic of cancer cells is an enhanced ability for cell division leading to uncontrolled proliferation. GLI1 has a proliferative-promoting effect when expressed in neural cells, evidenced by studies of GLI1 in the context of both cancer and embryonic development. For example, mice with Shh mutants leading to decreased GLI1 levels have brains with decreased proliferation (Dahmane et al., 2001). This same study found GLI1 could induce formation of several hyperplasias throughout the neural tube when GLI1 RNA was injected into tadpoles. Similarly, ectopic expression of GLI1 in embryonic mouse brain regions resulted in outgrowths of neural tissue (Hynes et al., 1997; Stecca and Ruiz i Altaba, 2009). The opposite also holds true in that inhibition of GLI1, either by cyclopamine or siRNA targeting, greatly reduces proliferation and tumor size (Clement et al., 2007; Wang et al., 2010). GLI1 has a similar effect on brain progenitor cells as GLI1 expression caused formation of tumor-like hyperplasia in the thalamus and cerebellum (Stecca and Ruiz i Altaba, 2009).
GLI1 expression in epithelial cells can induce cell transformation characterized by increased proliferation and anchorage-independent proliferation (Kimura et al., 2005; Li et al., 2006; Yoon et al., 2002). This is specific to GLI1 or GLI2 as GLI3 had no such effect (Kimura et al., 2005). These effects have been recapitulated in vivo as GLI1 induction in mouse epidermis led to hyperproliferative skin lesions (Li et al., 2006). In other epithelial cell types, GLI1 targeted expression in mammary glands led to mammary hyperplasia and tumor formation (Fiaschi et al., 2009). Interestingly, GLI1 was driven by a doxycycline promoter in this study and removal of doxycycline after GLI1 induction did not reduce mammary tumors, suggesting other cellular factors could replace GLI1 in sustaining tumor growth. In contrast, specific knockdown of GLI1 in inflammatory metastatic breast cancer cells resulted in decreased proliferation and increased apoptosis (Thomas et al., 2011). These data indicate GLI1 can induce a proliferative cell phenotype that can lead to tumor formation but may not be required for maintenance of a cancer phenotype in mammary tumors.
However, it is worth noting that siRNA-targeted expression knockdown would have silenced expression of both GLI1 and tGLI1, thus making it difficult to conclude that the growth inhibition is attributed to GLI1 suppression. This is a valid point as our studies have shown that tGLI1 may have stronger growth-stimulating effects on breast cancer cells (Cao et al., 2012). Using isogenic MDA-MB-231 cell lines stably expressing GLI1, tGLI1, or control vector, we observed that the cells with tGLI1 expression underwent significant anchorage-dependent and -independent growth than those with GLI1 and control cells (Cao et al., 2012). In the two GBM cell lines, we had examined to date, tGLI1 and GLI1 appeared to have similar effects on cell proliferation (Lo et al., 2009). Our most recent, unpublished results indicate that the in vivo growth of a GBM cell line, U87MG, was greatly enhanced by tGLI1 expression and to a lesser extent, by GLI1. The discrepancy between the in vitro and in vivo effects of tGLI1 on GBM growth is likely due to tumor microenvironment such as tumor vasculature.
GLI1ΔN did not have any significant impact on cell proliferation despite its ability to activate a similar gene set to that of GLI1 (Shimokawa et al., 2008). However, the induction of target genes by GLI1ΔN was significantly weaker than that of GLI1, indicating that GLI1ΔN may not be a strong promoter of cell proliferation. The ability of GLI1ΔN to regulate in vivo tumor growth is presently unknown.
B. Migration and invasiveness
Melanomas have been shown to express increased levels of GLI1 and osteopontin, along with enhanced ability to migrate, invade, and develop tumor xenografts and lung metastases (Das et al., 2009). This study found that osteopontin was a direct target of GLI1 and loss of either GLI1 or osteopontin resulted in decreased cell migration, invasion, and tumor growth, and reduced numbers of lung metastases. In breast cancer, increased GLI1 expression promotes cell motility (Thomas et al., 2011). Conversely, cyclopamine decreased the ability of melanoma cells to form lung metastases (Stecca et al., 2007), and also reduced invasiveness of prostate cancer cells (Sheng et al., 2004). In line with these findings, GLI1 has been shown to positively correlate with tumor grade and even lymph node status indicating it may play a role in progression of tumors toward metastasis (Stein et al., 1999; ten Haaf et al., 2009; Wang et al., 2010). Further, high GLI1 expression resulted in a less favorable prognosis (ten Haaf et al., 2009).
In light of the high homology between GLI1 and tGLI1, and the inability of antibodies and siRNA to selectively recognize tGLI1 versus GLI1, the reported positive correlations between GLI1 and tumor cell migration, invasion, and metastasis could be due to both GLI1 and tGLI1, or to a greater extent to tGLI1. This speculation is based on our results showing that tGLI1-expressing GBM and breast cancer cells migrated and invaded in significantly higher rates than those with GLI1 and control cells (Cao et al., 2012; Lo et al., 2009). When implanted in the flanks of nude mice, tGLI1-expressing GBM xenografts underwent a greater extent of infiltration into the skeletal muscles than those with GLI1 (Lo et al., 2009). In elucidating the molecular mechanisms underlying tGLI1-mediated migration and invasion, we conducted DNA microarray to compare the gene expression profiles of three isogenic U87MG GBM cell lines with control vector, GLI1 and tGLI1 (Lo et al., 2009). The results showed 75 genes to be expressed at a significantly higher level and 26 genes to be more suppressed in U87MG-tGLI1 cells compared to U87MG-vector and U87MG-GLI1 cells. Interestingly, the levels of well-known GLI1 target genes such as PTCH1 were higher in both U87MG-GLI1 and U87MG-tGLI1 cells compared to U87MG-vector cells. Importantly, the results of DNA microarray led us to identify and subsequently validate CD24 as a tGLI1-targeted gene and an important gene essential for tGLI1-associated migration and invasiveness in GBM cells. Notably, CD24 has been shown to recruit adhesion molecules to lipid rafts, thereby contributing to tumor cell migration, dissemination, and metastasis (Baumann et al., 2005; Runz et al., 2008). In addition to CD24, our results further indicated that tGLI1 induces expression of two invasion-associated genes, namely MMP-2 and MMP-9 (Cao et al., 2012). Ongoing efforts are investigating the role of tGLI1 in tumor metastasis, the leading cause of death for most cancers.
C. Epithelial–mesenchymal transition
Development of an invasive cell phenotype with enhanced ability for migration is necessary for progression to metastasis. In epithelial tumors, this is often accompanied by a dedifferentiaion program termed epithelial–mesenchymal transition (EMT), most notably characterized by loss of E-cadherin and increases in Snail, Twist, Vimentin, and other mesenchymal markers (Lo et al., 2007). There is also evidence that GLI1 may mediate EMT by increasing expression of Snail and repression of E-cadherin (Li et al., 2006; Louro et al., 2002). Expression of a dominant negative of Snail in GLI1-transformed epithelial cells decreased the ability of these cells to grow independent of attachment and led to increased E-cadherin levels (Li et al., 2006). This likely indicates GLI1 regulates EMT by induction of Snail and results were confirmed in vivo as targeted GLI1 expression in mouse epidermis caused hyperproliferative skin lesions along with increased Snail expression (Li et al., 2006). GLI1 has shown differing effects on E-cadherin as one study found GLI1 promoted E-cadherin redistribution toward the cell membrane (Neill et al., 2008) and another found increased E-cadherin expression induced by GLI1 (Liao et al., 2009). Despite showing increased E-cadherin, this last study also found increased expression of vimentin and Snail along with cells that had a greater capacity for proliferation, migration, and invasion (Liao et al., 2009). Inhibition of GLI1 in these cells with cyclopamine decreased proliferation, invasion, migration, E-cadherin expression, and Snail expression. This suggests GLI1 had a profound effect on the phenotype but perhaps did not induce a full EMT change. The impact of tGLI1 on EMT is still unknown.
D. Cancer stem cells
Cancer stem cells are thought to control tumor development and progression. In addition, cancer stem cells are thought to have limitless proliferation potential and mediate drug resistance. Stem cells are characterized by the presence of undifferentiated cell markers such as CD133 or Nestin. Considering that GLI proteins play a significant role in embryonic development and differentiation, it has been investigated whether GLI1 expression is associated with cancer stem cells.
Cells derived from GBM are found to express NANOG, a transcription factor that participates in regulation of self-renewal in stem cells (Zbinden et al., 2010). These cells show the ability to form gliomaspheres, which is exclusive to non-differentiated cells. NANOG appears to control the “stemness” of these cells as downregulation of NANOG reduced proliferation and the ability of these cells to form gliomaspheres and tumors. NANOG was found to be enriched in CD133+ cells along with GLI1, GLI2, Nestin, and OCT4. Loss of NANOG also resulted in decreased GLI1, Nestin, and OCT4 indicating a loss of stem cell markers. Enhancement of GLI1 levels by PTCH knockdown results in increased NANOG levels, which control the stemness and tumor formation abilities of these cells (Zbinden et al., 2010). Overexpression of GLI1 in normal mice brains also induces expression of stem cell markers such as SOX2, Nestin, Prominin1, and Bmi-1 that is accompanied by hyperproliferation and brain outgrowths (Stecca and Ruiz i Altaba, 2009). GLI1 is also involved in maintaining the stem cell phenotype in other cell types. Mammary stem cells form mammospheres and are enriched with greater than 20-fold higher levels of GLI1 compared to differentiated mammary cells (Liu et al., 2006). Shh enhanced mammosphere formation whereas cyclopamine suppresses mammosphere formation, further suggesting Shh signaling plays a role in the stem cell phenotype. For direct evidence for a role of GLI1, overexpression of GLI1 or GLI2 enhances the ability of these cells to form mammospheres (Liu et al., 2006). These data indicate the Shh pathway, and GLI1 in particular, play a significant role in cancer cells with stem-like characteristics. Future investigations are required to determine if some of the reported GLI1 activity is attributed to both tGLI1 and GLI1, or exclusively to tGLI1 or GLI1 alone.
The role of tGLI1 or GLI1ΔN in cancer stem cell marker expression or stem cell renewal has not been assessed. GLI1ΔN appears to have weaker transcriptional activity than GLI1 (Shimokawa et al., 2008) and likely will contribute to a stem cell phenotype to a lesser extent than GLI1. However, tGLI1 has a stronger ability to activate multiple genes compared to GLI1 including VEGF-A (Cao et al., 2012) that has been linked to stem cell phenotype (Oka et al., 2007; Ricci-Vitiani et al., 2010). In addition, cells expressing tGLI1 appear to have greater capabilities for migration and invasion, characteristics cancer stem cells purportedly have.
Our results indicate that CD24 is a transcriptional target of tGLI1, but not GLI1, in GBM and breast cancer (Cao et al., 2012; Lo et al., 2009). This finding is in agreement with our results showing that tGLI1 enhanced the propensity of cancer cells to migrate and invade both in vitro and in vivo. In line with our observations, CD24 has been shown to be involved in cancer cell motility, invasion, and metastasis (Baumann et al., 2005; Kristiansen et al., 2003; Runz et al., 2008) and shortened patient survival (Kristiansen et al., 2003; Lee et al., 2009). Also consistent with these reports are the facts that CD24 recruits adhesion molecules to lipid rafts (Runz et al., 2008) and that CD24 indirectly stimulates cell adhesion to fibronectin, collagens I/IV, and laminin through the activation of integrins (Baumann et al., 2005). Interestingly, several studies have shown that some breast cancer-initiating stem cells display the characteristic of being low/negative for CD24 expression but high for CD44 (Al-Hajj et al., 2003; Kim et al., 2007). This notion is not in agreement with our results showing that tGLI1 expression enables MDA-MB-231 cells to grow in an anchorage-independent fashion, a property commonly displayed by cancer stem cells. In support of our results, a recent report, however, showed that CD24 does not define breast cancer stem cells (Meyer et al., 2010). Also in agreement with our results, another recent study reported that pancreatic cancer stem cells demonstrate the CD44+CD24+ESA+phenotype (Li et al., 2007). In support of these notions, it has been suggested that the role of CD24 in cancer stemness may be tissue-specific (Keysar and Jimeno, 2010). Clearly, more work is needed to define the role that CD24 plays in both cancer progression and stem cell biology. Also needed is future research into the role of tGLI1 in mediating cancer stem cell properties will be interesting considering tGLI1 is expressed in multiple human cancers including GBM and breast cancers (Cao et al., 2012; Lo et al., 2009). More significantly, tGLI1 could be mistakenly detected as GLI1 by RT-PCR or western blotting.
E. Angiogenesis
Tumor growth beyond 2mm in diameter requires angiogenesis in order for tumors to receive nutrients and remove wastes to survive (Bergers and Benjamin, 2003). Angiogenesis is regulated by the balance of proangiogenic factors (e.g., VEGFs) and anti-angiogenic factors (e.g., Tsp-1). Tumor cells alter this balance by secreting proangiogenic factors, which draws growing microvessels toward the tumor in order to retrieve nutrients and discard wastes. Turning on the angiogenic switch is a crucial point in the progression of tumors toward metastasis as angiogenic tumors greatly increase the pace of tumor growth. In addition to increasing the rate of tumor growth, angiogenesis increases vascular density around the tumor providing a door for metastatic cells to enter the general circulation where they can find a new tissue for seeding secondary tumors. Thus, angiogenesis has attracted much attention in recent decades as a potential drug target for cancer.
Pancreatic adenocarcinoma cells have increased GLI1 levels whereas cyclopamine decreases GLI1 levels, xenograft tumor growth, and reduced vascularity within the tumor (Nakamura et al., 2010). Conditioned medium made from these cancer cells was mixed with matrigel and implanted in nude mice causing bone marrow-derived cells to be recruited and increasing vascular density. Cyclopamine decreased the number of bone marrow-derived cells that were recruited to the matrigel plug (Nakamura et al., 2010). Cyclopamine treatment to growing xenograft tumors decreased tumor angiogenesis and decreased tumor expression of several proangiogenic factors including VEGF-A, SDF-1, Ang-1, IGF-1, and PDGF-B (Nakamura et al., 2010). GLI1, specifically, can mediate these effects as glioma stem cells, which express GLI1, have increased production and release of VEGF in response to Shh and IGF-1 (Hsieh et al., 2011). However, knockdown of GLI1 significantly reduces VEGF production by these cells and reduces the ability of these cells to promote angiogenesis in vitro. Thus, GLI1 can have a proangiogenic effect with increased expression.
tGLI1 also induces a proangiogenic response. Expression of tGLI1 in breast cancer cells increased VEGF-A mRNA and protein production by these cells compared to cells expressing GLI1 (Cao et al., 2012). As mentioned above, GLI1 can promote an angiogenic response by induction of VEGF-A (Hsieh et al., 2011; Nakamura et al., 2010). The ability of tGLI1 to induce a much stronger level of VEGF-A production was found to be due to direct interaction with the VEGF-A promoter by tGLI1 whereas GLI1 had minimal binding (Cao et al., 2012). Medium from tGLI1-expressing cells also induced stronger endothelial cell tube formation compared to GLI1-expressing cells consistent with increased VEGF induction. This suggests that although GLI1 has a proangiogenic effect, tGLI1 may promote a stronger angiogenic response. Induction of angiogenesis by tGLI1 is consistent with its role in promoting a more aggressive cancer phenotype. There are currently no published data regarding the ability of GLI1ΔN to alter angiogenesis genes.
F. Drug resistance
Cancer cell resistance to chemotherapy is usually associated with more aggressive cell phenotypes that give a poor patient prognosis. Certain cancer cells adapt to the environment created by chemotherapy to escape toxicity induced by cancer killing therapies. GLI1 appears to have some association with the development of cellular resistance to chemotherapy. Patients with gliomas displayed a positive correlation between GLI1 expression and recurring tumors (Cui et al., 2010). Concurrent cell treatment with Shh and chemotherapy led to an increase in clonogenic survival of these cells whereas cells receiving chemotherapy alone had decreased survival (Cui et al., 2010). Adding to this, treatment of cancer cells with both chemotherapy and cyclopamine significantly increased cell death and apoptosis compared to either treatment alone (Cui et al., 2010). These data suggest upregulation of the Shh pathway, likely mediated by GLI1, provides protection for cells and resistance to chemotherapy. The precise mechanism for this protection was not elucidated. Considering tGLI1 is associated with a more aggressive cell phenotype, it is logical that tGLI1 could provide similar, if not greater, resistance to a chemotherapy challenge. Experiments to test this hypothesis have not been published. It will be interesting to see if the tGLI1 variant continues to show a more aggressive cell phenotype and what characteristics this splice variant can provide for cells. It will be equally important to determine if tGLI1-expressing tumor cells respond to Shh-targeted therapy that typically inhibits the Shh-SMO-GLI1 signaling axis.
V. Conclusion and Future Directions
Significance advances have been made in our understanding of the GLI1 family of proteins in the past few years. Most notably, the full-length GLI1 transcript is alternatively spliced to yield two shorter isoforms, namely GLI1ΔN and tGLI1. While the expression pattern of GLI1ΔN appears to be similar to GLI1 that is widely expressed in both normal and cancerous cells, tGLI1 has been shown to be expressed in a tumor-specific fashion in GBM and breast cancer. The unique pattern expression of tGLI1 is significant in the context of human cancers, in particular, tumorigenesis, tumor progression, and therapeutics. In line with these implications, tGLI1 has gained the ability to transcriptionally regulate several genes that are not regulated by GLI1 such as CD24, VEGF-A, MMP-2, and MMP-9, and potentially other genes identified via gene expression profiling. Interestingly, tGLI1 has retained its ability to undergo nuclear translocalization, activate GLI1-binding sequence, and increase PTCH1 expression. Consistent with the gained ability to upregulate these important cancer-related genes, tGLI1-expressing tumor cells display an increased propensity to migrate, invade, growth, and stimulate the proliferation of vascular endothelial cells. When implanted in the flanks of nude mice, tGLI1-expressing cells formed bigger xenografts compared to those with GLI1. These emerging evidences suggest that tGLI1 may be a stronger mediator of several malignant phenotypes of human cancer. They also present a new paradigm in which some of the known GLI1 functions may be attributed to both GLI1 and tGLI1, or to a greater extent to tGLI1. Future efforts are urgently needed to generate experimental evidence to establish this exciting new paradigm.
An important future direction is to further decipher the roles that the three GLI1 isoforms in human cancers. Given the minute difference between the GLI1 and tGLI1 transcripts and proteins, previous detection of GLI1 may have masked the existence of tGLI1. Also very likely, the siRNA knockdown studies may have downregulated expression of both GLI1 and tGLI1 rather than exclusively, that of GLI1. Further complicating the issue is the facts that the molecular difference between the tGLI1 and GLI1 is only about 4.5kDa (146kDa for tGLI1 and 150kDa for GLI1) and that commercially available antibodies appear to recognize both isoforms. Collectively, these notions and technical limitations have contributed to our inability to uncover the existence of tGLI1 for more than 20years and also call for a need to develop tools to selectively detect/silence tGLI1 or GLI1. For this, we have optimized two complementary methods to distinguish the two isoforms: (a) RNA extraction followed by RT-PCR to amplify the exons 1–4 of the GLI1 and tGLI1 transcripts, and then by DNA sequencing of the PCR product to confirm the deleted region within tGLI1 (Cao et al., 2012; Lo et al., 2009), and (b) a SDS-PAGE condition that can effectively resolve the mobility of the two GLI1 and tGLI1 proteins (Cao et al., 2012). Although these methods effectively and accurately detect tGLI1 versus GLI1 mRNAs and proteins, it is recognized that tGLI1-specific and GLI1-specific antibodies will be helpful as we decipher the functions of the two isoforms and the contributions of them to malignant biology of human cancers. This effort will also help determine the clinical correlations between tGLI1 and GLI1 expression profiles in patient tumors with patient prognosis and response to treatments.
The exciting discoveries of GLI1ΔN and tGLI1 also call for future investigations on several aspects of the two novel variants that are presently unknown. First, it is important to gain a greater understanding of the regulation of GLI1ΔN and tGLI1 by various components of the Hedgehog signaling pathway including SMO, SUFU, GLI2, and GLI3. Second, it is essential to characterize the extent to which the two new isoforms are regulated by the noncanonical pathways, in comparison to GLI1. Third, it is imperative to determine the molecular mechanisms by which tGLI1 regulates its target genes since the available evidence indicates that it can bind to the consensus GLI1-binding sequence and additionally bind to other gene promoters that do not have the sequence and are not regulated by GLI1. This effort will help identify additional molecular pathways and cellular processes that are subjected to tGLI1 regulation. Fourth, given the established facts that GLI1 is associated with tumor development and other important aspects of tumor biology, a critical future task is to determine the role that tGLI1 plays in these processes, in particular, its involvement in comparison to GLI1 and GLI1ΔN. Finally, in light of the emerging importance of the Hedgehog–GLI1 pathway as a target of anti-cancer therapy, it will be highly relevant to investigate whether tumors with different expression profiles of the three GLI1 isoforms differ in their sensitivity to Hedgehog-targeted therapy, in particular, those that are being evaluated in the clinical trials in cancer patients. The derived information will help stratify cancer patients for more effective treatments and also aid in identifying rationales for overcoming therapeutic resistance.
Acknowledgments
This study was supported by the NIH grant K01-CA118423, DOD grants W81XWH-07-1-0390 and W81XWH-11-1-0600, the Beez Foundation, the Pediatric Brain Tumor Foundation, and Intramural Division of Surgical Sciences Dani P. Bolognesi, Ph.D. Award (to H. W. L).
References
- Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, Oren M, Tanaka N. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci USA. 2008;105:4838–4843. doi: 10.1073/pnas.0712216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arheden K, Ronne M, Mandahl N, Heim S, Kinzler KW, Vogelstein B, Mitelman F. In situ hybridization localizes the human putative oncogene GLI to chromosome subbands 12q13.3-14.1. Hum Genet. 1989;82:1–2. doi: 10.1007/BF00288260. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, Yagita H, Sleeman JP. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–10793. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Bigner SH, Wong AJ, Mark J, Muhlbaier LH, Kinzler KW, Vogelstein B, Bigner DD. Relationship between gene amplification and chromosomal deviations in malignant human gliomas. Cancer Genet Cytogenet. 1987;29:165–170. doi: 10.1016/0165-4608(87)90045-8. [DOI] [PubMed] [Google Scholar]
- Cai Q, Li J, Gao T, Xie J, Evers BM. Protein kinase Cδ negatively regulates Hedgehog signaling by inhibition of Gli1 activity. J Biol Chem. 2009;284:2150–2158. doi: 10.1074/jbc.M803235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Geradts J, Dewhirst MW, Lo HW. Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene. 2012;31:104–115. doi: 10.1038/onc.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288:156–166. doi: 10.1016/j.jns.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i Altaba A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- Das S, Harris LG, Metge BJ, Liu S, Riker AI, Samant RS, Shevde LA. The hedgehog pathway transcription factor GLI1 promotes malignant behavior of cancer cells by up-regulating osteopontin. J Biol Chem. 2009;284:22888–22897. doi: 10.1074/jbc.M109.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Andre J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- Dennler S, André J, Verrecchia F, Mauviel A. Cloning of the human GLI2 promoter. J Biol Chem. 2009;284:31523–31531. doi: 10.1074/jbc.M109.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Marti E, Scott MP, McMahon AP. Antagonizing cAMP-dependent protein kinase A in the dorsal CNS activates a conserved Sonic hedgehog signaling pathway. Development. 1996;122:2885–2894. doi: 10.1242/dev.122.9.2885. [DOI] [PubMed] [Google Scholar]
- Fiaschi M, Rozell B, Bergstrom A, Toftgard R. Development of mammary tumors by conditional expression of GLI1. Cancer Res. 2009;69:4810–4817. doi: 10.1158/0008-5472.CAN-08-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forus A, Florenes VA, Maelandsmo GM, Meltzer PS, Fodstad O, Myklebost O. Mapping of amplification units in the q13-14 region of chromosome 12 in human sarcomas: Some amplica do not include MDM2. Cell Growth Differ. 1993;4:1065–1070. [PubMed] [Google Scholar]
- Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2000;28:220–226. [PubMed] [Google Scholar]
- Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol. 2011;226:1118–1127. doi: 10.1002/jcp.22433. [DOI] [PubMed] [Google Scholar]
- Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, Zhang K, Tang W, Stelter AA, Wang Q, Zhang H, Xie J. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006;27:1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M, Stone DM, Dowd M, Pitts-Meek S, Goddard A, Gurney A, Rosenthal A. Control of cell pattern in the neural tube by the zinc finger transcription factor and oncogene Gli-1. Neuron. 1997;19:15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates Hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: Mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Keysar SB, Jimeno A. More than markers: Biological significance of cancer stem cell-defining molecules. Mol Cancer Ther. 2010;9:2450–2457. doi: 10.1158/1535-7163.MCT-10-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim JB, Lee KM, Shin I, Han W, Ko E, Bae JY, Noh DY. Isolation of CD24(high) and CD24(low/−) cells from MCF-7: CD24 expression is positively related with proliferation, adhesion and invasion in MCF-7. Cancer Lett. 2007;258:98–108. doi: 10.1016/j.canlet.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Kristiansen G, Winzer KJ, Mayordomo E, Bellach J, Schluns K, Denkert C, Dahl E, Pilarsky C, Altevogt P, Guski H, Dietel M. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res. 2003;9:4906–4913. [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim SH, Lee ES, Kim YS. CD24 overexpression in cancer development and progression: A meta-analysis. Oncol Rep. 2009;22:1149–1156. doi: 10.3892/or_00000548. [DOI] [PubMed] [Google Scholar]
- Li X, Deng W, Nail CD, Bailey SK, Kraus MH, Ruppert JM, Lobo-Ruppert SM. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Liao X, Siu MK, Au CW, Wong ES, Chan HY, Ip PP, Ngan HY, Cheung AN. Aberrant activation of hedgehog signaling pathway in ovarian cancers: Effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30:131–140. doi: 10.1093/carcin/bgn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69:6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoRusso PM, Rudin CM, Borad MJ, Vernillet L, Darbonne WC, Mackey H, DiMartino JF, de Sauvage F, Low JA, Von Hoff DD. A first-inhuman, first-in-class, phase (ph) I study of systemic Hedgehog (Hh) pathway antagonist, GDC-0449, in patients (pts) with advanced solid tumors. J Clin Oncol. 2008;26:3516. (Meeting Abstracts) [Google Scholar]
- Louro ID, Bailey EC, Li X, South LS, McKie-Bell PR, Yoder BK, Huang CC, Johnson MR, Hill AE, Johnson RL, Ruppert JM. Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res. 2002;62:5867–5873. [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen K, Huang S, Zhang X, Adegboyega PA, Evers BM, Zhang H, Xie J. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis. 2005;26:1698–1705. doi: 10.1093/carcin/bgi130. [DOI] [PubMed] [Google Scholar]
- Ma X, Sheng T, Zhang Y, Zhang X, He J, Huang S, Chen K, Sultz J, Adegboyega PA, Zhang H, Xie J. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer. 2006;118:139–148. doi: 10.1002/ijc.21295. [DOI] [PubMed] [Google Scholar]
- Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, Wu G, Wu D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70:4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Sasajima J, Mizukami Y, Sugiyama Y, Yamazaki M, Fujii R, Kawamoto T, Koizumi K, Sato K, Fujiya M, Sasaki K, Tanno S, et al. Hedgehog promotes neovascularization in pancreatic cancers by regulating Ang-1 and IGF-1 expression in bone-marrow derived pro-angiogenic cells. PLoS One. 2010;5:e8824. doi: 10.1371/journal.pone.0008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill GW, Harrison WJ, Ikram MS, Williams TD, Bianchi LS, Nadendla SK, Green JL, Ghali L, Frischauf AM, O’Toole EA, Aberger F, Philpott MP. GLI1 repression of ERK activity correlates with colony formation and impaired migration in human epidermal keratinocytes. Carcinogenesis. 2008;29:738–746. doi: 10.1093/carcin/bgn037. [DOI] [PubMed] [Google Scholar]
- Ng JM, Curran T. The Hedgehog’s tale: Developing strategies for targeting cancer. Nat Rev Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka N, Soeda A, Inagaki A, Onodera M, Maruyama H, Hara A, Kunisada T, Mori H, Iwama T. VEGF promotes tumorigenesis and angiogenesis of human glioblastoma stem cells. Biochem Biophys Res Commun. 2007;360:553–559. doi: 10.1016/j.bbrc.2007.06.094. [DOI] [PubMed] [Google Scholar]
- Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001;106:107–117. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: New perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Wild A, Grzeschik KH, Antonarakis SE. Mutation in GLI3 in postaxial polydactyly type A. Nat Genet. 1997;17:269–271. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- Raffel C, Jenkins RB, Frederick L, Hebrink D, Alderete B, Fults DW, James CD. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57:842–845. [PubMed] [Google Scholar]
- Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010;96(2):169–179. doi: 10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: Preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res. 1994;54:4299–4303. [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts WM, Douglass EC, Peiper SC, Houghton PJ, Look AT. Amplification of the gli gene in childhood sarcomas. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runz S, Mierke CT, Joumaa S, Behrens J, Fabry B, Altevogt P. CD24 induces localization of beta1 integrin to lipid raft domains. Biochem Biophys Res Commun. 2008;365:35–41. doi: 10.1016/j.bbrc.2007.10.139. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, Eberhart CG. The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: A potential mechanism of therapeutic resistance. Clin Cancer Res. 2010;16:6060–6070. doi: 10.1158/1078-0432.CCR-10-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K, Mohri D, Tanaka Y, Ijichi H, Tateishi K, Kanai F, Kawabe T, et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol Carcinog. 2009;48:703–712. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- Sheng T, Li C, Zhang X, Chi S, He N, Chen K, McCormick F, Gatalica Z, Xie J. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng T, Chi S, Zhang X, Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. J Biol Chem. 2006;281:9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgard R, Zaphiropoulos PG. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J Biol Chem. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz IAA. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein U, Eder C, Karsten U, Haensch W, Walther W, Schlag PM. GLI gene expression in bone and soft tissue sarcomas of adult patients correlates with tumor grade. Cancer Res. 1999;59:1890–1895. [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- ten Haaf A, Bektas N, von Serenyi S, Losen I, Arweiler E, Hartmann A, Knuchel R, Dahl E. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer. 2009;9:298. doi: 10.1186/1471-2407-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ZI, Gibson W, Sexton JZ, Aird KM, Ingram SM, Aldrich A, Lyerly HK, Devi GR, Williams KP. Targeting GLI1 expression in human inflammatory breast cancer cells enhances apoptosis and attenuates migration. Br J Cancer. 2011;104:1575–1586. doi: 10.1038/bjc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67:1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang Z, Xu Z, Yin H, Bai L, Ma Z, Decoster MA, Qian G, Wu G. Activation of the sonic hedgehog signaling controls human pulmonary arterial smooth muscle cell proliferation in response to hypoxia. Biochim Biophys Acta. 2010;1803:1359–1367. doi: 10.1016/j.bbamcr.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Xie J, Aszterbaum M, Zhang X, Bonifas JM, Zachary C, Epstein E, McCormick F. A role of PDGFRα in basal cell carcinoma proliferation. Proc Natl Acad Sci USA. 2001;98:9255–9259. doi: 10.1073/pnas.151173398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kwon YJ, Frolova N, Steg A, Yuan K, Johnson M, Grizzle W, Desmond R, Frost A. Gli1 promotes cell survival and is predictive of a poor outcome in ERα-negative breast cancer. Breast Cancer Res Treat. 2010;123:59–71. doi: 10.1007/s10549-009-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA, Jacob H, Walterhouse D, Iannaccone P. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277:5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Lo HW. The human glioma-associated oncogene homolog 1 (GLI1) family of transcription factors in gene regulation and diseases. Curr Genomics. 2010;11:238–245. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]