Abstract
Functions of p53 during mitosis reportedly include prevention of polyploidy and transmission of aberrant chromosomes. However, whether p53 plays these roles during genomic surveillance in vivo and, if so, by direct or indirect means remain unknown. The ability of normal, mature hepatocytes to respond to stimuli, reenter cell cycle and regenerate liver mass offers an ideal setting to assess mitosis in vivo. In quiescent liver, normally high ploidy levels in adult mice increased with loss of p53. Following partial hepatectomy, p53−/− hepatocytes exhibited early entry into cell cycle and prolonged proliferation with an increased number of polyploid mitoses. Ploidy levels increased during regeneration of both WT and p53−/− hepatocytes, but only WT hepatocytes were able to dynamically resolve ploidy levels and return to normal by the end of regeneration. We identified multiple cell cycle and mitotic regulators, including Foxm1, Aurka, Lats2, Plk2 and Plk4, as directly regulated by chromatin interactions of p53 in vivo. Over a time course of regeneration, direct and indirect regulation of expression by p53 is mediated in a gene-specific manner.
Conclusion
Our results show that p53 plays a role in mitotic fidelity and ploidy resolution in hepatocytes of normal and regenerative liver.
Keywords: regeneration, mitosis, chromatin, transcription, polyploid
Chromosomal polyploidy presents a considerable challenge to the orderly process of mitosis. There are normal tissues and cells in both vertebrates and invertebrates that display polyploidy during development or as fully differentiated tissues. How mitotic fidelity is maintained in these cells is a question of considerable interest. Recent studies in Drosophila establish that polyploid chromosomes of larval rectal cells are faithfully duplicated and segregated through multiple cell cycles during the course of normal development (1). Although the division of these polyploid cells progresses through normal, recognizable stages, the time course of each is extended and the process is highly error-prone. Genome instability and aneuploidy may be one cost of maintenance and proliferation of polyploid cells, as a substantial number of chromosomal abnormalities arise in these cells.
Hepatocytes of the mammalian liver develop polyploidy and aneuploidy over the lifespan of the organism. Hepatocytes can be mononucleated or binucleated, and each nucleus can have diploid, tetraploid, octaploid or higher nuclear content (2). Polyploidization occurs via failed cytokinesis or endoreduplication (2). Moreover, proliferating polyploid hepatocytes undergo chromosome segregation errors, generating a high degree of aneuploidy. Approximately 60% of adult WT mouse hepatocytes are aneuploid and 30–90% of hepatocytes in humans are aneuploid (3, 4). Hepatocytes are highly tolerant of nuclear alterations, undergoing cycles of ploidy expansion, ploidy reversal, and aneuploidy, described as the “ploidy conveyor” (3). Hepatocyte polyploidy may be further expanded during liver regeneration induced by a two-thirds partial hepatectomy (PH) in mice (5, 6). Given that a polyploid mitotic division may lead to increased aneuploidy and possibly tumor development (7, 8), it remains unclear how these hepatocytes remain mitotically active and accumulate chromosomal instability without becoming tumorigenic.
Genome integrity is protected in normal cells by various means, including cell cycle checkpoints, DNA repair mechanisms and the induction of apoptosis. A critical player in each of these processes is the p53 tumor suppressor, which provides surveillance against cellular insults of many types and may induce G1 arrest and cellular senescence in response to tetraploidy or missegregated chromosomes (9, 10). p53 and its close relative, p73, are also linked to the mitotic spindle assembly checkpoint, as both proteins interact with kinetochore and spindle checkpoint proteins (7, 11, 12). In fact, combined loss of p53 and p73 leads to increased polyploidy and aneuploidy in primary cultured cells (9) and results in a higher incidence of tumor development in mouse liver (13).
The striking tolerance of the liver for altered ploidy leads to consideration of whether a “tetraploid” checkpoint exists in hepatocytes. We addressed this question by analysis of checkpoint mediator p53, and characterized the synchronized process of cellular proliferation and growth that occurs to regenerate the liver in response to PH in both wild type and p53-null mice. Our results reveal that p53 alters levels of hepatocyte ploidy during liver regeneration and aging. Although chromosome segregation errors are common in WT hepatocytes expressing p53, these errors (e.g., abnormal mitotic figures and lagging chromosomes) are even more frequent in hepatocytes deficient for p53. Since p53’s effects may be mediated by context-specific, mitotic regulators (14), we examined whether p53 regulated expression of mediators of hepatic cell division in normal and regenerating liver. We identified Aurora kinase A (Aurka), Forkhead-box transcription factor Foxm1, regulator of cytokinesis Lats2, and Polo-like kinases (Plk2 and Plk4) as directly regulated by p53 in quiescent liver, a mitotic transcription program that is altered during liver regeneration. Thus, our findings suggest that p53 plays a role in controlling levels of hepatic polyploidy/aneuploidy by direct transcription regulation of multiple downstream effectors.
MATERIALS AND METHODS
Liver regeneration in mice
PH to remove 70% of total liver tissue, or Sham surgery was performed using isoflurane anesthesia, as described (24). 5–7 C57Bl6/Sv129 F1 mice, WT or p53−/−, 2 months of age, were used for each experimental condition according to the MD Anderson Cancer Center Institutional Animal Care and Use Committee guidelines. p53 knock-out mice were sacrificed 0.5, 1, 1.5, 2, 3, 3.5, 4, or 7 days following PH and Sham surgeries; remnant liver tissue was harvested, flash-frozen, and processed for RNA, immunoblot, and chromatin immunoprecipitation (ChIP) analyses. Liver/body weight ratios were calculated to determine the recovery of liver mass.
ChIP and RNA analyses
ChIP analyses were performed as previously described (24). Briefly, chromatin lysate was precleared and incubated overnight with the following specific antibodies for ChIP: p53 (Novocastra) and normal sheep IgG (Upstate/Millipore). To analyze protein-bound DNA, primers for real-time quantitative PCR (qPCR) were used (Supplementary Table). The percentage of the input that was bound was calculated by the dCt method and averaged for at least three biological replicates.
Primers and reverse transcription-PCR determinations of RNA expression were performed as previously described. Briefly, total RNA from homogenized mouse liver was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was obtained by reverse transcription of 2 ug of RNA using the SuperScript system (Invitrogen). Real-time PCR was carried out using primers for indicated genes (Supplementary Table).
Histology and Immunohistochemistry
Collected liver samples were fixed in 10% Neutral Buffered Formalin (Fisher), embedded in wax paraffin and sectioned at 5–6 mm by the MD Anderson Department of Veterinary Medicine and Surgery. Sections were rehydrated and stained with hematoxylin and eosin (H&E) or with anti-Ki67 (Abcam) marker and then counterstained with hematoxylin following the manufacturers’ recommended protocols. Antigen retrieval and immunodetection was performed using solutions from Vector Labs per manufacturer’s instructions (Vector Laboratories). Endogenous peroxidase activity was quenched by incubating slides in methanol/H2O2 solution.
Western Blot
Immunoblotting was performed using standard SDS-PAGE and Western blot methodology (24). Protein extracts from liver tissue collected at 0, 12, 24, 48, 72, 84, 96, and 168 hours following PH and 12h following Sham surgery were prepared using T-PER buffer (Pierce/Thermoscientific) per manufacturer instructions.
Flowcytometry analysis
Hepatocytes were isolated from adult 129 p53+/+, p53+/− and p53−/− littermates by collagenase perfusion (15). Live hepatocytes were loaded with Hoechst33342 (Invitrogen) and DNA content determined with an InFlux flow cytometer (Beckton Dickinson) using a 150 um nozzle, as previously described (3). The Institutional Animal Care and Use Committee of Oregon Health & Science University approved these experiments.
To quantify DNA content (ploidy profiling), hepatocytes were isolated from livers after PH and Sham surgeries by collagenase perfusion (15). Cells were fixed by gentle vortexing in ice-cold 80% (v/v) ethanol and stained with 50 μg/mL propidium iodide (PI) in 0.2% Tween in PBS supplemented with 1 μg/mL (w/v) RNAse A (Molecular Probes). Cytofluorometric acquisitions were performed on a FACSCalibur (BD Biosciences). Statistical analysis was performed by using the CellQuest software (BD Biosciences), upon gating on the events characterized by normal forward scatter and side scatter parameters.
Statistical analysis
Results are expressed as means standard error of the mean (SEM). Statistical analyses were performed by Students t-test, and p values <0.05 were considered significant.
RESULTS
Lack of p53 disrupts ploidy control in proliferating hepatocytes
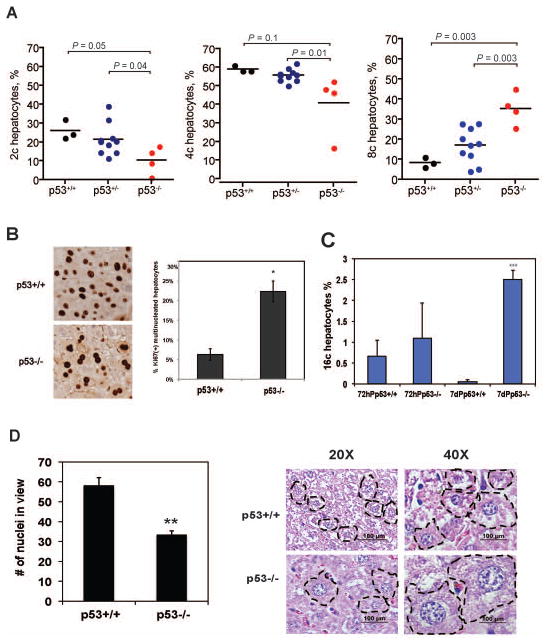
The ploidy of murine hepatocytes increases during development, liver disease, aging and regeneration (6, 16). This tolerance of polyploidy suggests that specific checkpoints, which either maintain a diploid state and/or eliminate cells that exhibit altered ploidy, may be lacking in hepatic tissue. Although p53 is implicated in mitotic surveillance of cultured immortalized and tumor-derived cells (17, 18), this has not been assessed during normal development or under conditions of induced cellular proliferation and tissue regeneration. To address this, we determined the ploidy status of live WT (p53+/+) and p53-null hepatocytes during normal development by flow cytometry and analysis of DNA content (Fig. 1A). Consistent with previous studies of p53+/+ mouse liver, we observed that 60% of total hepatocytes in quiescent, 4–5 month old p53+/+ liver are tetrapoloid (4c), with a second, major population of diploid cells (2c, ~30%) and a smaller fraction of octaploid cells (8c, ~10%). However, in quiescent p53−/− mouse liver of the same age, less than 50% of hepatocytes are tetraploid and many are octaploid (>30%). Concomitantly, there is a significantly reduced number of diploid cells. This distribution in ploidy was dependent on p53 dosage, as indicated by an intermediate ploidy phenotype in heterozygous, p53+/− hepatocytes. These data suggest that hepatocyte ploidy, during normal growth and development of the liver, is monitored by a p53-dependent process.
Figure 1. p53 controls parenchymal cell ploidy, size, and cell number in the liver.
(A) Distribution of hepatic ploidy populations in p53+/+, p53+/− and p53−/− mice. The frequency of 2c, 4c, and 8c populations was determined in live hepatocytes from p53+/+(n=3), p53+/− (n=9) and p53−/− (n=4) littermates aged 4–5 months by FACS analysis. P values are indicated. (B) Ki67(+) hepatocytes with nuclei that failed to form completely separated nuclear envelopes where counted in 10 fields at 48h post-PH. The percentage of binucleated hepatocytes was significantly higher in p53−/− compared to the wt liver sections, as determined by t-test (*p < 0.05). (C) The frequency of hepatocytes with 16c nuclear content was determined in fixed cells isolated from livers of p53+/+ and p53−/− mice at 72h and 7 days following PH. Significant difference in number of 16c hepatocytes was determined by t-test (***p<0.001). (D) H&E staining of liver tissue sections collected at 7 days following PH in p53+/+ and p53−/− mice was used to count hepatocyte nuclei. 10 fields per sample were counted, and difference in number of hepatocyte nuclei in view between p53+/+ and p53−/− was determined by t-test (** p < 0.01). Representative photographs of H&E stained p53+/+ and p53−/− liver tissue at 7 days following PH were taken at 20X and 40X magnification. Notice similar size of p53−/− hepatocytes at 20X and of p53+/+ hepatocytes at 40X magnification, suggesting that p53−/− hepatocytes have at least 2X larger nuclei.
To determine whether p53 acts in mitotic surveillance during acute injury response, we utilized a model of surgically induced growth and replacement of liver tissue. We compared 2-month old p53+/+ and p53−/− mice, which have fewer differences in ploidy at t=0 than 4–5-month old mice (data not shown and Fig. 1A). Two-thirds PH of mouse liver elicits a synchronized wave of cell cycle re-entry, proliferation, mitosis and growth in the remnant liver, to regenerate and restore the size of the liver (liver/body weight ratio or liver index) to its pre-surgical set point (Suppl. Fig. 1A) (19). In situ staining of the DNA replication marker Ki67 revealed dividing hepatocytes at 48h after 2/3 PH in p53+/+ and p53−/− mice (Fig. 1B, left panel). Strikingly, binucleated Ki67(+) p53−/− hepatocytes were present at 4-fold higher numbers than in p53+/+ liver (Fig. 1B, right panel), suggesting enhanced proliferation and/or cytokinesis failure. To examine ploidy, we analyzed nuclear content at various times following PH. Whereas nuclear content is equivalent to ploidy class in quiescent adult livers (e.g., 4c DNA = tetraploid cell) (3), nuclear content in regenerating livers is complicated by ploidy class and cell cycle status. For instance, in proliferating hepatocytes, 4c DNA content indicates either a tetraploid cell in G0/G1 or a diploid cell in G2. Therefore, to focus exclusively on polyploid hepatocytes, we examined cells with nuclear content of 8c or higher. Compared to p53+/+ mice, the frequency of 8c hepatocytes was higher in p53−/− Sham livers, and the population expanded by 48h post-PH (Suppl. Fig. 1B). Even more dramatic changes were seen in hepatocytes with 16c DNA content. In p53+/+ mice, regenerative proliferation after PH led to a small 16c population (Fig. 1C). However, the population of 16c hepatocytes in p53+/+ mice, which was clearly observed 72h after PH, diminished with restoration of liver mass (seven days post-PH) (Fig. 1C). In contrast, the number of 16c hepatocytes in p53−/− regenerating liver continued to increase over time, resulting in a 50-fold increase in 16c hepatocytes compared to p53+/+ at the termination stage of liver regeneration (Fig. 1C). These data suggest that p53 regulates the formation and maintenance of polyploidy even in cells that are highly tolerant of polyploidy and aneuploidy.
To fully characterize cellular changes associated with hepatocyte proliferation, we also examined cellular and nuclear size. Analysis of sections of liver tissue isolated at the end of regeneration (day 7) revealed major differences between p53+/+ and p53−/− mice (Fig. 1D). p53−/− hepatocytes were significantly larger, resulting in fewer cells per field-of-view (Fig. 1D, left panel). Moreover, larger hepatic nuclei were observed in p53−/− mice (Fig. 1D, right panel), which is consistent with the high degree of polyploidy in p53−/− mice.
A lack of uniformity in increased cell size at day 7 post-PH in p53−/− mice (Fig. 1D) suggests a possibility of liver over-growth. However, in response to the challenges of cell division and growth after PH, both p53+/+ and p53−/− hepatocytes achieved a similar recovery of liver mass (Suppl. Fig. 1A), despite their differences in cell size and ploidy (Fig. 1C and 1D). These data extend a recent report, showing the impact of increased hypertrophy of WT hepatocytes during liver regeneration (20), and link increases in ploidy to hypertrophy in p53−/− mice after PH.
p53 regulates mitotic entry and exit during liver regeneration
To determine whether p53+/+ and p53−/− hepatocytes had comparable levels of proliferation after PH, we performed in situ staining of Ki67 over a time course of regeneration (Fig. 2A). Consistent with previous measurements by BrdU incorporation (19), p53+/+ mouse liver entered an initial period of cellular proliferation after 24h, reached a maximum at day 2 (48h), and engaged more than 80% of all remnant hepatocytes. In addition, we observed a second round of DNA synthesis that occurred at day 4 post-PH, which involved 46% of hepatocytes in p53+/+ liver (Fig. 2A). In comparison, p53−/− liver had an earlier onset of cellular proliferation, less than 24h after PH, followed by a broadened span of proliferation over 2–3.5 days that involved only 63% of remnant hepatocytes. In p53−/− liver, a second, less distinct peak of proliferation occurred 12 hours earlier than the 2nd proliferation wave in p53+/+ liver, followed by a significant number of p53−/− hepatocytes exiting mitosis at day 4 post-PH (Fig. 2A).
Figure 2. Proliferation of hepatocytes during liver regeneration is controlled by p53.

(A) Tissue sections from livers collected at indicated time points following PH and Sham surgeries were stained with Ki67 antibody, followed by hematoxylin staining to reveal non-proliferating nuclei. Positive Ki67(+) staining was determined by immunohistochemistry. Ki67(+) hepatocyte nuclei and (hematoxylin-only stained) hepatocyte nuclei were counted in 10 fields for each time point. Significant difference between Ki67(+) hepatocytes between p53+/+ and p53−/− was determined by t-test separately at each time point (*p < 0.05; **p < 0.01, ***p < 0.001). Sham livers did not show any Ki67(+) cells. (B) Representative photographs of mitotic figures in p53+/+ and p53−/− regenerating livers collected at indicated time points following PH. H&E staining of liver tissue sections collected at 48, 72 and 96 hours following PH was used to count mitotic figures. 10 fields per sample were counted, and lagging chromosomes and multipolar mitotic spindles were determined visually based on criteria discussed elsewhere (3, 10) and shown as a percent of total mitotic figures in the field. Increase in number of dividing cells with lagging chromosomes in p53−/− livers compared to p53+/+ is statistically significant at 48h (p < 0.01) and 72h (p < 0.05); number of p53−/− hepatocytes with multipolar spindles is significantly higher at 48h (p < 0.05).
To assess whether p53−/− hepatocytes were progressing through mitosis over a time course of regeneration or, alternatively, undergoing endoreduplication where cells undergo multiple rounds of DNA replication without entering mitosis, which may lead to formation of polyploidy (21), we determined gain or loss of specific phosphorylation sites that mark activated Cyclin-dependent kinase 1 (CDK1 or Cdc2) during mitotic transition (2). This analysis revealed similar profiles in WT and p53−/− liver; supporting S-phase replication followed by mitotic entry/transition, and not endoreduplication, during regeneration (Suppl. Fig. 2A).
p53 deficiency leads to increased mitotic errors
We next compared the number and appearance of mitotic figures in regenerating livers from p53+/+ and p53−/− livers. As previously reported, p53+/+ hepatocytes display multipolar spindles and lagging chromosomes during regenerative proliferation (3). During normal growth and in response to PH, approximately 95% of multipolar spindles resolve into bipolar spindles in polyploid WT hepatocytes, generating daughter cells with ploidy levels equivalent to the parental cell. Furthermore, cell divisions by polyploid hepatocytes can generate daughter cells with reduced ploidy ((3, 5); Fig. 1). Similar to WT livers, regenerating p53−/− livers also had abnormal mitotic figures and lagging chromosomes, but the frequency of these events was higher (Fig. 2B and Suppl. Fig. 2B). Together, these data indicate that increases in nuclear segregation errors by p53−/− hepatocytes correlate with the altered ploidy levels seen in p53−/− livers.
p53 binds to consensus motifs in mitotic targets and regulates their expression in liver
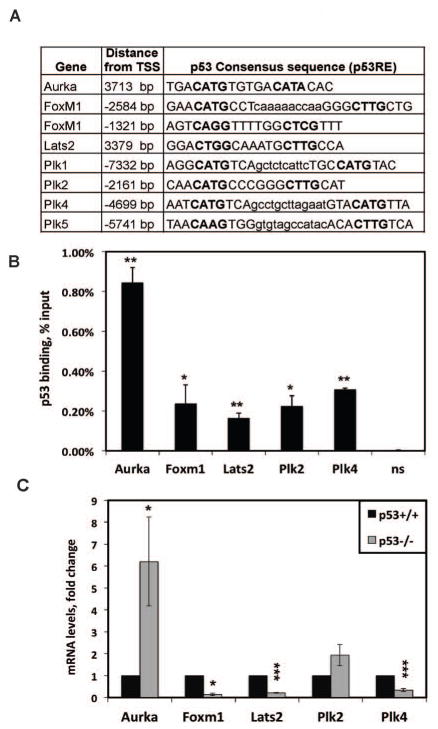
The majority of p53 functions are attributed to its ability to regulate transcription of target genes. p53 has transcriptional activity in quiescent liver (22, 23), but direct target genes involved in hepatic cell division are unknown. Using a previously determined consensus DNA sequence for p53 binding (p53 response element, p53RE) (24), we assessed genes implicated in the regulation of mitotic progression and fidelity for potential p53 binding sites within 10 kilobases upstream and downstream of transcription start sites (TSS). We identified p53REs in seven genes encoding major mitotic regulators: Aurora kinase A (Aurka), Forkhead-box transcription factor Foxm1, regulator of cytokinesis Lats2 and Polo-like kinases (Plk1, Plk2, Plk4 and Plk5) (Fig. 3A). ChIP with a p53 antibody detected significant binding of p53 to five p53REs of these genes in WT liver: Aurka, Foxm1, Lats2, Plk2 and Plk4 (Fig. 3B). Interestingly, motif analysis of the p53REs of these genes revealed general but not perfect agreement with the ‘canonical’ consensus of p53-bound response elements, derived primarily from in vitro studies (24) (Suppl. Fig. 3A).
Figure 3. p53 binds the p53REs of mitotic target genes and regulates their expression in normal quiescent liver.
(A) p53-DNA binding motifs (p53 response elements, p53REs) were found upstream of the transcription start site (TSS) of seven target genes using an algorithm (37). Two half-sites are shown in the uppercase letters with conserved core (CA/TG) in bold and a spacer sequence is in the lowercase (if present). Locations of the identified p53REs are shown as a distance from TSS. (B) p53 binds to the p53RE of Aurka, Foxm1, Lats2, Plk2, and Plk4 in quiescent liver. ChIP from adult mouse liver tissue was performed using antibodies against p53. Primers used for consequent ChIP experiments were specific to the p53RE sites, indicated in (A). Binding of p53 to the previously identified p53RE of Foxo3 served as a positive control (data not shown). NS region located 2kb away from Plk1 p53RE was used as a negative control. Average of at least three independent ChIPs is shown as percentage of input bound. The difference between p53 binding to p53REs as compared to NS site is statistically significant when marked with asterisks (* p<0.05; ** p<0.01). (C) Aurka, Foxm1, Lats2, Plk2, and Plk4 mRNA levels in p53+/+ and p53−/− mice were compared by real-time PCR. The statistically significant difference between p53+/+ and p53−/− is indicated with asterisks (* p<0.05; ** p<0.01, ***p<0.001).
Since p53 may regulate transcription of target genes as either a direct repressor or activator (25), we compared expression of the p53-bound genes in p53+/+ and p53−/− liver (Fig. 3C). Expression of Aurka was up regulated 6-fold in p53−/− liver compared to p53+/+, suggesting p53 repressed Aurka expression in normal quiescent liver. In contrast, Foxm1, Lats2, and Plk4 were strongly down regulated in p53−/− mice, thus, p53 activated expression of these genes in normal quiescent liver. Plk2 mRNA levels did not change significantly in p53−/− livers compared to p53+/+, indicating that p53 binding to Plk2 p53RE may not regulate basal expression of this gene in the quiescent liver.
To determine whether p53 regulates each of these genes during the process of liver regeneration, we examined expression at time points following PH (Fig. 4). First, expression of Aurka in the WT liver dramatically increased during the first round of mitosis at 24h and 48h after PH while p53 was still bound to the Aurka p53RE (Fig. 4A and 4B). These results suggest that p53 deficiency could lead to elevated Aurka expression (i.e., loss of p53-mediated Aurka inhibition) during regeneration. However, we found reduced Aurka levels in p53−/− livers compared to WT at 24h and 48h post-PH, indicating that Aurka expression is independent of p53 through the first round of mitosis (Fig. 4B). Interestingly, expression of Aurka was significantly up regulated at the end of liver regeneration in p53−/− liver (day 7, Fig. 4B). This finding supports p53-mediated repression of Aurka expression, observed in normal quiescent liver (Fig. 3), was reestablished with cessation of liver regeneration. Indeed, we observed an increase in p53 binding to the Aukra p53RE at the later time points following PH (72–96h, Fig. 4A).
Figure 4. p53 regulates mitotic genes during liver regeneration.

(A) Levels of p53 binding to the target genes during proliferative phase of liver regeneration (24h, 48h, 72h, and 96h) compared to Sham (as described in Figure 3). (B) Expression of Aurka, (C) Foxm1 and (D) Plk2, (E) Plk4, and (F) Lats2 in p53+/+ and p53−/− regenerating livers. RNA was isolated from mouse liver tissue at indicated time points following the PH and Sham surgeries. mRNA levels were measured by real-time PCR and calculated as a fold change over Sham for each time point. Time points with statistically significant difference in gene expression between p53+/+ and p53−/− regenerating livers are marked with asterisks * p<0.05; ** p<0.01, ***p<0.001.
Second, expression analysis of Foxm1, previously reported as a critical regulator of the G2-M transition in regenerating liver (26), showed p53-dependant and -independent changes over a time course of regeneration (Fig. 4A and C). p53-independent, compensatory mechanisms activated expression of Foxm1 during the onset of the 1st cell cycle. At the onset of the 2nd wave of hepatic proliferation, these mechanisms do not provide compensation and significantly decreased Foxm1 expression occurs in p53−/− liver (Fig. 4C). We observed binding of p53 to the Foxm1 p53RE at 72h post-PH (Fig. 4A), together with high levels of expression of Foxm1 at 72–84h post-PH (2nd round of mitosis). The increase in Foxm1 expression was not detected in p53−/− liver after the 1st round of mitosis (Fig. 4C) when hepatocyte proliferation was significantly lower compared to the wt mice (Fig. 2A). Thus, p53-mediated activation of Foxm1 may be necessary for the onset of the 2nd cell cycle during liver regeneration.
Third, Polo-like kinases are known positive regulators of proliferation at all stages of the cell cycle (27). p53 binding to the Plk2 and Plk4 p53REs at 24h, 48h, 72h, and 96h after PH was measurable (Fig. 4A), but Plk2 and Plk4 levels changed inconsistently with p53 deficiency, suggesting that p53 is not a primary driver of hepatic Plk2 and Plk4 expression during liver regeneration (Fig. 4D).
Finally, Lats2 is a negative regulator of cell growth and a tissue-specific inhibitor of cell size (28–30). Similar to Foxm1, p53 remained bound to a Lats2 p53RE at 24h post-PH (Fig. 4A). But, p53-independent mechanisms compensate for loss of p53 regulation of Lats2 during regeneration of liver (Fig. 4F). In quiescent liver, the data indicate that both mitotic activators and negative regulators of cell growth are directly regulated by p53, as either a gene-specific activator or repressor of transcription. During liver regeneration, however, regulation of specific p53 target genes and mitotic regulators, Cyclin B and Aurora kinase B that lack p53REs (Suppl. Fig. 3B), is complex and p53-dependent or -independent at specific time points.
Our results demonstrate that p53 directly binds the p53REs of genes regulating the cell cycle in regenerating mouse liver. While Plk2, Plk4, and Lats2 may be additionally regulated by a p53-independent mechanism, expression of Aurka and Foxm1 is directly regulated by p53 at specific time points following PH. Importantly, p53-mediated activation of Foxm1 occurs at the onset of the 1st as well as the 2nd mitotic division of hepatocytes following PH. p53-mediated repression of Aurka in quiescent hepatocytes is diminished at the peak of proliferative stage, and partially regained at the cessation of liver regeneration.
DISCUSSION
Aneuploidy and polyploidy are detected in multiple types of cancer, and they are clearly linked to neoplasia and tumor development (7). However, recent studies show that evolutionarily conserved, but poorly understood, mechanisms allow replication of polyploid chromosomes without generation of genomic instability in “ploidy tolerant” cells (1, 3). Such cells are found in significant numbers, which increase with age, in normal liver and exist in a polyploid state without progression to a dysplasia. Polyploid hepatocytes use dynamic aneuploidization and polyploidization to support homeostasis or overcome environmental stress by a process termed the ploidy conveyor (3). Likely candidates for regulation of the ploidy conveyor are members of the p53 family, such as p53 and p73 isoforms, which can prevent propagation of aneuploid and polyploid cultured cells (7, 9). We found that p53 impacts ploidy levels in the liver: normal ratios of diploid, tetraploid and octaploid hepatocytes in adult mouse liver were significantly skewed towards octaploidy in the absence of p53, at the expense of diploid and tetraploid hepatocyte populations. Intriguingly, PH-induced liver regeneration, which challenges the normally quiescent, polyploid hepatocytes to undergo cell cycle reentry, mitosis and growth, accelerates the ability of oncogenes to induce HCC and may predispose liver to cancer development (31). Despite known deletions and mutations of p53 in hepatocellular carcinoma (HCC) (32), it was previously unknown whether p53 played any role in liver regeneration linked to genomic instability. Our findings demonstrate that absence of p53 reduces ploidy resolution and regeneration of p53−/− liver relies more on increased cell size, due to over-representation of polyploid hepatocytes. Thus, loss of p53 functions and accumulative effects on ploidy during cycles of regenerative repair may accelerate liver tumorigenesis and decrease time of progression to HCC.
Polyploid WT hepatocytes form multipolar spindles and have lagging chromosomes during mitotic divisions Here, we show that this process involves p53-dependent regulation of transcription during normal liver development and regeneration. In response to regenerative signaling, multipolar spindles and lagging chromosomes were seen in both WT and p53−/− hepatocytes, but these abnormal mitotic figures were observed in higher numbers in p53−/− mice. We speculate that elevated frequency of nuclear segregation errors in p53−/− hepatocytes contributed to cytokinesis failure and, therefore, enhanced polyploidization (10). Our observation of an orderly progression of mitosis, as marked by comparable activation of Cdk1/cdc2 in WT and p53−/− hepatocytes, suggests that endoreduplication does not contribute to higher polyploidy with p53 deficiency.
Questions arise whether a mitotic checkpoint exists in hepatocytes, in light of this fluidity of ploidy numbers. Although the liver exhibits dynamic changes in levels of polyploidy during aging and regeneration, it is clear that p53 exercises some level of control over this process. We uncovered a network of ploidy determinants, which are direct gene targets of p53, regulated in quiescent liver and responsive in a gene-specific manner to regenerative signaling. To our knowledge, this report is the first description of p53 binding to these newly identified p53REs and, more specifically, to cell cycle regulators during mitotic division in vivo. Our results reveal p53-dependent differences in the expression of genes that regulate mitotic entry and progression (Aurka, Foxm1), division (Plk2 and Plk4), and exit back to the G0 (Lats2). Importantly, we identified Foxm1 and Aurka genes as new direct transcriptional targets of p53. Our data demonstrate that binding of p53 to Foxm1 p53RE occurs specifically at the onset of the 1st and the 2nd round of mitosis (24h and 72h post-PH) resulting in the robust activation of the Foxm1 expression, which is essential for DNA replication and mitosis in regenerating hepatocytes (26). The direct repression of Aurka by p53 in quiescent liver may be necessary to suppress the tumor-promoting consequences of the overexpression of Aurora kinase A in liver (33, 34). The overall results of our p53 ChIP and target gene expression analyses demonstrate that transcriptional regulation by p53 is necessary, whether by direct or indirect means, for timely activation or repression of specific target genes at different stages of the cell cycle (Figure 5).
Figure 5. Modes of p53-dependent regulation of mitotic target genes and outcomes of p53 loss during regenerative proliferation of hepatocytes.

Aurora A is frequently over expressed in HCC in correlation with mutation of p53 and poor prognosis (33). Lats2 is associated with regulated cytokinesis: depletion of Lats2 induces accumulation of polyploid cells (28). The Foxm1 transcription factor is activated during liver regeneration (26), to impact timely mitotic entry and accurate chromosome segregation (38). Plk4+/− liver has lower activation of p53, aberrant mitotic figures after PH, and develops HCC (39). Plk2 shows loss of heterozygocity in HCC and is a critical regulator of G1-S transition and mitosis during cell proliferation (27, 40). Aurora A, Foxm1, Lats2 and specific Plk’s are regulators of mitosis that impact ploidy levels in a p53-dependent manner.
Cell division is a highly conserved process, but there are clearly tissue-specific modes of regulation. In other cell systems (i.e., non-hepatocytes), p53-mediated suppression of Aurka prevents pre-mitotic arrest (34), and basal activation of Lats2 minimizes aneuploidy/polyploidy. In hepatocytes, cell division is complex because polyploidy and aneuploidy are extremely high in p53+/+ livers from mice (3) and humans (4). Nonetheless, disruption of normal Aurka and Lats2, to a lesser extent Foxm1 and Plk4, expression partially accounts for enrichment in mitotic segregation errors and enhanced polyploidy seen in p53-deficient liver. Following PH, transcriptional activity of p53 and how it contributes to activation or repression of mitotic or cell cycle regulators is more difficult to interpret. There may be a partial compensation by TA-p73, which has been shown to play a role in liver tumor suppression in combination with p53 (35).
A fully delineated story of how hepatocytes survive, and even thrive, in spite of high levels of polyploidy and aneuploidy is not yet clear. p53 and its downstream effectors contribute to polyploidization and mitotic fidelity, as shown here in vivo. Whether p53 regulation is connected to activation of the insulin receptor and AKT signaling, implicated in cytokinesis failure and formation of polyploid hepatic cells (36), is unknown. Further characterization of new hepato-specific cell cycle pathways and definition of regulatory mechanisms are critical to understanding development, homeostasis, regeneration and pathology of the liver.
Supplementary Material
Acknowledgments
Financial support: This work was supported by funds from the National Institutes of Health (DK078024) to MCB, the William Randolph Hearst Foundation to ZC, and the NCI Cancer Center Support Grant to the University of Texas M.D. Anderson Cancer Center.
Abbreviations
- PH
Partial hepatectomy
- WT
Wild type
- p53RE
p53 response element
- ChIP
chromatin immunoprecipitation
- PH
partial hepatectomy
- HA
hemagglutinin
- TSS
transcription start site
- qPCR
real-time quantitative PCR
- H&E
hematoxylin and eosin
- PI
propidium iodide
References
- 1.Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes & development. 2010;24:2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. The Journal of biological chemistry. 2003;278:19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 3.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faktor VM, Uryvaeva IV. Progressive polyploidy in mouse liver following repeated hepatectomy. Tsitologiia. 1975;17:909–916. [PubMed] [Google Scholar]
- 6.Uryvaeva IV, Factor VM. Liver growth fraction, its composition with regard to cell ploidy and changes with aging. Ontogenez. 1975;6:458–465. [PubMed] [Google Scholar]
- 7.Aylon Y, Oren M. p53: guardian of ploidy. Molecular oncology. 2011;5:315–323. doi: 10.1016/j.molonc.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 9.Talos F, Nemajerova A, Flores ER, Petrenko O, Moll UM. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol Cell. 2007;27:647–659. doi: 10.1016/j.molcel.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernole P, Neale MH, Barcaroli D, Munarriz E, Knight RA, Tomasini R, Mak TW, et al. TAp73alpha binds the kinetochore proteins Bub1 and Bub3 resulting in polyploidy. Cell Cycle. 2009;8:421–429. doi: 10.4161/cc.8.3.7623. [DOI] [PubMed] [Google Scholar]
- 12.Gao F, Ponte JF, Levy M, Papageorgis P, Cook NM, Ozturk S, Lambert AW, et al. hBub1 negatively regulates p53 mediated early cell death upon mitotic checkpoint activation. Cancer biology & therapy. 2009;8:548–556. [PubMed] [Google Scholar]
- 13.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Fang X, Zhang P. Aneuploidy and tumorigenesis. Seminars in cell & developmental biology. 2011;22:595–601. doi: 10.1016/j.semcdb.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nature genetics. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S. Hepatic polyploidy and liver growth control. Seminars in cancer biology. 2000;10:161–171. doi: 10.1006/scbi.2000.0317. [DOI] [PubMed] [Google Scholar]
- 17.Gjoerup OV, Wu J, Chandler-Militello D, Williams GL, Zhao J, Schaffhausen B, Jat PS, et al. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8334–8339. doi: 10.1073/pnas.0703164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nature reviews Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 20.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Current biology: CB. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Ullah Z, Lee CY, Depamphilis ML. Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell division. 2009;4:10. doi: 10.1186/1747-1028-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui R, Nguyen TT, Taube JH, Stratton SA, Feuerman MH, Barton MC. Family members p53 and p73 act together in chromatin modification and direct repression of alpha-fetoprotein transcription. J Biol Chem. 2005;280:39152–39160. doi: 10.1074/jbc.M504655200. [DOI] [PubMed] [Google Scholar]
- 23.Kurinna S, Stratton SA, Tsai WW, Akdemir KC, Gu W, Singh P, Goode T, et al. Direct activation of forkhead box O3 by tumor suppressors p53 and p73 is disrupted during liver regeneration in mice. Hepatology. 2010;52:1023–1032. doi: 10.1002/hep.23746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 25.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nature reviews Molecular cell biology. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 28.Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes & development. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. The Journal of biological chemistry. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 30.Mao H, Zhang L, Yang Y, Sun J, Deng B, Feng J, Shao Q, et al. RhoBTB2 (DBC2) functions as tumor suppressor via inhibiting proliferation, preventing colony formation and inducing apoptosis in breast cancer cells. Gene. 2011;486:74–80. doi: 10.1016/j.gene.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS biology. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz SF, Lechel A, Obenauf AC, Begus-Nahrmann Y, Kraus JM, Hoffmann EM, Duda J, et al. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229–1239. e1223. doi: 10.1053/j.gastro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:2065–2071. doi: 10.1158/1078-0432.ccr-1057-03. [DOI] [PubMed] [Google Scholar]
- 34.Li CC, Chu HY, Yang CW, Chou CK, Tsai TF. Aurora-A overexpression in mouse liver causes p53-dependent premitotic arrest during liver regeneration. Mol Cancer Res. 2009;7:678–688. doi: 10.1158/1541-7786.MCR-08-0483. [DOI] [PubMed] [Google Scholar]
- 35.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 36.Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. The Journal of clinical investigation. 2009;119:1880–1887. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci U S A. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature cell biology. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 39.Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nature genetics. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 40.Pellegrino R, Calvisi DF, Ladu S, Ehemann V, Staniscia T, Evert M, Dombrowski F, et al. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.