Conspectus
Many peptides and proteins self-assemble into amyloid fibrils, including polypeptides that are associated with human amyloid diseases, mammalian and fungal prion proteins, and proteins that are believed to have biologically functional amyloid states. Proper understanding of the common propensity for polypeptides to form amyloid fibrils depends on elucidation of the molecular structures of these fibrils, as does rational design of amyloid inhibitors and imaging agents. Whereas amyloid fibril structures were largely mysterious 15 years ago, a considerable body of reliable structural information now exists, with important contributions from solid state nuclear magnetic resonance (NMR) measurements. This article reviews results from our laboratories and discusses several structural issues that have been sources of controversy.
In many cases, the molecular structures of amyloid fibrils are not determined uniquely by their amino acid sequences. Self-propagating, molecular-level polymorphism complicates the structure determination problem and can lead to apparent disagreements between results from different laboratories, when in fact different laboratories are simply studying different polymorphs. For 40-residue β-amyloid (Aβ1–40) fibrils associated with Alzheimer’s disease, we have developed detailed structural models from solid state NMR and electron microscopy data for two polymorphs, which we found to have similar peptide conformations, identical in-register parallel β-sheet organizations, but different overall symmetry. Other polymorphs have also been partially characterized by solid state NMR, and appear to have similar structures. In contrast, cryo-electron microscopy studies that use significantly different fibril growth conditions have identified structures that appear (at low resolution) to be different from those examined by solid state NMR.
The in-register parallel β-sheet organization found in β-amyloid fibrils has also been found in many other fibril-forming systems by solid state NMR and electron paramagnetic resonance (EPR), and is attributable to stabilization of amyloid structures by intermolecular interactions among like amino acids, including hydrophobic interactions and polar zippers. Surprisingly, antiparallel β-sheets have been identified and characterized by solid state NMR in certain fibrils formed by the D23N mutant of Aβ1–40, which is associated with early-onset, familial neurodegenerative disease. Antiparallel D23N-Aβ1–40 fibrils are metastable with respect to conversion to parallel structures, and therefore represent an off-pathway intermediate in the amyloid fibril formation process. Evidence for antiparallel β-sheets in other amyloid-formation intermediates has been obtained recently by other methods.
As an alternative to simple parallel and antiparallel β-sheet structures, β-helical structural models have been proposed for various fibrils, especially those formed by mammalian and fungal prion proteins. Solid state NMR and EPR data show that fibrils formed in vitro by recombinant PrP have in-register parallel β-sheet structures, but the structure of infectious PrP aggregates is not yet known definitively. The fungal HET-s prion protein has been shown by solid state NMR to have a β-helical structure, but all yeast prions studied by solid state NMR (i.e., Sup35p, Ure2p, and Rnq1p) have in-register parallel β-sheet structures, with the fibril core being formed by their Gln- and Asn-rich N-terminal segments.
Introduction
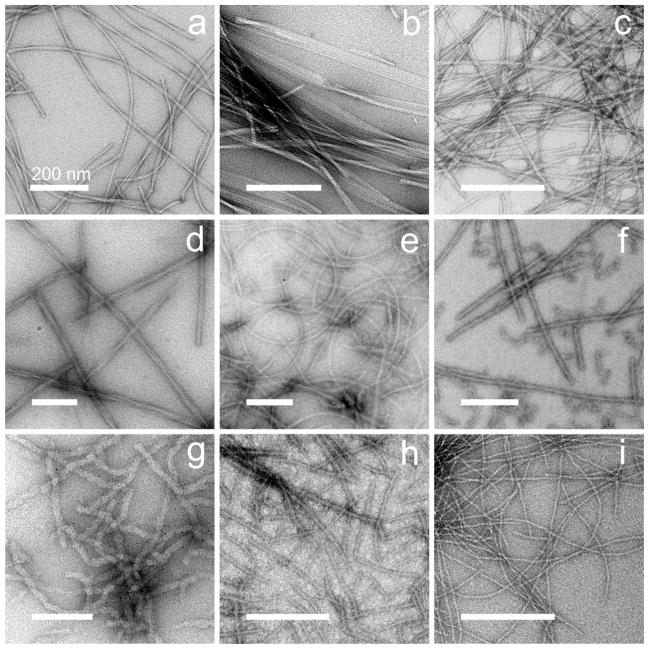
Many polypeptides can self-assemble into filamentous entities known as amyloid fibrils. Amyloid-forming polypeptides include disease-associated molecules such as the β-amyloid peptide of Alzheimer’s disease, the α-synuclein protein of Parkinson’s disease, and the islet amyloid polypeptide (IAPP, or amylin) of type 2 diabetes. Prion proteins, responsible for transmissible spongiform encephalopathies in mammals and transmissible traits and diseases in yeast and other fungi, form amyloid fibrils that encode the prion state. Amyloid fibrils with essential biological functions have also been identified. Amyloid fibrils formed by polypeptides with quite diverse amino acid sequences have similar appearances in transmission electron microscope (TEM) images (Figure 1), suggesting that their underlying molecular structures are similar.
Figure 1.
Negatively stained TEM images of amyloid and prion fibrils, illustrating the diversity of fibril morphologies and fibril-forming polypeptides. (a) 40-residue β-amyloid (Aβ1–40) fibrils with “twisted” morphology.2,10 (b) Aβ1–40 fibrils with “striated ribbon” morphology.3,10 (c) 37-residue islet amyloid polypeptide (IAPP or amylin) fibrils.14 (d) Recombinant PrP fibrils with “R” morphology,16 Syrian hamster sequence, residues 23–231. (e) PrP fibrils with “S” morphology.16 (f) Pmel17 repeat domain fibrils.15 (g) Ure2p prion fibrils, full-length S. cerevisiae sequence.43 (h) Sup35NM prion fibrils.41 (i) HET-s prion fibrils, P. anserina sequence, residues 218–289.4,19 All scale bars are 200 nm.
The defining molecular structural property of an amyloid fibril, distinguishing it from non-amyloid filaments formed by proteins such as actin and hemoglobin S, is that an amyloid fibril contains a “cross-β” motif. A cross-β motif is a ribbon-like β-sheet, comprised of β-strand segments running nearly perpendicular to the fibril growth direction, linked by inter-strand hydrogen bonds running nearly parallel to the growth direction. The presence of cross-β motifs in amyloid fibrils was established well before 1990.1 All other aspects of amyloid fibril structures were unclear, however, largely because amyloid fibrils are inherently noncrystalline and insoluble and hence are not amenable to direct structure determination by x-ray crystallography or multidimensional solution nuclear magnetic resonance (NMR) methods. For example, the identities of the β-strand segments, the organization of the β-sheets (parallel vs. antiparallel), the number of β-sheets, and the conformations of non-β-strand segments within amyloid fibrils were unknown. It was not even clear that amyloid fibrils contain well-defined molecular structures, rather than being highly disordered assemblies with cross-β character.
In the 1990s, new experimental approaches began to shed light on the molecular-level details of of amyloid structures. Solid state NMR techniques have been especially important, because these techniques provide detailed information about molecular and supramolecular structure without requiring crystallinity or solubility. By now, full structural models for fibrils formed by a variety of polypeptides have been developed from solid state NMR data.2–6 These models reveal the principal intermolecular interactions that stabilize fibril structures, help explain the nearly generic nature of amyloid formation, and provide a basis for future progress towards the discovery of new amyloid inhibitors and imaging agents. Related information has also been obtained from electron paramagnetic resonance (EPR).7–9
Despite dramatic advances over the past 15 years, an impression that amyloid structures are “controversial” persists in the biology, biochemistry, and chemistry communities. This article discusses some of the issues that have generated controversy, especially issues related to our own work. Although important structural questions remain to be answered, it is our view that consensus currently outweighs controversy.
Self-propagating polymorphism in β-amyloid fibrils: sequence does not determine structure
TEM images of amyloid fibrils formed by polypeptides with a given amino acid sequence typically show a variety of morphologies, differing in the diameter, twist period, curvature, or other properties (Figures 1a, b and 1d, e). When we began our studies in 1998, the molecular structural significance of these morphological variations was unclear. Our initial solid state NMR spectra of 40-residue β-amyloid (Aβ1–40) fibrils showed multiple sets of 13C NMR chemical shifts for individual 13C-labeled residues, varying in their relative intensities from sample to sample. Subsequent experiments showed that the chemical shifts correlated with the fibril morphologies, that the predominant chemical shifts and morphology could be controlled reproducibly by subtle variations in Aβ1–40 fibril growth conditions, and that both chemical shifts and morphologies were self-propagating in seeded fibril growth.10 Thus, fibrils with different morphologies have distinct molecular structures, i.e., the amino acid sequence alone does not uniquely determine the molecular structure in amyloid fibrils. Subsequent studies have provided additional support for molecular-level polymorphism in β-amyloid fibrils2, 11–13 and other fibrils.14–16 Polymorphism has also been observed in crystal structures of peptide fragments derived from amyloid-forming proteins.17
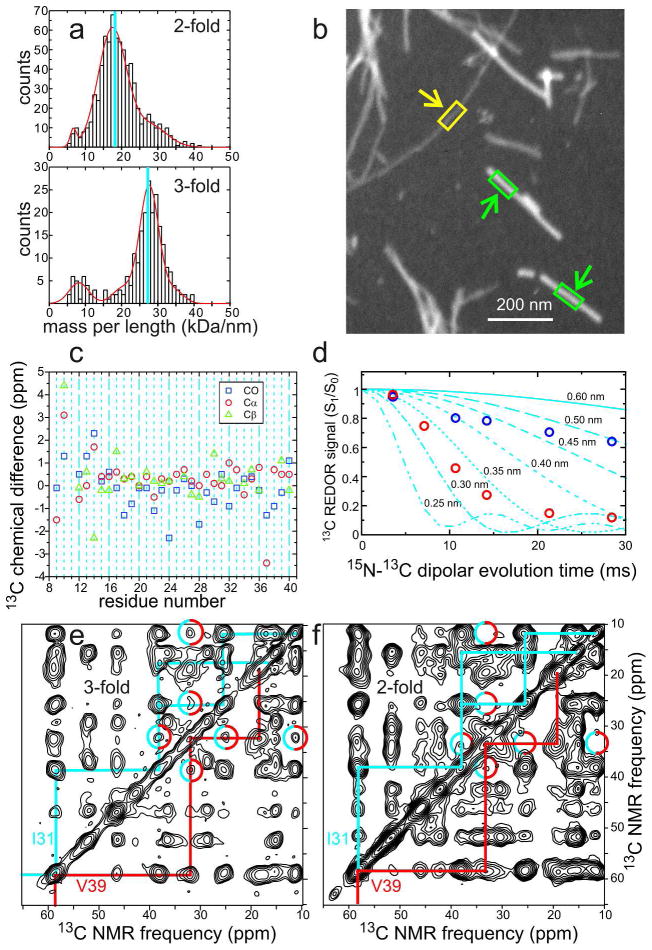
The mass-per-length (MPL) of a fibril is an important structural characteristic that can be measured by various dark-field electron microscopy techniques.18,19 Early measurements on polymorphic Aβ1–40 fibrils by Goldsbury et al. showed a distribution of MPL values, with peaks near 21, 31, and 42 kDa/nm.18 In our own measurements, we observed values around 18 kDa/nm in some samples and 27 kDa/nm in others. We interpreted these MPL values as evidence for structures with either two or three cross-β units, since a value near 9 kDa/nm is expected for one cross-β unit comprised of 4.3 kDa peptides with the standard 4.7–4.8 Å interstrand spacing of a β-sheet. Since solid state NMR spectra of isotopically labeled, morphologically homogeneous Aβ1–40 fibrils show a single set of 13C NMR chemical shifts, these structures must have approximate two-fold or three-fold symmetry about the long fibril axis.
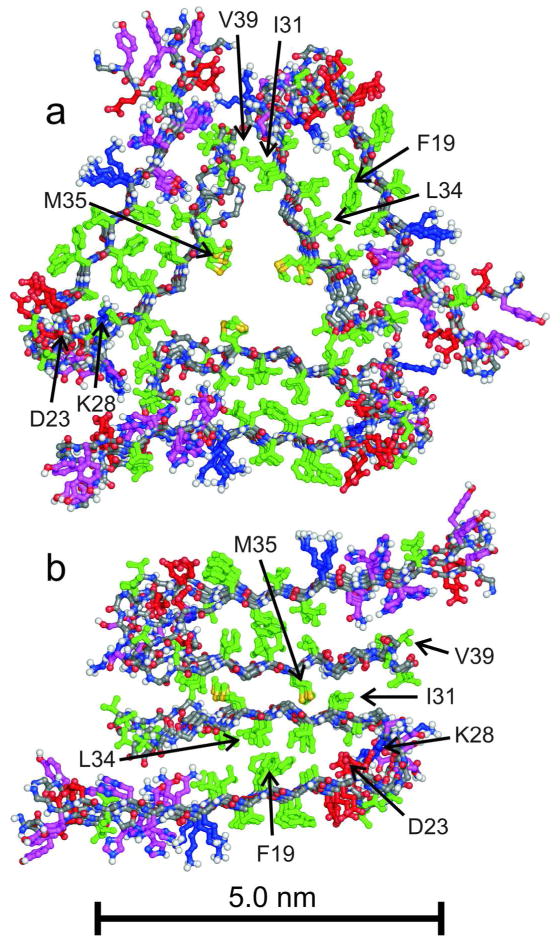
Molecular models for specific Aβ1–40 polymorphs with two-fold and three-fold symmetry (Figure 2) were developed from sets of restraints that include conformation-dependent 13C chemical shifts, quantitative measurements of internuclear magnetic dipole-dipole couplings that restrain intramolecular and intermolecular distances,20,21 and semi-quantitative measurements of dipole-dipole couplings that reveal the proximities of specific pairs of amino acid sidechains. Root-mean-squared deviations for backbone and heavy atom coordinates in these models (see Protein Data Bank files 2LMN, 2LMO, 2LMP, and 2LMQ) are about 2.1 Å and 2.7 Å, respectively. Figure 3 shows representative experimental data that distinguish the two Aβ1–40 fibril structures. The models in Figure 2 illustrate some of the structural variations that underlie fibril polymorphism. Aβ1–40 adopts a U-shaped conformation in both models, with β-strands formed by residues 10–22 and 30–40 separated by a bend or loop that allows the two β-strands to fold back on one another. In both structures, the two β-strands form a double-layered cross-β unit, with a parallel β-sheet in each layer and with similar hydrophobic contacts between the two layers. The bend conformation, the number of cross-β units (either two or three), the contacts between cross-β units, and the overall symmetry are different in the two models.
Figure 2.
Structural models for Aβ1–40 fibrils developed from solid state NMR and electron microscopy data.2,3 Models with three-fold (a) and two-fold (b) symmetry correspond to “twisted” fibrils and to individual filaments within “striated ribbon” fibrils in Figures 1a and 1b, respectively. Models include residues 9–40 and are viewed in cross-section, with the long fibril axis perpendicular to the page. Four repeats of the parallel cross-β structures are shown, with sidechains of hydrophobic residues in green, polar residues in magenta, negatively charged residues in red, and positively charged residues in blue.
Figure 3.
Examples of experimental data that lead to the structural models for Aβ1–40 fibrils in Figure 2. (a) Mass-per-length (MPL) histograms derived from dark-field transmission electron microscopy,19 showing the predominant values of 18 kDa/nm and 27 kDa/nm expected for twofold and three-fold symmetric cross-β structures. (b) Example of an unstained dark-field TEM image, showing a three-fold Aβ1–40 fibril (yellow arrow) and tobacco mosaic virus particles (green arrows) that serve as MPL standards. Fibril MPL values are determined from the ratios of integrated intensities within yellow and green rectangles. (c) Differences in 13C NMR chemical shifts between two-fold and three-fold Aβ1–40 fibrils, for backbone carbonyl, α-carbon, and β-carbon sites. (d) Quantitative measurements of magnetic dipole-dipole couplings between K28 ζ-nitrogen and D23 γ-carbon sites in two-fold (red circles) and three-fold (blue circles) fibrils, using a frequency-selective rotational echo double-resonance technique.21 These data demonstrate the presence of D23-K28 salt bridge interactions in the two-fold fibrils3 and the absence of these interactions in three-fold fibrils.2 (e) Aliphatic region of a two-dimensional 13C-13C NMR spectrum of three-fold fibrils, prepared with uniform 13C labeling of I31, G33, M35, G37, and V39, recorded with a 1.5 s mixing period that allows long-range crosspeaks to develop. Intra-residue crosspeaks for I31 and V39 are identified by cyan and red assignment paths, respectively. Cyan/red circles indicate inter-residue crosspeaks that imply proximity of I31 and V39 sidechains,2 as in Figure 2a. (f) Similar two-dimensional spectrum of two-fold fibrils. Absence of clear crosspeaks in cyan/red circles implies a greater I31-V39 separation, as in Figure 2b.
Molecular-level polymorphism implies that different research groups are likely to study different molecular structures, unless their fibril growth conditions are identical, their fibrils are indistinguishable in TEM images, and their fibrils have the same NMR chemical shifts. Aβ1–40 is capable of forming at least 5–10 structurally distinct polymorphs.5,6 Bertini et al. have recently reported a two-fold symmetric Aβ1–40 fibril model, based on solid state NMR data, that resembles the model in Fig. 2b but differs in the details of contacts within and between cross-β units.22 Sample preparation conditions used by Bertini et al. were significantly different from our conditions, and 13C chemical shifts are different.
Reif and coworkers have reported 2D solid state NMR spectra of Aβ1–40 fibrils that show two sets of chemical shifts, suggesting a repeat unit containing two structurally inequivalent peptide molecules.23 Although fibril structures with two inequivalent molecules in the repeat unit are possible,24,25 it is difficult to rule out the alternative explanation that two polymorphs with different NMR spectra coexist in the NMR samples, unless nuclear spin polarization transfers between the two sets of chemical shifts are observed unambiguously.25
Cryo-electron microscopy (cryoEM) studies of Aβ1–40 and Aβ1–42 fibrils have produced models for the cross-sectional density of the fibrils that seem inconsistent with the models in Fig. 2.12,26,27 Again, sample preparation conditions for cryoEM studies were significantly different from our conditions, and the fibrils are clearly different in negatively-stained TEM images. For example, one Aβ1–40 fibril polymorph described by Grigorieff and coworkers, prepared in 50 mM borate buffer at pH 7.8 and 4° C, has a width of roughly 18 nm and approximate two-fold symmetry about the long fibril axis. This polymorph has MPL ≈ 46 kDa/nm, corresponding to five Aβ1–40 molecules per 4.8 Å repeat,26 a value that is difficult to reconcile with a two-fold symmetric, cross-β structure. Frieden and coworkers describe a two-fold symmetric, approximately tubular Aβ1–42 fibril polymorph, prepared at pH 2 and 37° C.27
Hydrogen/deuterium (H/D) exchange rates for backbone amide sites have been used to identify the structurally ordered or β-sheet-forming segments within amyloid fibrils. For Aβ1–40 and Aβ1–42 fibrils, most H/D exchange studies suggest that the ordered segments comprise a smaller fraction of the peptide sequence than is indicated by solid state NMR measurements,11,28 although some studies suggest otherwise.29 Although certain segments in amyloid fibrils exhibit extremely slow H/D exchange (< 10−5 s−1), segments with much faster H/D exchange (≥10−3 s−1) may still be part of the ordered structure and rigid from the standpoint of solid state NMR. Thus, apparent discrepancies between H/D exchange and solid state NMR results may simply reflect differences in the sensitivities of the two techniques to minor or infrequent structural fluctuations. Note that H/D exchange rates for intrinsically disordered proteins30 are typically greater than 10−2 s−1 near pH 7 and 5° C. Smaller rates at higher temperatures therefore indicate at least some degree of structural involvement or rigidity.
β-sheets in amyloid fibrils: parallel, antiparallel, or both?
Prior to 1998, amyloid fibrils were often assumed to contain antiparallel cross-β structures. Solid state NMR measurements on fibrils formed by residues 34–42 of the 42-residue β-amyloid peptide (Aβ34–42) supported this assumption.5 Subsequent solid state NMR measurements on residues 10–35 (Aβ10–35) by Lynn, Meredith, Botto, and coworkers6 provided the first evidence that amyloid fibrils could contain parallel cross-β structures, especially “in-register” parallel β-sheets in which backbone amide and carbonyl groups of residue n of one peptide chain are hydrogen-bonded to carbonyl and amide groups of residues n−1 and n+1 of a neighboring chain, respectively. By now, solid state NMR2,10,14 and EPR measurements7,8 on numerous systems have identified in-register parallel cross-β structures as the most common structures in amyloid fibrils. In-register parallel β-sheets appear to maximize intermolecular hydrophobic interactions and “polar zipper” interactions,31 accounting for the experimental results. Antiparallel β-sheets have been found in fibrils5,24,32 and amyloid-like crystals17 formed by short fragments of full-length amyloid-forming proteins, especially peptides that contain a single hydrophobic β-strand, so that either parallel or antiparallel alignments can produce favorable intermolecular hydrophobic interactions. Antiparallel alignment may then produce more favorable electrostatic interactions.
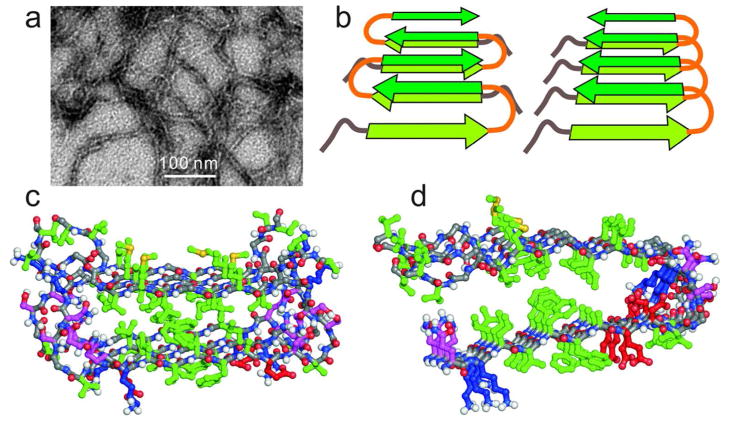
Surprisingly, the Asp23-Asn mutant of Aβ1–40 (D23N-Aβ1–40) was recently found to form both straight fibrils with the expected parallel cross-β structure and more curved and shorter fibrils with an unanticipated antiparallel cross-β structure,13,33 as shown in Figure 4. The structural model for antiparallel D23N-Aβ1–40 fibrils in Figure 4c, developed from solid state NMR data,13 shows how favorable hydrophobic interactions in an amyloid fibril core can be achieved with antiparallel intermolecular alignments, using a U-shaped peptide conformation that is similar to the peptide conformations in Figure 2.
Figure 4.
(a) Negatively stained TEM image of D23N-Aβ1–40 fibrils, prepared by a method that selects metastable, antiparallel β-sheet structures.13 (b) Schematic comparison of antiparallel (left) and parallel (right) cross-β structures that can be constructed from similar U-shaped peptides. (c) Structural model for metastable D23N-Aβ1–40 fibrils developed from solid state NMR data,13 viewed in cross-section. Two repeats (four D23N-Aβ1–40 chains, residues 15–40 only) are shown, with sidechains colored as in Figure 2. (d) Residues 15–40 of one cross-β unit within a two-fold Aβ1–40 fibril. Comparison with panel c shows how similar sets of hydrophobic interactions in the core of the cross-β unit can stabilize both parallel and antiparallel structures.
Interestingly, antiparallel D23N-Aβ1–40 fibrils are metastable, slowly converting to parallel structures when both structures are initially present.13 For D23N-Aβ1–40, antiparallel cross-β structures therefore represent “off-pathway” intermediates in the process of parallel cross-β fibril formation. These observations suggest that the worm-like “protofibrils” that have been described as neurotoxic intermediates in wild-type Aβ1–40 and Aβ1–42 fibril formation34 may have similar antiparallel structures. Alternatively, Härd and coworkers have suggested that protofibrils are comprised of β-hairpins,35 which are distinct structures in that they contain intramolecular hydrogen bonds. β-hairpin structures have been observed in crystal structures of small oligomeric states of amyloid-forming peptides.36
Structures of prion fibrils: parallel β-sheets, β-helices, or something else?
A β-helix (or β-solenoid) is a structural motif in which short β-strand segments alternate with bends to create multiple coils in a solenoid-like rod. The β-strand segments are perpendicular to the long axis of the rod, as in a cross-β motif. A single polypeptide chain contributes many coils to the β-helix. Early on, β-helical structures were proposed for amyloid fibrils,35 and more recently for mammalian and fungal prions.36–38
Definitive evidence for a β-helical structure in the case of the HET-s prion of Podospora anserina has been obtained by Meier and coworkers, who have developed a very precise structural model for HET-s fibrils from a large set of solid state NMR measurements.4 In HET-s fibrils, each protein molecule contributes two coils to the β-helix, so that hydrogen bonds between β-strand segments are both intramolecular and intermolecular. HET-s fibrils have a number of properties that distinguish them from disease-associated fibrils, especially an absence of polymorphism at biologically relevant pH. In contrast to HET-s fibrils, fibrils formed by the yeast prion proteins Sup35p, Ure2p, and Rnq1p exhibit both structural heterogeneity and biological heterogeneity (multiple prion strains on infection of yeast).
Whether β-helices exist in other amyloid and prion fibrils remains an open question. β-helical models for mammalian PrP fibrils have been proposed, originally based on electron microscopy of PrP constructs in two-dimensional crystalline form.37 Direct measurements on recombinant PrP fibrils indicate an in-register parallel β-sheet core structure, formed by a C-terminal segment that is roughly 60 residues in length.8 Brain-derived, infectious PrP aggregates exhibit strong protection from H/D exchange in a larger segment of the protein sequence, spanning approximately 145 residues.38 Additional experimental data are clearly needed before firm conclusions can be drawn about the molecular structures in infectious PrP aggregates.
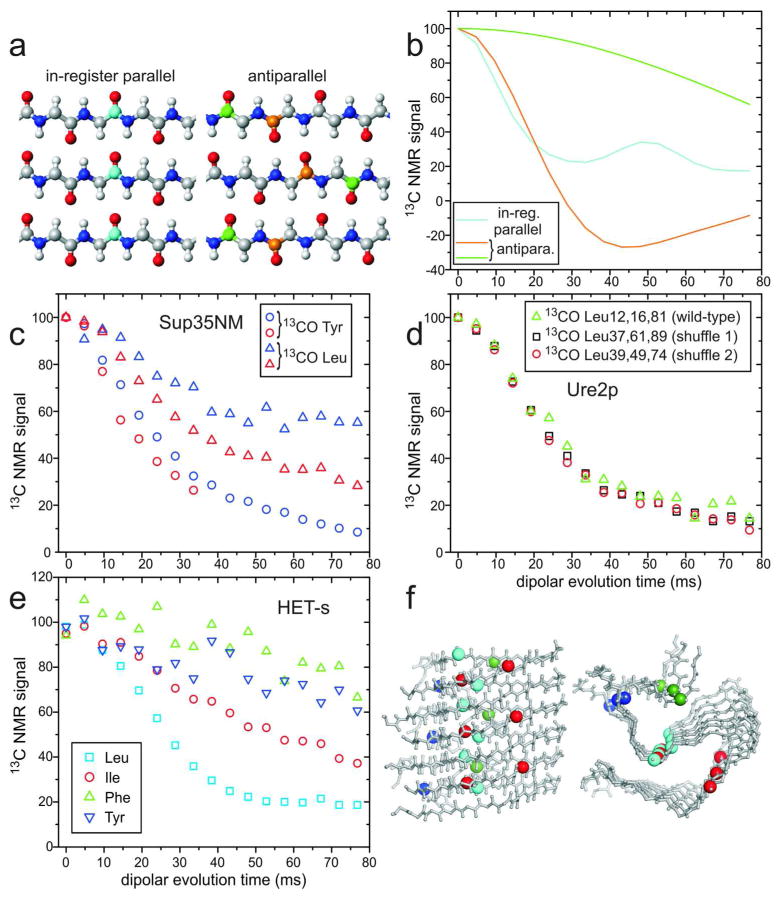
Extensive solid state NMR measurements on yeast prion fibrils, including fibrils formed by the prion domains of Sup35p,39–41 Ure2p,40,42,43 and Rnq1p,44 support in-register parallel β-sheet structures, probably stabilized by polar zippers31 among the many Gln and Asn residues in these domains. Evidence for in-register parallel structures comes from multiple measurements of intermolecular dipole-dipole interactions among multiple 13C-labeled sites, in samples that were lyophilized,39,40,42 lyophilized and then rehydrated,41,44 and never lyophilized,41,43 as well as in samples that were shown to have biological activity41 (i.e., prion infectivity in yeast). Some of these data are shown in Figure 5, including data for HET-s fibrils (not previously published) that illustrate the difference in β-sheet organization.
Figure 5.
Investigations of β-sheet structures in prion fibrils, through measurements of intermolecular 13C-13C magnetic dipole couplings with the PITHIRDS-CT technique.20 (a) Ideal in-register parallel and antiparallel β-sheets. (b) Ideal PITHIRDS-CT curves for an in-register parallel β-sheet with a single carbonyl 13C label at any residue in each peptide chain (cyan) and for antiparallel β-sheets with a single carbonyl 13C label at either of two residues (orange and green). (c) PITHIRDS-CT data for Sup35NM fibrils, prepared with 13C labels at backbone carbonyl sites of all Tyr (circles) or Leu (triangles) residues. Data were obtained from lyophilized (blue) or fully hydrated (red) samples.41 All 20 Tyr residues are in the N domain of Sup35NM, while 7 of 8 Leu residues are in the M domain. (d) PITHIRDS-CT data for wild-type Ure2p1–89 fibrils (triangles), prepared with 13C labels at backbone carbonyl sites of all Leu residues, and for two “shuffled prion” fibrils40 with Leu at the indicated residue numbers (squares and circles). (e) PITHIRDS-CT data for HET-s fibrils, residues 218–289, prepared with 13C labels at backbone carbonyl sites of the indicated residues, in separate samples. (f) Side and cross-sectional views of the HET-s fibril model developed by van Melckebeke et al. (PDB 2KJ3).4 Residues 224–248 and 259–289 are shown, corresponding to the most highly ordered and rigid segments. Labeled carbonyl sites are represented by large spheres, colored according to panel e. In the β-helical core structure of HET-s fibrils, only Leu carbonyls have ~5 Å nearest-neighbor distances. Signal contributions from labeled sites outside the core are uncertain. (All PITHIRDS-CT data are corrected for natural-abundance 13C contributions.41)
Random shuffling of amino acids within a yeast prion domain (Fig. 5d) does not abrogate prion formation, a finding that is consistent with an in-register parallel β-sheet structure stabilized by polar zippers,45 but is more difficult to reconcile with β-helices that depend on specific sets of intramolecular and intermolecular interactions. MPL measurements indicate one molecule per 4.7–4.8 Å repeat,19 consistent with the in-register parallel architecture. Lateral association of specific numbers of β-helices would be required to explain the MPL data. Moreover, in-register parallel β-sheet structures provide a natural explanation for the existence and self-propagation of prion strains (i.e., biologically distinct manifestations of prion infection, caused by the same prion protein). Whether prion strains are due to variations in the number of cross-β units and symmetry, as in Figure 2, variations in the identities or lengths of β-sheet-forming segments,46 or variations in the locations of folds in the β-sheets,45 the favorable interactions between identical sidechains in a parallel structure force monomers that add to a growing fibril to adopt the same conformation as molecules in the bulk.
Sup35NM is a commonly studied construct of Sup35p, the prion protein responsible for the [PSI+] trait in yeast, containing the Gln- and Asn-rich N-terminal prion (N) domain and the Glu-rich “middle” (M) domain, but not the C-terminal globular domain. Data from two types of experiments have been presented as evidence for a β-helical structure in Sup35NM fibrils: (i) Kishimoto et al. reported that the 8–10 Å equatorial scattering peak in x-ray fiber diffraction data, attributed to stacking of β-sheets perpendicular to the long fibril axis, was not detectable in fully hydrated samples (from which the total x-ray scattering intensity due to fibrils was also weak).47 While these data argue against a structure that contains many stacked β-sheets, they do not imply a β-helical structure. Calculated fiber diffraction patterns for realistic in-register parallel β-sheet structures show relatively weak and broad equatorial scattering features, due to the limited number β-sheet layers within a single fibril48; (ii) Krishnan and Lindquist reported measurements of fluorescence emission spectra from pyrene-labeled Sup35NM fibrils, showing strong red-shifted excimer fluorescence characteristic of pyrene-pyrene proximity only when the labels were in residues 25–38 and 91–106. From these data, they suggested a “head-to-head, tail-to-tail” β-helical structure for Sup35NM fibrils.49 Although one might naively expect the excimer fluorescence to be uniform across the sequence in an in-register parallel β-sheet, the phenomenon of excimer fluorescence requires a specific geometry of pyrene-pyrene interactions that is likely to be inaccessible in the relatively rigid and crowded core of a fibril. Unlike solid state NMR measurements of intermolecular distances, which require only isotopic labeling, pyrene labeling requires introduction of a pyrene maleimide group that is larger than any amino acid sidechain. Therefore, the absence of strong excimer fluorescence for residues between 38 and 91 does not imply absence of in-register parallel β-sheets. Moreover, excimer fluorescence ratios for samples in which pyrene-labeled Sup35NM was diluted in unlabeled Sup35NM appear to support an in-register parallel structure, in that the excimer fluorescence is reduced by only a factor of two with 1:3 dilution.49 (Within an in-register parallel β-sheet, each labeled site has two intermolecular neighbors, each of which could be labeled.)
Solid state NMR data indicate that portions of the M domain, in addition to the N domain, participate in the in-register parallel β-sheet structure of Sup35NM fibrils. Specifically, measurements of dipole-dipole couplings among 13C labels at carbonyl sites of leucine residues (Fig. 5c) indicate that at least four of the eight leucine carbonyls have nearest-neighbor distances of approximately 5 Å to other leucine carbonyls, although only one leucine is in the N domain.39,41 Participation of the M domain in β-sheet structure is unexpected because it is highly charged. Although the N domain is sufficient for propagation of many [PSI+] strains, others require proximal parts of M for faithful propagation.50 Also, barriers to transmission of [PSI+] between sequence polymorphs depend in part on differences in the M domain.51 Moreover, solution NMR data suggest that four leucine residues are at least partially immobilized in Sup35NM fibrils, and that portions of the M domain remain partially protected from H/D exchange.46 It seems that the structural and dynamical properties of the M domain may vary among [PSI+] strains.
In the case of Ure2p, responsible for the [URE3] trait in yeast, solid state NMR spectra reported by Loquet et al. reveal that the C-terminal enzymatic domain retains its globular structure but becomes immobilized in Ure2p fibrils.52 This observation rules out structural models in which the C-terminal domain is loosely tethered to a cross-β core. Loquet et al. initially suggested that their data support a model for Ure2p fibril structure proposed by Melki and coworkers, in which the C-terminal domain forms the fibril core, without an N-terminal cross-β structure.53 Our subsequent solid state NMR measurements confirmed immobilization of the C-terminal domain, but additionally showed that the N-terminal domain does indeed form an in-register parallel β-sheet structure in the context of full-length Ure2p fibrils.43 The C-terminal domain is selectively removed by proteinase K treatment, supporting a model in which dimerized C-terminal domains form a helical shell around the cross-β core.43 Fibrils formed by a recombinant N-terminal segment of Ure2p are infectious in yeast, and the N-terminal domain alone (but not the C-terminal domain) is sufficient for prion propagation in vivo.54,55
Concluding remarks
Our understanding of amyloid and prion fibrils structures has made remarkable progress in the past 15 years, with major contributions from solid state NMR measurements. While important questions remain about the structures of fibrils (and prefibrillar intermediates) formed by specific proteins, existing data for well-studied systems do not contain real contradictions. Where different laboratories have obtained different results for the same protein or peptide, apparent inconsistencies are largely attributable to the highly polymorphic nature of most amyloids, or to differences in the structural information content of various experimental techniques. Future work is likely to focus on the development of detailed structural models for fibrils formed by larger proteins, on further clarification of the specific structural variations that underlie prion strains, and on characterization of the specific structural states that are important in amyloid and prion diseases.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Biographies
Robert Tycko is a Senior Investigator in the NIH Intramural Research Program, where he has been since 1994. His background includes A. B. and Ph.D. degrees in chemistry, and eight years as a Member of Technical Staff at AT&T Bell Laboratories. His research centers on solid state NMR and its applications in biophysics and biophysical chemistry.
Reed Wickner is a Distinguished NIH Investigator. His background includes a B.A. in mathematics and an M.D. His research has focused on genetic, biochemical and biophysical studies of viruses and prions of Saccharomyces cerevisiae.
References
- 1.Eanes ED, Glenner GG. X-Ray Diffraction Studies on Amyloid Filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 2.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular Structural Basis for Polymorphism in Alzheimer’s β-Amyloid Fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petkova AT, Yau WM, Tycko R. Experimental Constraints on Quaternary Structure in Alzheimer’s β-Amyloid Fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Melckebeke H, Wasmer C, Lange A, Eiso AB, Loquet A, Bockmann A, Meier BH. Atomic-Resolution Three-Dimensional Structure of Het-s(218–289) Amyloid Fibrils by Solid state NMR Spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 5.Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RGS, Tidor B, Griffin RG. Structural Model for the β-Amyloid Fibril Based on Interstrand Alignment of an Antiparallel-Sheet Comprising a C-Terminal Peptide. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 6.Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating Structure of Alzheimer’s β-Amyloid(10–35) Is Parallel β-Sheet with Residues in Exact Register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margittai M, Langen R. Fibrils with Parallel In-Register Structure Constitute a Major Class of Amyloid Fibrils: Molecular Insights from Electron Paramagnetic Resonance Spectroscopy. Q Rev Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 8.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular Architecture of Human Prion Protein Amyloid: A Parallel, In-Register β-Structure. Proc Natl Acad Sci U S A. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedrood S, Li YY, Isas JM, Hegde BG, Baxa U, Haworth IS, Langen R. Fibril Structure of Human Islet Amyloid Polypeptide. J Biol Chem. 2012;287:5235–5241. doi: 10.1074/jbc.M111.327817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s β-Amyloid Fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 11.Kodali R, Williams AD, Chemuru S, Wetzel R. Aβ1–40 Forms Five Distinct Amyloid Structures Whose β-Sheet Contents and Fibril Stabilities Are Correlated. J Mol Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Aβ1–40 Fibril Polymorphism Implies Diverse Interaction Patterns in Amyloid Fibrils. J Mol Biol. 2009;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiang W, Yau WM, Luo YQ, Mattson MP, Tycko R. Antiparallel β-Sheet Architecture in Iowa-Mutant β-Amyloid Fibrils. Proc Natl Acad Sci U S A. 2012;109:4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luca S, Yau WM, Leapman R, Tycko R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu KN, McGlinchey RP, Wickner RB, Tycko R. Segmental Polymorphism in a Functional Amyloid. Biophys J. 2011;101:2242–2250. doi: 10.1016/j.bpj.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarava N, Baskakov IV. The Same Primary Structure of the Prion Protein Yields Two Distinct Self-Propagating States. J Biol Chem. 2008;283:15988–15996. doi: 10.1074/jbc.M800562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic Structures of Amyloid Cross-β Spines Reveal Varied Steric Zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury CS, Wirtz S, Muller SA, Sunderji S, Wicki P, Aebi U, Frey P. Studies on the In Vitro Assembly of Aβ1–40: Implications for the Search for Aβ Fibril Formation Inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 19.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of Amyloid Fibril Mass-Per-Length by Tilted-Beam Transmission Electron Microscopy. Proc Natl Acad Sci U S A. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tycko R. Symmetry-Based Constant-Time Homonuclear Dipolar Recoupling in Solid State Nmr. J Chem Phys. 2007:126. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 21.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency Selective Heteronuclear Dipolar Recoupling in Rotating Solids: Accurate 13C-15N Distance Measurements in Uniformly 13C, 15N-Labeled Peptides. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 22.Bertini I, Gonnelli L, Luchinat C, Mao JF, Nesi A. A New Structural Model of Aβ40 Fibrils. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 23.del Amo JML, Schmidt M, Fink U, Dasari M, Fandrich M, Reif B. An Asymmetric Dimer as the Basic Subunit in Alzheimer’s Disease Amyloid-β Fibrils. Angew Chem-Int Edit. 2012;51:6136–6139. doi: 10.1002/anie.201200965. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen JT, Bjerring M, Jeppesen MD, Pedersen RO, Pedersen JM, Hein KL, Vosegaard T, Skrydstrup T, Otzen DE, Nielsen NC. Unique Identification of Supramolecular Structures in Amyloid Fibrils by Solid state NMR Spectroscopy. Angew Chem-Int Edit. 2009;48:2118–2121. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- 25.Lewandowski JR, van der Wel PCA, Rigney M, Grigorieff N, Griffin RG. Structural Complexity of a Composite Amyloid Fibril. J Am Chem Soc. 2011;133:14686–14698. doi: 10.1021/ja203736z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt M, Sachse C, Richter W, Xu C, Fandrich M, Grigorieff N. Comparison of Alzheimer Aβ1–40 and Aβ1–42 Amyloid Fibrils Reveals Similar Protofilament Structures. Proc Natl Acad Sci U S A. 2009;106:19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Hu XY, Khant H, Ludtke SJ, Chiu W, Schmid MF, Frieden C, Lee JM. Interprotofilament Interactions between Alzheimer’s Aβ1–42 Peptides in Amyloid Fibrils Revealed by CryoEM. Proc Natl Acad Sci U S A. 2009;106:4653–4658. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D Structure of Alzheimer’s Amyloid-β1–42 Fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide Solvent Protection Analysis Demonstrates That Amyloid-β1–40 and Amyloid-β1–42 Form Different Fibrillar Structures under Identical Conditions. Biochem J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Structure and Properties of α-Synuclein and Other Amyloids Determined at the Amino Acid Level. Proc Natl Acad Sci U S A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine Repeats as Polar Zippers: Their Possible Role in Inherited Neurodegenerative Diseases. Proc Natl Acad Sci U S A. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid State NMR Reveals a pH-Dependent Antiparallel β-Sheet Registry in Fibrils Formed by a β-Amyloid Peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 33.Tycko R, Sciarretta KL, Orgel J, Meredith SC. Evidence for Novel β-Sheet Structures in Iowa Mutant β-Amyloid Fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams AD, Sega M, Chen ML, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. Structural Properties of Aβ Protofibrils Stabilized by a Small Molecule. Proc Natl Acad Sci U S A. 2005;102:7115–7120. doi: 10.1073/pnas.0408582102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandberg A, Luheshi LM, Sollvander S, de Barros TP, Macao B, Knowles TPJ, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, Lannfelt L, Dobson CM, Hard T. Stabilization of Neurotoxic Alzheimer Amyloid-β Oligomers by Protein Engineering. Proc Natl Acad Sci U S A. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao ML, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for Assembly of Prions with Left-Handed β3-Helices into Trimers. Proc Natl Acad Sci U S A. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural Organization of Brain-Derived Mammalian Prions Examined by Hydrogen-Deuterium Exchange. Nat Struct Mol Biol. 2011;18:504–506. doi: 10.1038/nsmb.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shewmaker F, Wickner RB, Tycko R. Amyloid of the Prion Domain of Sup35p Has an In-Register Parallel β-Sheet Structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of Shuffled Prion Domains That Form Prions Have a Parallel In-Register β-Sheet Structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 41.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two Prion Variants of Sup35p Have In-Register Parallel β-Sheet Structures, Independent of Hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R. Characterization of β-Sheet Structure in Ure2p1–89 Yeast Prion Fibrils by Solid state Nuclear Magnetic Resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 43.Kryndushkin DS, Wickner RB, Tycko R. The Core of Ure2p Prion Fibrils Is Formed by the N-Terminal Segment in a Parallel Cross-β Structure: Evidence from Solid state Nmr. J Mol Biol. 2011;409:263–277. doi: 10.1016/j.jmb.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the Basis of the [PIN+] Prion, Has a Parallel In-Register β-Sheet Structure. Proc Natl Acad Sci U S A. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemecek J, McGlinchey R, Bateman D, Winchester CL. Prion Amyloid Structure Explains Templating: How Proteins Can Be Genes. FEMS Yeast Res. 2010;10:980–991. doi: 10.1111/j.1567-1364.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyama BH, Kelly MJS, Gross JD, Weissman JS. The Structural Basis of Yeast Prion Strain Variants. Nature. 2007;449:233–U8. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 47.Kishimoto A, Hasegawa K, Suzuki H, Taguchi H, Namba K, Yoshida M. β-Helix Is a Likely Core Structure of Yeast Prion Sup35 Amyloid Fibers. Biochem Biophys Res Commun. 2004;315:739–745. doi: 10.1016/j.bbrc.2004.01.117. [DOI] [PubMed] [Google Scholar]
- 48.McDonald M, Box H, Bian W, Kendall A, Tycko R, Stubbs G. Fiber Diffraction Data Indicate a Hollow Core for the Alzheimer’s Aβ 3-Fold Symmetric Fibril. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.08.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnan R, Lindquist SL. Structural Insights into a Yeast Prion Illuminate Nucleation and Strain Diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley ME, Liebman SW. The Sup35 Domains Required for Maintenance of Weak, Strong or Undifferentiated Yeast [PSI+] Prions. Mol Microbiol. 2004;51:1649–1659. doi: 10.1111/j.1365-2958.2003.03955.x. [DOI] [PubMed] [Google Scholar]
- 51.Bateman DA, Wickner RB. [PSI+] Prion Transmission Barriers Protect Saccharomyces Cerevisiae from Infection: Intraspecies ‘Species Barriers’. Genetics. 2012;190:569–U463. doi: 10.1534/genetics.111.136655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, Habenstein B, Schutz A, Meier BH, Melki R, Bockmann A. Prion Fibrils of Ure2p Assembled under Physiological Conditions Contain Highly Ordered, Natively Folded Modules. J Mol Biol. 2009;394:108–118. doi: 10.1016/j.jmb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 53.Bousset L, Thomson NH, Radford SE, Melki R. The Yeast Prion Ure2p Retains Its Native α-Helical Conformation Upon Assembly into Protein Fibrils In Vitro. EMBO J. 2002;21:2903–2911. doi: 10.1093/emboj/cdf303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masison DC, Maddelein ML, Wickner RB. The Prion Model for [URE3] of Yeast: Spontaneous Generation and Requirements for Propagation. Proc Natl Acad Sci U S A. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brachmann A, Baxa U, Wickner RB. Prion Generation In Vitro: Amyloid of Ure2p Is Infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]