Abstract
Eukaryotic genomes are intricately arranged into highly organized yet dynamic structures that underlie patterns of gene expression and cellular identity. The recent adaptation of novel genomic strategies for assaying nuclear architecture has significantly extended and accelerated our ability to query the nature of genome organization and the players involved. In particular, recent explorations of physical arrangements and chromatin landscapes in higher eukaryotes have demonstrated that chromatin insulators, which mediate functional interactions between regulatory elements, appear to play an important role in these processes. Here we reflect on current findings and our rapidly expanding understanding of insulators and their role in nuclear architecture and genome function.
Keywords: Chromatin, Epigenetics, CTCF, TFIIIC, Nuclear Organization
INTRODUCTION
The identity and developmental potential of any given cell begins within the cell nucleus, where spatiotemporal regulation of genome organization and expression underlie the transformation from a totipotent cell into a complex system of tissues and differentiated cell types. This incredible pathway is largely accomplished through the action of functional non-coding regulatory elements, which include enhancers, silencers, promoters, and insulators. Chromatin insulators were first discovered for their ability to protect genes from position effects in transgene assays, and have since been characterized as multi-protein DNA complexes capable of facilitating long-range inter- and intra-chromosomal interactions. More importantly, interactions facilitated by insulator proteins typically underlie functional contacts between regulatory elements, such as enhancers and promoters, or chromatin domain organization conducive to coregulation of active or silent genes. These features, combined with microscopy-based and biochemical studies suggesting insulators target associated loci to specific nuclear subcompartments, suggest that insulators are crucial players in constructing appropriate three-dimensional nuclear architecture. Though much of our current understanding of how insulators function comes from studies of CTCF, a highly conserved zinc finger protein capable of insulator activity in vertebrates, studies in other model systems have provided concordant evidence that insulators function by bridging together distant loci, yet the composition and proteins required for activity varies [1,2]. Nevertheless, pinpointing the exact purpose of insulators in genome biology has proven to be difficult, as insulator proteins appear to be involved in a multitude of diverse, context-dependent biological activities. In this respect, we consider recent developments in our understanding of insulators and their roles in nuclear organization and cell differentiation.
DISTRIBUTION, CORRELATION, AND ORGANIZATION OF INSULATORS THROUGHOUT THE GENOME
The occupancy landscape of insulator proteins has been mapped genome-wide by combining chromatin immunoprecipitation with microarray hybridization (ChIP-chip) [3–6], high throughput sequencing (ChIP-seq) [7–9], and more recently with even higher precision using ChIP-exo [10]. Insulator proteins localize to thousands of sites characterized by conserved target sequences, wherein differences in DNA motifs can influence protein occupancy levels and features of insulator function [9,11,12]. The CTCF insulator protein localizes to DNase I-hypersensitive sites, characteristic of “open chromatin”, that are generally common across cell types [13]. Detailed comparison of CTCF binding sites across 38 human cell lines suggests that while a majority of insulators are indeed invariant between cell types, thousands of cell-type specific CTCF sites are also present [14]. Variable CTCF binding sites are associated with differential DNA methylation within the CTCF recognition sequence [15], and whereas ubiquitous CTCF sites predominantly map to intergenic regions, cell-type specific CTCF sites are enriched within the introns of genes [14]. In addition to CTCF, RNA polymerase III (RNAP III) transcription factor TFIIIC is capable of insulator activity in both yeast and humans [16], at tRNA genes and RNAP III independent sites where TFIIIC is recruited to highly conserved B-box elements. TFIIIC and CTCF sites associate with the cohesin complex [17–19], which likely stabilizes long-range interactions and is essential for insulator activity [20,21].
The distribution of insulator proteins initially provided a certain degree of support to previously proposed models, wherein chromatin insulators function as heterochromatin barriers. For example, insulators localize to the borders of some repressive chromatin domains in yeast [20,22], Drosophila [23], and mammals [8,14], suggesting they might establish a roadblock to prevent the spread of gene silencing, consistent with their ability to protect transgenes from position effects. However, these correlations do not account for a majority of insulator-binding sites and do not explain why insulators only delimit a subset of repressed loci. In light of this discrepancy, recent exploration of the nature and function of insulator proteins in their endogenous contexts point to a role beyond barrier function. For one, depletion of Drosophila insulator proteins does not lead to substantial changes in the distribution of H3K27me3, an epigenetic signature of Polycomb (Pc)-mediated repression, at most domain borders [24]. Meanwhile, mapping of all CTCF-mediated interactions in mouse embryonic stem (ES) cells has offered a complex picture of chromatin domain organization, characterized by distinct underlying epigenetic states and governed by functional long-range insulator-insulator interactions [25]. Together, these studies would suggest that insulators are involved in establishing the structural arrangement of chromatin domains, but do not actively participate in the delineation of epigenetic status.
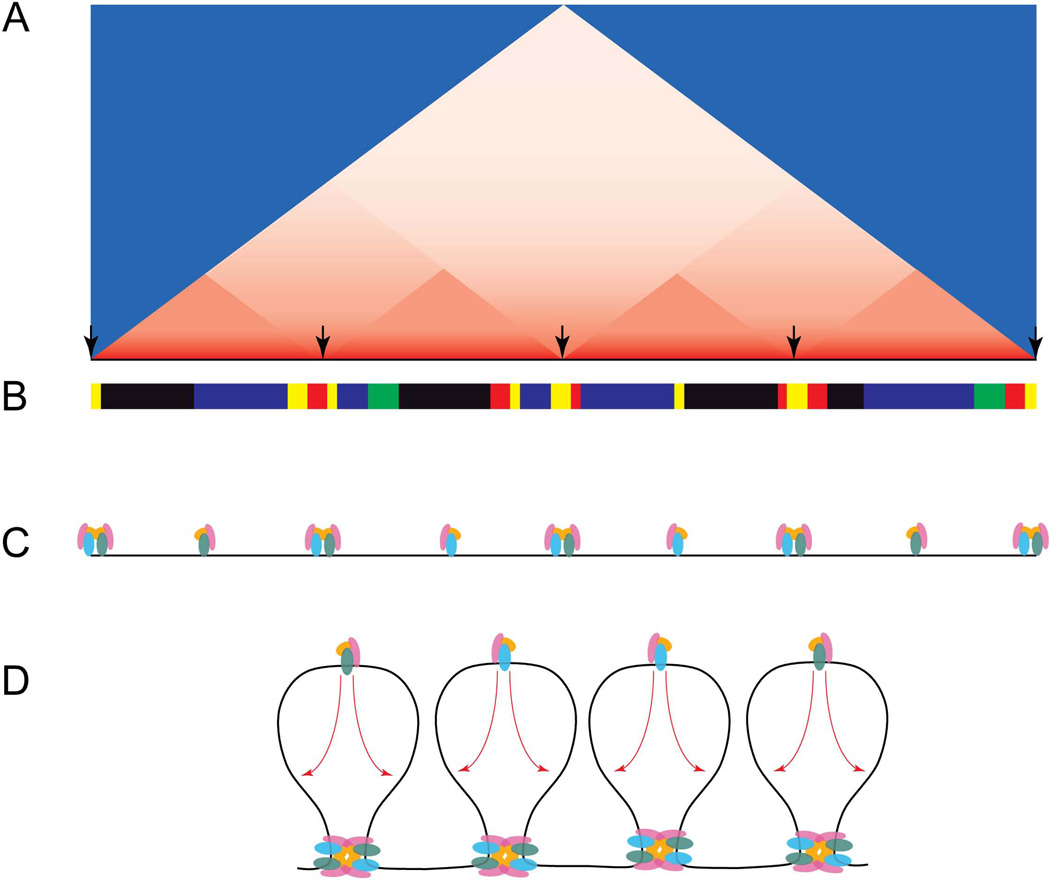
Chromosome conformation capture (3C)-based genome-wide interaction studies in Drosophila, mice, and human cell lines have also uncovered the structural organization of interphase chromosomes in unprecedented detail [26–28], providing new insight into the correlation and possible role of insulators in nuclear architecture. Genomes appear spatially segregated into topological chromatin domains defined by strong interaction frequencies and separated by domain borders characterized by a dramatic decrease in short-range interactions (Fig. 1). By integrating the generated contact maps with genome-wide histone and chromatin-associated protein mapping data, these studies demonstrate that physical domains are related to [26], but clearly independent of epigenetically defined chromatin domains [27,28]. Domain boundaries are enriched for combinations of insulator proteins [26–28], and correlate most strongly with gene density and transcription status, suggesting that active transcription may drive the formation of physical domains [28] (Fig. 1). Supporting evidence for this scenario comes from analysis of the chromatin composition and gene orientation at domain boundaries. In particular, genes that flank domain borders are predominantly active and preferentially oriented towards the domain boundary, with insulator proteins positioned distally from the domain boundary [28]. Nevertheless, to what degree insulator-mediated interactions contribute to the spatial segregation of discrete physical domains remains unclear. Deletion of a 58 kb boundary element containing CTCF-binding sites between domains harboring Xist and Tsix was shown to cause aberrant inter-domain chromosomal contacts and transcriptional misregulation [29]. However, the two neighboring domains retain a certain level of spatial segregation, suggesting multiple factors contribute to boundary function.
Figure 1.
Topological domain structure and correlation with epigenetic profiles. A) Cartoon heatmap representation of physical interactions. Interactions frequencies are represented from high (red) to low (blue). Physical domains are identified as having strong interaction frequencies, separated by sites with a dramatic decrease in short-range interactions (arrows – domain boundaries). B) Principle chromatin types defined by the presence of specific chromatin components. Comparison between physical domains (A) with chromatin profiles (B) reveals that physical domains are independent of epigenetic signatures. Sites of active chromatin (yellow, red) correlate with domain boundaries, whereas silent chromatin domains (black, blue, green) are enriched within the interior of physical domains. C) Domain boundaries also correlate with combinations of insulator proteins, whereas thousands of independent insulator sites localize within physical domains. D). Model integrating the identified topological domains with studies examining the nature and function of insulator proteins. Aligned insulator proteins are enriched at chromatin domain boundaries, establishing the framework for chromatin looping. Interior independent insulators presumably mediate short-range interactions within individual physical domains.
Further detailed analysis of chromatin insulators in D. melanogaster has also revealed that insulator proteins, previously broken into separate classes based on localization and associated gene ontologies [3,4], actually cluster together often in a manner suggestive of cooperative function [12,24]. For example, the Drosophila orthologoue of CTCF tandemly aligns with other unique DNA-binding insulator proteins BEAF-32 and Suppressor of Hairy-wing (Su(Hw)) [12]. Aligned insulators are enriched for additional insulator-associated proteins, raising the possibility that by clustering, insulators efficiently recruit essential co-factors necessary to stabilize long-range interactions. Compared to independent insulator sites, tandemly aligned chromatin insulators also appear to be less susceptible to disruption of post-translational modifications that regulate insulator function [30], suggesting that these sites may have evolved to resist regulatory mechanisms that otherwise modulate independent insulator function. Whether CTCF alignment is a conserved feature of insulator function in humans remains unknown. However, physical domains in mammalian chromosomes are associated with tRNA genes and CTCF [27], and recent genome-wide mapping of TFIIIC in both mouse and humans has uncovered an association with CTCF-binding sites [17,31], together providing preliminary evidence that insulator alignment may indeed be evolutionarily conserved [2].
EMERGING ROLES OF INSULATORS IN NUCLEAR ARCHITECTURE AND GENOME FUNCTION
Though insulators have been extensively characterized by their ability to influence gene expression in transgenic reporter assays, mapping of insulator protein-binding sites has allowed recent studies to probe the function of chromatin insulators in their normal context. Concurrent mapping of physical interactions and the three-dimensional organization of eukaryotic genomes have also allowed these studies to effectively address the role of insulators in nuclear organization. Results consistently support a role for chromatin insulators in facilitating long-range interactions important for both gene expression and repression, and recent findings demonstrate the importance of insulators in cell signaling, cellular development, and the cell cycle.
Long-range interactions and gene regulation
Functional long-range interactions between non-coding regulatory elements have been well established and extensively studied in the context of looping between promoters and distal enhancers [32]. Interactions between genomic elements are a hallmark of both gene expression, where coregulated genes are dynamically recruited to transcription factories, and gene silencing, where genes targeted by Pc and characterized by H3K27me3 colocalize to repressive Pc bodies [1]. Whereas insulators were previously proposed to prevent the promiscuity of enhancers through enhancer-blocking activities, new studies instead suggest that insulators direct the interaction and specificity between enhancers and promoters [33]. For one, CTCF underlies cell-type specific chromatin architecture conducive to enhancer-promoter interactions at numerous developmental loci [34]. CTCF mediated interactions in mouse ES cells include functional long-range contacts between enhancer-binding protein p300 and the promoters of genes whose expression is reduced following CTCF knockdown [25]. Furthermore, the presence of CTCF does not impede looping between promoters and distal elements mapped in humans [35], and transgenic analyses for different subclasses of insulators in Drosophila also demonstrate the absence of enhancer-blocking activity for most insulator classes under the conditions employed in the assays [24].
Insulators also appear to play an important role in bridging functional interactions between Pc-domains [36], rather than the barrier activity for which they were defined. For example, interactions between transgenes containing Mcp or Fab-7 Polycomb response elements (PREs) from the BX-C Hox cluster were shown to be insulator-dependent, suggesting long-range interactions are mediated by chromatin insulators rather than PcG complexes [37]. Disruption of CTCF insulator activity in Drosophila was also shown to induce a reduction in H3K27me3 levels within associated Pc-domains, rather than a spread into adjacent chromatin domains [12].
The reduction in H3K27me3 suggests that maintenance, perhaps facilitated by PRC1-mediated compaction of Pc domains, is negatively affected by loss of CTCF. Meanwhile, insulators in D. melanogaster are also capable of bridging PREs with target gene promoters [38], analogous to enhancer-promoter communication, thereby facilitating targeted gene repression. The co-localization of mammalian CTCF to both RNAP II transcription factories and Pc bodies [39,40] underscores the apparent role of insulator-mediated interactions in gene regulation and its relationship to nuclear architecture.
Cell signaling and development
The participation of chromatin insulators in facilitating both active and repressive gene regulation has led to current speculation that insulators ultimately direct the localization of specific loci to discrete nuclear subcompartments, and that this process may be regulated via post-translational modification of insulator proteins [36]. In particular, insulator activities appear to be modulated through development, and recent studies have uncovered an apparent role for CTCF in cell signaling processes required for cell differentiation. In one study, computational analysis of spatial binding constraints for pairs of transcription factors identified pair-wise binding between CTCF and Early growth response protein 1 (Egr1) [41], a protein implicated in multiple signaling pathways in humans. Intact CTCF-binding has also been shown to be essential for the recruitment of receptor-regulated SMAD proteins (R-SMADs) to specific loci in response to TGF-β signaling [42,43]. Though CTCF appears to physically interact with specific R-SMADs [42], whether CTCF directly recruits SMADs or simply provides an accessible landscape for DNA-binding of SMADs and other cell signaling effectors, and whether CTCF re-directs the localization of signaling response genes, will require future exploration. Nevertheless, the relationship between cell signaling and CTCF-binding sites presents an attractive possibility for how environmental and developmental stimuli might direct changes in gene expression through insulator-mediated nuclear architecture. Supporting evidence comes from reports that insulator function and the recruitment of insulator proteins are regulated in response to developmental cues. In D. melanogaster, extensive changes in gene expression induced by 20-hydroxyecdysone, a steroid hormone that regulates various developmental processes, are accompanied and accomplished to some degree by changes in the occupancy of DNA-binding insulator proteins and recruitment of co-factors essential for insulator activity [7]. Similarly, insulator-mediated nuclear architecture was recently shown to be developmentally regulated at the conserved HOXA locus in mammals, wherein the recruitment of cohesin to CTCF sites, and consequently the organization and expression pattern at HOXA, is controlled by pluripotency factor OCT4 [44]. CTCF/cohesin complexes also associate with TATA-binding protein associated factor 3 (TAF3) in embryonic stem cells, where TAF3 is involved in long-range interactions important for the specification of endoderm lineages [45].
Regulatory mechanisms and signaling pathways aside, the embryonic lethality of CTCF-null mice testifies to the relative importance of chromatin insulators in development [46,47]. Pre-implantation development of Ctcf nullizygous embryos prior to early embryonic lethality was recently shown to depend on the presence of maternal Ctcf mRNA [48], further demonstrating that expression of CTCF is absolutely essential for early embryonic development. CTCF also plays a critical role in neuronal development and blood cell differentiation. In neurons, CTCF and cohesin help to establish cell identity by binding to variable exon promoters within the protocadherin (Pcdh) gene clusters [49,50]. CTCF-deficient neurons show decreased expression of Pcdh genes, resulting in neuronal growth defects and abnormal behavior in conditional knockout mice [51]. Interestingly, CTCF and cohesin exhibit paired binding at variable exons within the protocadherin α complex in a manner suggestively similar to a recently discovered role for chromatin insulators in alternative splicing in immune cells [49,52]. CTCF also plays a key role in B-cell and T-cell differentiation through regulation of long-range interactions and appropriate chromatin structures involved in V(D)J recombination [53,54].
Cell cycle: DNA replication and mitosis
The role of insulators in nuclear organization and cellular development would suggest that insulators might also represent epigenetic marks for inheriting the blueprints of appropriate higher-order chromatin architecture and relevant gene expression patterns from one cell generation to the next [55]. In support of this possibility, DNA-binding insulator proteins, including CTCF, appear to remain associated with condensed mitotic chromosomes [56,57]. However, the dynamic association of insulator proteins at certain individual loci suggests that not all chromatin insulators are maintained throughout mitosis, and in some cases must be reassembled by the next cell generation [58]. Comparison of insulator protein-binding sites mapped by ChIP-seq in populations of interphase and mitotic cells reveals that only a subset of Drosophila insulators are retained during mitosis [59]. Mitotic insulators are enriched for the tandemly aligned insulator subclass, and commonly associate with DREF, a transcription factor involved in regulating the expression of DNA replication and cell cycle genes. Moreover, inherited insulators are enriched for the Orc2 origin recognition complex, suggesting these sites may provide a bookmark for assembly of pre-replication complexes. Mapping of CTCF across several human cell types also supports a role for chromatin insulators in DNA replication. In particular, cell-type specific CTCF-binding sites were found to be enriched within early- and middle replication time zones, and positively correlated with early- and middle- replication timing [14]. BORIS, an additional nuclear factor that shares high homology with the central DNA-binding domain of CTCF, also appears to play a role in the coordination of DNA replication events and cell cycle progression [60]. Elucidating the functional relationship between CTCF and BORIS, the nature of insulator protein dynamics throughout the cell cycle, and the exact role of chromatin insulators in mitosis remain poorly characterized yet important issues to be addressed by future studies.
CONCLUSIONS
Our understanding of how non-coding regulatory elements effectively coordinate the genome for efficient transcription, replication, DNA repair, and cell division remains far from complete. However, genome biologists are now equipped with improved technologies and novel assays for querying the fundamental nature of nuclear organization and genome function in unprecedented detail. As a consequence, our understanding of how chromatin insulators are involved in nuclear architecture and the role of insulator proteins in cellular processes has rapidly evolved within just the past year. In particular, insulators have outgrown the simple enhancer-blocking and barrier activities for which they were first defined, and instead play an active role in facilitating long-range interactions that direct enhancer-promoter communication, co-localization of Pc domains, and contacts between PREs and target genes. Recent studies have also extended our understanding of where insulators fit into cellular differentiation and the cell cycle, together suggesting that cell growth and development is accompanied by changes in insulator-mediated nuclear architecture and epigenetic inheritance of insulator complexes at specific sites. Nevertheless, how insulators mechanistically direct the specificity and influence the stability of long-range interactions and to what degree insulator-mediated chromatin structure actively drives cellular development remain some of the key questions for future research.
ACKNOWLEGMENTS
Work in the authors’ laboratory is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM035463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
* of special interest
** of outstanding interest
- 1.Van Bortle K, Corces VG. Nuclear Organization and Genome Function. Annu Rev Cell Dev Biol. 2012 doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Bortle K, Corces V. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012;3 doi: 10.4161/trns.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000814. e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soshnev AA, He B, Baxley RM, Jiang N, Hart CM, Tan K, Geyer PK. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi J, Corces V. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012 doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive Identification and Annotation of Cell Type-Specific and Ubiquitous CTCF-Binding Sites in the Human Genome. PLoS One. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. * Comparison of CTCF binding sites across the genome of 38 Human cell lines. This study provides an in-depth look at ubiquitous and cell-type specific CTCF sites and their relationship to gene structure, chromatin domain boundaries, and DNA replication.
- 15.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. 2011 doi: 10.1038/emboj.2011.406. **The first study to identify tRNA genes as chromatin insulators in humans, suggesting tDNAs and transcription factor TFIIIC play a highly conserved role in genome biology from yeast to humans
- 17.Carriere L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, Jounier S, Aude JC, Keime C, Murvai J, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:270–283. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 22.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 23.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012 doi: 10.1101/gr.138156.112. * A Comprehensive analysis of distinct classes of insulator proteins in D. melanogaster. Findings suggest endogenous insulator elements are not acting as enhancer-blockers or barrier elements as previous characterized in transgenic reporter assays. Also provides a look at the relationship between different insulator proteins by ChIP-chip analyses after RNAi
- 25. Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. ** Identification of cis- and trans-interactions facilitated by CTCF binding in mouse ES cells. Correlational analyses between interaction sites and chromatin profiling suggest CTCF functionally organizes coregulated genes into distinct chromatin domains, and that chromatin loops include functional interactions between enhancers and target gene promoters
- 26. Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012 doi: 10.1016/j.cell.2012.01.010. ** First genome-wide map of physical domains in Drosophila, with comparisons to epigenetic signatures. This study also demonstrates the correlational relationship between insulators and physical domain separation.
- 27. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. ** Genome-wide mapping of physical domains in humans and mice. This study also emphasizes the correlation between insulators, house-keeping genes, and tDNAs at chromatin boundaries
- 28. Hou C, Li L, Qin Z, Corces V. Gene density, transcription and insulators contribute to partitioning the Drosophila genome into physical domains. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.08.031. In press. ** Independent mapping of physical domains and epigenetic associations in Drosophila. This study provides valuable insight into the relationship between "epigenetic domains" and physical domains, suggesting physical domains are independent of specific chromatin profiles, and that gene density, orientation, and active transcription strongly correlate with and thus may drive the separation of topological domains
- 29.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong CT, Ramos E, Van Bortle K, Corces V. Poly(ADP-ribosyl)ation regulates insulator function and intra-chromosomal interactions in Drosophila. Submitted Manuscript. 2012 doi: 10.1016/j.cell.2013.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev. 2012;22:79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.11.004. * A comprehensive review of the relationship between chromatin insulators and long-range interactions between Pc domains. This review brings together numerous findings that ultimately suggest that insulator elements, rather than PREs, direct the co-localization of Pc targets. Provides attractive model in which insulators dynamically direct the localization of associated loci to transcription factories for gene activation, or Pc bodies for gene repression.
- 37. Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. * This study demonstrates that interactions between DNA elements derived from the bithorax complex, previously thought to rely on Pc for co-localization, depend on inherent insulator elements, rather than PREs.
- 38.Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci U S A. 2011;108:2294–2299. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Methods. 2011;8:963–968. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002638. e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bergstrom R, Savary K, Moren A, Guibert S, Heldin CH, Ohlsson R, Moustakas A. Transforming growth factor beta promotes complexes between Smad proteins and the CCCTC-binding factor on the H19 imprinting control region chromatin. J Biol Chem. 2010;285:19727–19737. doi: 10.1074/jbc.M109.088385. * This study provides compelling evidence that CTCF is necessary for the localization of SMAD proteins to specific loci bound by CTCF, in reponse to TGF-β signaling.
- 43.Burton T, Liang B, Dibrov A, Amara F. Transforming growth factor-beta-induced transcription of the Alzheimer beta-amyloid precursor protein gene involves interaction between the CTCF-complex and Smads. Biochem Biophys Res Commun. 2002;295:713–723. doi: 10.1016/s0006-291x(02)00725-8. [DOI] [PubMed] [Google Scholar]
- 44.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore JM, Rabaia NA, Smith LE, Fagerlie S, Gurley K, Loukinov D, Disteche CM, Collins SJ, Kemp CJ, Lobanenkov VV, et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One. 2012;7:e34915. doi: 10.1371/journal.pone.0034915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dekker J. CTCF and cohesin help neurons raise their self-awareness. Proc Natl Acad Sci U S A. 2012;109:8799–8800. doi: 10.1073/pnas.1206195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF Is Required for Neural Development and Stochastic Expression of Clustered Pcdh Genes in Neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribeiro de Almeida C, Stadhouders R, Thongjuea S, Soler E, Hendriks RW. DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation. Blood. 2012;119:6209–6218. doi: 10.1182/blood-2012-03-402586. [DOI] [PubMed] [Google Scholar]
- 55.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 58.Komura J, Ikehata H, Ono T. Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc Natl Acad Sci U S A. 2007;104:15741–15746. doi: 10.1073/pnas.0702363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurudatta BV, Sung ER, Yang J, Bryant D, Donlin-Asp PG, Corces V. DREF and other Drosophila insulator proteins remain bound to mitotic chromosomes at sites of pre-replication complex assembly. Submitted Manuscript. 2012 [Google Scholar]
- 60.Rosa-Garrido M, Ceballos L, Alonso-Lecue P, Abraira C, Delgado MD, Gandarillas A. A cell cycle role for the epigenetic factor CTCF-L/BORIS. PLoS One. 2012;7:e39371. doi: 10.1371/journal.pone.0039371. [DOI] [PMC free article] [PubMed] [Google Scholar]