Abstract
Advances in cancer genomics have been propelled by the steady evolution of molecular profiling technologies. Over the past decade, high-throughput sequencing technologies have matured to the point necessary to support disease-specific shotgun sequencing. This has compelled whole-genome sequencing studies across a broad panel of malignancies. The emergence of high-throughput sequencing technologies has inspired new chemical and computational techniques enabling interrogation of cancer-specific genomic and transcriptomic variants, previously unannotated genes, and chromatin structure. Finally, recent progress in single-cell sequencing holds great promise for studies interrogating the consequences of tumor evolution in cancers presenting with genomic heterogeneity.
Keywords: next-generation sequencing, cancer genomics, transcriptomics, chromosomal conformation sequencing, bioinformatics, tumor heterogeneity
1. Introduction
Cancer is often described as a population of cells that have either lost genomic integrity or have altered the way they manifest their genome. Another way to say this is that cancer is a genomic and epigenomic disease. Advances in genomic profiling have accordingly benefitted our understanding of cancer [1, 2]. Molecular aberrations identified in cancer using genomic profiling predict therapeutic response, providing a suite of diagnostic features that may help inform treatment selection and patient care [3, 4]. Cancer genomics is thus at the heart of both basic and translational research aiming to eradicate this deadly disease.
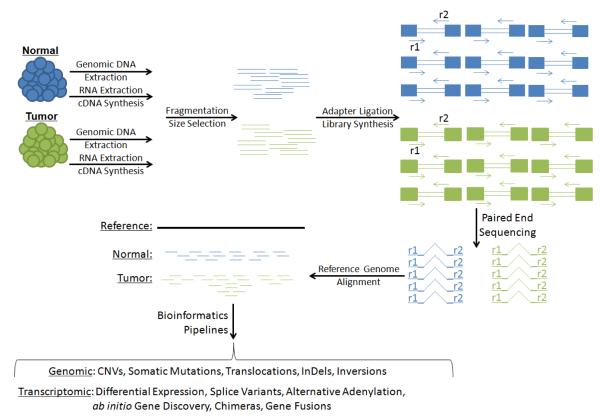
A major limitation of the previous generation of genomics technologies was its reliance on hybridization and probe-based techniques. While highly informative with respect to copy number variations (CNVs) [5], single-nucleotide polymorphisms (SNPs) [6], and differential expression of known transcripts [7], microarrays by design capture only information about genomic variation that can be assessed by probes with predetermined sequences. Reviewed here are advances reflected in the current generation of cancer genomics, which are being propelled by the rapid evolution of high-throughput sequencing technology and concomitant bioinformatics [8]. This generation of genomic technologies is furthering our understanding of the cancer genome by providing insight into cancer-associated somatic sequence variants, genomic conformations, and expression of previously unannotated transcripts (Figure 1).
Figure 1. Shotgun Sequencing.
Genomic DNA and cDNA synthesized from RNA are fragmented into a size range compatible with the specifications of massively parallel high-throughput sequencing machines. During repair of fragmented ends, distinct adapters are ligated to each end, permitting paired-end sequencing of DNA fragments. Read pairs are then aligned computationally against a reference genome. Read depth at specific loci can be used to assess genomic copy number or RNA expression of genes. Bioinformatic comparisons of normal, tumor, and reference genomes identify sequence variants associated with disease. Discordant read pairs where the ends of a pair align to separate genomic loci nominate structural genomic rearrangements and expressed chimeras. Evidence of junction spanning read pairs supports ab initio annotation of novel genes in previously unannotated loci.
Several cancers have been studied extensively using high-throughput sequencing. The Cancer Genome Atlas (TCGA) is an ongoing publicly funded endeavor invested in comprehensively studying the cancer genomes across a broad panel of cancer types [9]. Several whole-exome and a few whole-genome sequencing studies independent of the TCGA have also been reported [10-14].
2. Highly Parallel Shotgun Sequencing
Shotgun sequencing is defined by the practice of sequencing many overlapping genomic fragments while relying on computational algorithms exploiting regions of sequence overlap to assemble fragments into a whole genome. This differs from clone-based sequencing where defined segments of the genome are cloned into and sequenced from bacterial or yeast artificial chromosomes [15]. A significant debate in the late 1990s over the feasibility of applying shotgun-sequencing to the genomes of higher order organisms focused on major concerns such as cost, logistics, and the computational challenges inherent to assembling large genomes from relatively short reads [16,17]. Nevertheless, a first draft of the human genome developed by using shotgun sequencing was published in 2001 [18].
While this draft was a critical step toward enabling cancer genome sequencing, the National Human Genome Research Institute estimated the cost of shotgun sequencing in 2001 at nearly $100 million per genome. During the decade since, sequencing technologies have significantly improved their throughput, reducing this cost into the range of thousands of dollars per genome [19]. These advances have made cancer genome sequencing studies economically feasible.
3. Genomic Sequencing
Genomic sequencing can be divided into two major categories: whole-genome sequencing and targeted sequencing. Whole-genome sequencing refers to sequencing the full length of the genome, whereas targeted sequencing enables greater depth of coverage for higher resolution analysis of sequence variation within specific genomic elements. The most frequent case of targeted sequencing is whole-exome sequencing, where known exons are enriched and analyzed for mutations. In contrast, whole-genome sequencing identifies structural changes in the cancer genome, such as inversions, deletions, and other genomic rearrangements, in addition to mutations [8].
Targeted exome sequencing efforts have identified recurrent somatic mutations in diseases where oncogenic drivers were previously not well known. For instance, it was recently reported that the most frequent class of genomic aberrations in malignant melanomas is activating events in the MAPK pathway [20]. Interestingly, the same study showed differences between cutaneous, acral, uveal, and mucosal melanomas with respect to their proclivities for accumulating specific aberrations within this pathway. This suggests exploitation of a common pathway across known anatomical subtypes of disease while also laying a foundation for investigating differences in the molecular etiologies of each type. Similarly, the mutational landscape as determined through exome sequencing of high-grade serous ovarian cancer reveals that, while more than 95% of sequenced tumors demonstrate inactivating mutations in participants of the tumor-suppressive TP53-pathway, activating alterations (mutational and amplification) of oncogenes were diverse and rarely recurrent in more than 10% of samples [21]. In a study of colorectal carcinomas, more than 90% of tumor samples presented with activating events in the WNT signaling pathway, while the specific events responsible occurred at low frequencies across a broad panel of genes involved in the WNT pathway [22].
An added layer of complexity is emerging from early whole-genome studies. While the existence of recurrent copy-number alterations in cancer has been well documented using array–comparative genomic hybridization (CGH) studies [23], whole-genome sequencing studies have also revealed a breadth of genomic rearrangements previously underappreciated in many solid tumors [14, 21, 24]. While accepted as drivers of oncogenesis in hematologic malignancy, recurrent genomic rearrangements have recently also come to the fore as oncogenic events in solid tumors where recurrent functional gene fusions are being identified by sequencing [25].
The emerging theme of complex genetic backgrounds within cancers and differing mechanisms of oncogenesis across cancer subtypes has prompted creative computational biology efforts to understand the structural genomic rationale behind the tissue specificity of oncogenic events [for example, 26]. This study in particular identified a significant association between the breakpoints of recurrent somatic copy-number alterations and guanine-rich DNA stretches capable of adopting G-quadraplex structure. A separate study identified a strong association between the genomic endpoints involved in somatic copy-number changes in cancer and genomic loci involved in chromosomal contact, thus lending further support to the hypothesis that recurrent rearrangements are structurally predisposed by genomic conformation [27].
4. Transcriptome Sequencing and Genome Annotation
The use of expressed sequence tags as a method for identifying genes has a long history in the molecular biology of gene discovery and the annotation of the human genome [28]. The technique involves cloning-based sequencing of cDNA fragments produced from RNA extracts and cloned into bacterial artificial chromosomes. Much like shotgun genome sequencing accelerated genomics, shotgun transcriptome sequencing (RNAseq) also overcomes the throughput limitations of cloning-based methods in transcriptomics. The result has been RNAseq-driven discovery of cancer-associated functional gene-fusion transcripts [29] and in some cases cancer-associated transcripts from genes annotated for the first time [30].
Inspired by advances in transcriptome sequencing, bioinformatics techniques for ab initio transcriptome assembly were developed with the intent of resolving alternatively spliced isoforms and were applied to questions in developmental biology [31]. However, this creative suite of assembly tools has also led to discovery of transcripts derived from genomic loci previously considered gene deserts. Specifically, ab initio assembly of RNAseq reads generated from libraries developed from 16 normal human tissues as part of the Illumina BodyMap Project reported as many as 8000 long non-coding RNA (lncRNA) genes. These RNAs are >200bp in length and are transcribed from intergenic loci previously underappreciated as possible gene sites because of their lack of protein-coding potential [32]. Similar ab initio assembly also identified functional lncRNAs associated with prostate cancer progression [30]. It will be interesting to see how annotation of the genome with new genes discovered by RNAseq will lead to reinterpretation of prior genome-wide association studies and CGH studies reporting recurrent but overlooked cancer-associated SNPs and CNVs in genomic regions dismissed as gene deserts.
In addition to finding new genes, RNAseq also permits sequence analysis of transcript variants of known genes arising from alterations in posttranscriptional RNA processing. Such studies have provided insight into mechanisms leading to oncogene dysregulation in the absence of genomic aberrations. For instance, it was recently reported that a chimeric transcript generated by cis-splicing of the adjacent genes SLC45A3 and ELK4 results in expression of a proliferation-promoting fused gene product despite there being no chromosomal rearrangement at this locus [33]. Likewise, alternative polyadenylation leading to truncation of the 3 -untranslated region and concomitant escape of microRNA-mediated transcript decay was observed as a posttranscriptional mechanism for elevated expression of known oncogenes [34]. Alternative splice variants of the androgen receptor identified by RNAseq analysis are also thought to contribute to prostate cancer progression from a hormone therapy–responsive to a treatment-resistant state [35]. Together these results suggest that posttranscriptional sequence variants in the transcriptome may constitute an underappreciated class of molecular events contributing to oncogenesis and tumor progression.
The ability to call sequence variants from RNAseq data also presents the potential of using transcriptome sequencing as a method of identifying genomic events. For instance RNAseq calls identifying tumor-specific single-nucleotide variants and chimeras could, respectively, nominate somatic mutations and putative gene-fusions. However, because RNA processing can alter transcripts independent of genomic events, an orthogonal method of validating calls using genomic DNA would be required to confirm the genomic origins of nominated alterations. Recent work comparing sample-matched genomic and transcriptomic mutation calls demonstrated the value of calls performed on transcriptome sequencing data in interpreting somatic mutations identified by genomic sequencing [36]. Specifically, the study demonstrated the value of integrating RNAseq data with somatic mutation calls from whole-genome sequencing to categorize mutations into four expressional categories: silent, expressed-variant, wild-type biased, and mutant-biased. Such categorization helps separate mutations of functional consequence from bystander events that although present in the cancer genome are not expressed or are biased against at the RNA level.
5. Sequencing Malignant Epigenomes
While tumor genome and cancer transcriptome sequencing have helped identify molecular aberrations leading to altered gene function in malignant disease, the field of epigenetics has focused on understanding how heritable patterns of genomewide expression are encoded and passed on between generations of cells. This “code above the code” consists of chemical changes to the DNA and the proteins it interacts with to form transcriptionally-accessible and transcriptionally-repressed chromatin states. Epigenetic alterations therefore do not require alterations to the coding sequence of the genome to promote pathogenesis. Instead changes in the epigenome alters the selective accessibility of genomic elements to transcriptional machinery, a process thought to be responsible for allowing the genome of a single zygote to manifest the variety of stable tissue specific cell-lineages seen in mammalian development [37, 38]. Advances in high-throughput sequencing enable analysis of genomewide landscapes for histone modifications by ChIP-seq [39], DNA-methylation by high-throughput sequencing of bisulfite-converted [40] or methyl-enriched genomic libraries [41], and transcriptionally-accessible chromatin regions by the DNAse-seq method of identifying DNAseI digestion hypersensitive sites (DHS) in the genome [42].
Cancer epigenome studies have exploited these technologies to expand our understanding of epigenetic mechanisms of tumor development and progression. Recent work in colorectal carcinoma integrated H3K4me1 ChIPseq with DNAse-seq to identify variant enhancer loci associated with known genetic risk alleles for colorectal cancer, implying a role for epigenetically active loci in the pathogenesis of this disease. [43] A study of prostate cancer DNA-methylation using MethylPlex-NGS revealed regions of cancer specific DNA methylation despite no significant difference with respect to the total number of methylated regions between benign and malignant tissue, suggesting a role for the distribution of this epigenetic mark in prostatic malignancy. [41] Additionally DNA-methylation has established roles in spontaneous deamination of cytosine causing C to T mutations and the development of microsatellite instability, both of which provide a causal mechanism linking changes in the epigenome to potential downstream genetic events. [44] As these technologies mature and become more widely implemented, we are likely to learn much about the etiology of cancers driven by specific molecular lesions from the integration of epigenomic and genomic sequencing findings.
6. HiC: Understanding Genomic Conformations
Although genomic sequences are frequently represented as linear character strings, it would not be physically possible for the human genome to be contained within the nuclei of cells in such form. Instead the genome is densely packaged into a structurally complex conformation allowing it to fit within the nucleus of somatic cells [45]. The mechanics of such packaging necessarily induces proximity of linearly distant elements of the genome causing both interchromosomal and long-range intrachromosomal contacts [46]. Emerging epigenetic studies suggest that the structural conformation of the genome is carefully regulated as a mechanism for defining transcriptional assemblies responsible for coordinated regulation of gene-networks [47].
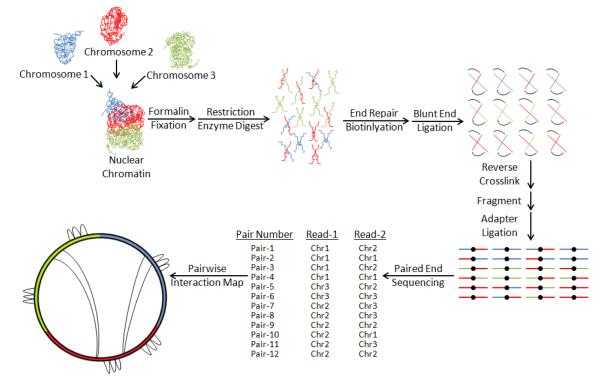
HiC is a high-throughput sequencing method that utilizes mate-pair sequencing of crosslinked and fragmented chromatin extracts processed by restriction enzymes such that each end represents a different member of two DNA segments engaged in structural interaction. The result of this unique sequencing modality is an unbiased map of pairwise interactions between any two points in the genome [48] (Figure 2). Recently, HiC delineation of ERG-induced chromosomal conformations associated with coordinated gene expression in prostate cancer provided proof-of-principle that the technique may prove valuable in discovering conformational genomic interactions underlying transcription factor–driven oncogenic activity [49].
Figure 2. HiC Method for Chromosomal Conformation Sequencing.
Long-range interchromosomal and intrachromosomal interactions implicated in regulation of epigenetically coordinated gene networks can be identified by HiC. Briefly, chromosomal interactions are preserved by formalin fixation, while restriction enzyme digestion cuts the genome into fragments containing sites of formalin fixation between genomic segments. Sticky ends are then repaired using biotinylated nucleotides and the repaired ends are ligated as blunt-ends introducing biotin at the site of ligation before crosslinks are reversed. DNA is then sheared to a fragment size compatible with high-throughput sequencing, biotin bearing fragments are isolated, and a paired-end sequencing library is made where each end represents a partner in a pairwise interaction. Pairs are then mapped by sequence alignment to a reference genome, creating a genome-wide atlas of structural interactions.
In addition to HiC, 5C [50] and ChIA-PET [51] assays have also been developed to map structural interactions between genomic loci. The advantage of ChIA-PET is its inclusion of a chromatin immunopreciptation step allowing for identification of genomic interacts specifically associated with a defined protein [51]. While there is no chromatin immunoprecipitation step in 5C, the method has the advantage of flexibility due the ability to use either high-throughput sequencingor genomic microarrays to generate readouts [50]. Despite methodological differences between the two techniques a comparison of interaction maps generated by 5C and ChIA as part of the ENCODE project demonstrates high concordance between interactions identified by the two assays [53].
6. Integrative Analysis: Inferring Disease-Relevant Events and Interactions Across Profiles
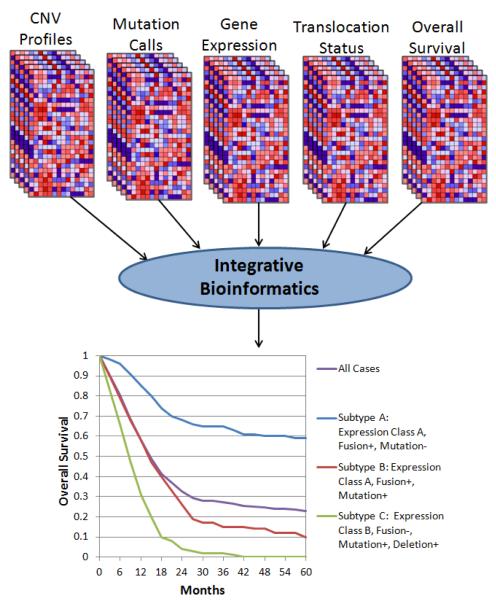
Integrative analysis has proven to be a powerful approach to ascertaining information across multiple platforms of genomic profiles. Such analysis aims to infer associations that are not apparent when analyzing a single form of genomic profile (e.g., copy number only) but become apparent when reading one genomic profile in light of another (e.g., copy number alterations cooperating with a defined mutation status). The assumption inherent to such analysis is that some genomic aberrations are stochastic events, resulting from genomic instability, that do not contribute to disease biology. Integrative analysis attempts to enrich for clinically relevant functional events by identifying aberrations supported by data across multiple genomic analysis platforms (Figure 3).
Figure 3. Integrative Analysis.
Emerging bioinformatics pipelines utilize a combination of graph theory, statistical enrichment, and correlation algorithms to identify associations between distinct categories of genomic aberrations and clinical outcome. The goal of these pipelines is to push past the noise of single-platform analysis by nominating potentially functional genomic attributes supported across a panel of analyses.
An early opportunity for integrative bioinformatics arose from the evolution of datasets generated by screening the NCI60 panel of cell lines for both drug sensitivity and genomic attributes [53]. While many bioinformatics toolkits were initially developed to address profile queries within each screening modality (reviewed in [53]), over time the desire for analysis across the modalities compelled the development of CellMiner [54]. The tool is structured as a relational database of the individual NCI60 molecular profiles, permitting extraction of complex associations across the compendia of genomic studies performed on this cell-line panel.
Mutually Exclusive Modules in Cancer (MEMo) is a tool recently developed to use a combination of correlation analysis and graph theory to integrate multiplatform data with the intention of identifying gene sets nominating dysregulation of a pathway. When applied to data from the TCGA characterizing glioblastoma multiforme, the method homed in on two regulatory networks identified by a total of six genes [55]. This represents a significant reduction in the number of genes nominated as putative functional drivers for interrogation in follow-up studies. The bioinformatics package integrOmics is another example. This tool was written in R to facilitate integration between any pair of –omics datasets using correlation analysis [56]. Given the creative approach taken by existing and admittedly early algorithms, it will be interesting to see what new strategies for multiplatform genomic data integration emerge as this field matures.
Advances in the analysis of data-rich genomic profiles have spawned the emerging field of cancer systems biology. While integrative genomics emphasizes the identification of distinct genomic aberrations, cancer systems biology takes analysis a step further by examining regulatory interactions responsible for sustaining malignant pathobiology [57]. Many methods have been built on top of networks modeled from known biological interactions detailed in pathway databases. For example, the ARACNe algorithm was recently used to uncover regulatory interactions driving epithelial-to-mesenchymal transformation in glioblastoma, a critical step in cancer progression [58]. Similarly the recently developed tool PARADIGM integrates genomic profiles into a model of transcriptional, posttranscriptional, and cell-signaling interactions to infer activity profiles for interacting molecules [59]. The algorithm recently nominated activation of the FOXM1 signaling network as a critical contributor to ovarian cancer [21].
High-throughput sequencing datasets present several challenges for integrative and systems analysis not faced by early algorithms developed for use with microarrays. Two technical challenges are the computational power and algorithmic efficiency needed when processing datasets of the scale produced by sequencing. This challenge in particular will be important to overcome for integrative analysis to play a role in molecular diagnostics as clinical decisions will require a more rapid turnaround time than many current algorithms provide [60]. Analytically, sequencing provides information about changes in both relative abundance (copy number, expression) and sequence (alternative adenylation, mutations, rearrangements) between tumors and normal specimens. Thus, computational methodologies systematically accounting for changes in profile as well as changes in sequence are needed to fully capitalize on cancer genome sequencing data.
7. Single-Cell Sequencing: Interrogating Tumor Heterogeneity and Evolution
While genomic sequencing is helping to identify somatic aberrations unique to cancer that may serve as drug targets, clinical observations from targeted therapy trials have raised critical questions. Specifically, innate and acquired resistance to targeted therapy is a challenge shared across cancer types [61-64]. This has led some to develop dynamic models of treatment response, applying basic principles from evolutionary biology [65]. This view holds that tumors likely possess a plurality of clones that initiate, compete with each other to sustain, and regenerate overt lesions despite the selective pressure presented by various microenvironmental and therapeutic stresses. From this perspective, tumors are populations that evolve, and understanding this process requires an appreciation for both molecular aberrations and clonality.
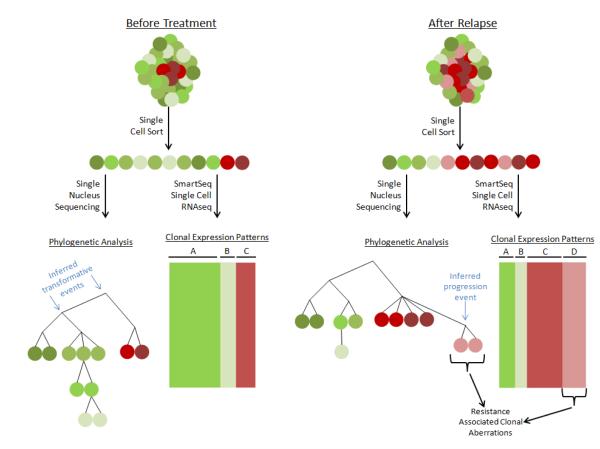
The sequencing modalities described so far do not directly address the heterogeneity of human cancer. However, the next generation of cancer genomics is emerging with sequencing technology permitting –omic scale analysis of single cells (Figure 4). Navin et al. presented single-nucleus sequencing as a powerful technique to assess CNVs between individual cells with sufficient resolution to enable evolutionary inferences with respect to the metastatic process [66]. Inspired by questions in developmental biology, Tang and associates developed methods for single-cell RNAseq [67], which they later applied to investigate transcriptional processes regulating the earliest stages of differentiation occurring in embryonic inner mass cells [68]. Sandberg and associates recently furthered single-cell RNAseq technology by developing Smart-Seq, which prepares sequencing libraries representing full-length RNAs through template switching during cDNA preparation prior to library amplification [69]. In this study, the group provided proof-of-principle by comparing the transcriptomes of circulating tumor cells to patient-matched melanoma tumors, indicating that the technique could be applied to study small quantities of cancer cells. Although very nascent, these techniques may help identify, characterize, and quantify the relative abundance of genomic and transcriptomic clones composing heterogeneous tumor cell populations.
Figure 4. Single-Cell Sequencing.
Emerging techniques for genome and transcriptome sequencing of single cells permit identification of cancer clones, their lineages, and clonal gene-expression programs in heterogeneous tumors. These technologies have ramifications for future studies of acquired therapeutic resistance and disease progression through tumor evolution.
The teleological thread held in common by cancer genomics efforts deploying single-cell sequencing is the belief that cancer is more than an altered normal genome. On the contrary, cancer from this perspective is viewed as a collection of independent genomes that evolve separately from and more quickly than the heritable germline of the host in which the tumor forms. Thus, while the past and present of cancer genomics has focused on identifying functional genomic aberrations in cancer cells, the next wave of cancer genomics is primed to study the evolution of whole genomes and the role of intratumoral evolutionary forces in driving the biology of heterogeneous tumors. It is an exciting time to be involved in cancer genome sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement None
References
- [1].Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- [2].Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- [3].Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Davies JJ, Wilson IM, Lam WL. Array CGH technologies and their applications to cancer genomes. Chromosome Res. 2005;13:237–248. doi: 10.1007/s10577-005-2168-x. [DOI] [PubMed] [Google Scholar]
- [6].Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr Opin Oncol. 2007;19:43–49. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- [7].Schulze A, Downward J. Navigating gene expression using microarrays - a technology review. Nat Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- [8].Metzker ML. Applications of next-generation sequencing: sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- [9].Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies, Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- [10].Stephens PJ, Tarpey PS, Davies H, Loo P. Van, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, Macdonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wei XM, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, Robinson W, Robinson S, Rosenberg SA, Samuels Y, N.C.S. Progra Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ellis MJ, Ding L, Shen D, Luo JQ, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du FY, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Adams J. Complex genomes: shotgun sequencing. Nat Educ. 2008;1 [Google Scholar]
- [16].Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- [17].Green P. Against a whole-genome shotgun. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- [18].Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Miklos G.L. Gabor, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Francesco V. Di, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- [19].Wetterstrand KA. [Accessed [Aug 2012]];DNA sequencing costs: data from the NHGRI Large-Scale Genome Sequencing Program. Available at: www.genome.gov/sequencingcosts.
- [20].Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- [21].Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, Godwin AK, Gross J, Hartmann L, Huang M, Huntsman DG, Iacocca M, Imielinski M, Kalloger S, Karlan BY, Levine DA, Mills GB, Morrison C, Mutch D, Olvera N, Orsulic S, Park K, Petrelli N, Rabeno B, Rader JS, Sikic BI, Smith-McCune K, Sood AK, Bowtell D, Penny R, Testa JR, Chang K, Dinh HH, Drummond JA, Fowler G, Gunaratne P, Hawes AC, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Santibanez J, Reid JG, Trevino LR, Wu YQ, Wang M, Muzny DM, Wheeler DA, Gibbs RA, Getz G, Lawrence MS, Cibulskis K, Sivachenko AY, Sougnez C, Voet D, Wilkinson J, Bloom T, Ardlie K, Fennell T, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, Koboldt DC, McLellan MD, Wylie T, Walker J, O’Laughlin M, Dooling DJ, Fulton L, Abbott R, Dees ND, Zhang Q, Kandoth C, Wendl M, Schierding W, Shen D, Harris CC, Schmidt H, Kalicki J, Delehaunty KD, Fronick CC, Demeter R, Cook L, Wallis JW, Lin L, Magrini VJ, Hodges JS, Eldred JM, Smith SM, Pohl CS, Vandin F, Raphael BJ, Weinstock GM, Mardis R, Wilson RK, Meyerson M, Winckler W, Getz G, Verhaak RGW, Carter SL, Mermel CH, Saksena G, Nguyen H, Onofrio RC, Lawrence MS, Hubbard D, Gupta S, Crenshaw A, Ramos AH, Ardlie K, Chin L, Protopopov A, Zhang JH, Kim TM, Perna I, Xiao Y, Zhang H, Ren G, Sathiamoorthy N, Park RW, Lee E, Park PJ, Kucherlapati R, Absher DM, Waite L, Sherlock G, Brooks JD, Li JZ, Xu J, Myers RM, Laird PW, Cope L, Herman JG, Shen H, Weisenberger DJ, Noushmehr H, Pan F, Triche T, Berman BP, Berg D.J. Van den, Buckley J, Baylin SB, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Jakkula LR, Durinck S, Han J, Dorton S, Marr H, Choi YG, Wang V, Wang NJ, Ngai J, Conboy JG, Parvin B, Feiler HS, Speed TP, Gray JW, Levine DA, Socci ND, Liang Y, Taylor BS, Schultz N, Borsu L, Lash AE, Brennan C, Viale A, Sander C, Ladanyi M, Hoadley KA, Meng S, Du Y, Shi Y, Li L, Turman YJ, Zang D, Helms EB, Balu S, Zhou X, Wu J, Topal MD, Hayes DN, Perou CM, Getz G, Voet D, Saksena G, Zhang JNH, Zhang H, Wu CJ, Shukla S, Cibulskis K, Lawrence MS, Sivachenko A, Jing R, Park RW, Liu Y, Park PJ, Noble M, Chin L, Carter H, Kim D, Karchin R, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Durinck S, Han J, Korkola JE, Heiser LM, Cho RJ, Hu Z, Parvin B, Speed TP, Gray JW, Schultz N, Cerami E, Taylor BS, Olshen A, Reva B, Antipin Y, Shen R, Mankoo P, Sheridan R, Ciriello G, Chang WK, Bernanke JA, Borsu L, Levine DA, Ladanyi M, Sander C, Haussler D, Benz CC, Stuart JM, Benz SC, Sanborn JZ, Vaske CJ, Zhu J, Szeto C, Scott GK, Yau C, Hoadley KA, Du Y, Balu S, Hayes DN, Perou CM, Wilkerson MD, Zhang N, Akbani R, Baggerly KA, Yung WK, Mills GB, Weinstein JN, Penny R, Shelton T, Grimm D, Hatfield M, Morris S, Yena P, Rhodes P, Sherman M, Paulauskis J, Millis S, Kahn A, Greene JM, Sfeir R, Jensen MA, Chen J, Whitmore J, Alonso S, Jordan J, Chu A, Zhang JH, Barker A, Compton C, Eley G, Ferguson M, Fielding P, Gerhard DS, Myles R, Schaefer C, Shaw KRM, Vaught J, Vockley JB, Good PJ, Guyer MS, Ozenberger B, Peterson J, Thomson E, C.G.A.R. Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Reid JG, Santibanez J, Shinbrot E, Trevino LR, Wu YQ, Wang M, Gunaratne P, Donehower LA, Creighton CJ, Wheeler DA, Gibbs RA, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, Stojanov P, McKenna A, Lander ES, Gabriel S, Getz G, Ding L, Fulton RS, Koboldt DC, Wylie T, Walker J, Dooling DJ, Fulton L, Delehaunty KD, Fronick CC, Demeter R, Mardis ER, Wilson RK, Chu A, Chun HJE, Mungall AJ, Pleasance E, Robertson AG, Stoll D, Balasundaram M, Birol I, Butterfield YSN, Chuah E, Coope RJN, Dhalla N, Guin R, Hirst C, Hirst M, Holt RA, Lee D, Li HI, Mayo M, Moore RA, Schein JE, Slobodan JR, Tam A, Thiessen N, Varhol R, Zeng T, Zhao Y, Jones SJM, Marra MA, Bass AJ, Ramos AH, Saksena G, Cherniack AD, Schumacher SE, Tabak B, Carter SL, Pho NH, Nguyen H, Onofrio RC, Crenshaw A, Ardlie K, Beroukhim R, Winckler W, Getz G, Meyerson M, Protopopov A, Zhang J, Hadjipanayis A, Lee E, Xi R, Yang L, Ren X, Zhang H, Sathiamoorthy N, Shukla S, Chen PC, Haseley P, Xiao Y, Lee S, Seidman J, Chin L, Park PJ, Kucherlapati R, Auman JT, Hoadley KA, Du Y, Wilkerson MD, Shi Y, Liquori C, Meng S, Li L, Turman YJ, Topal MD, Tan D, Waring S, Buda E, Walsh J, Jones CD, Mieczkowski PA, Singh D, Wu J, Gulabani A, Dolina P, Bodenheimer T, Hoyle AP, Simons JV, Soloway M, Mose LE, Jefferys SR, Balu S, O’Connor BD, Prins JF, Chiang DY, Hayes DN, Perou CM, Hinoue T, Weisenberger DJ, Maglinte DT, Pan F, Berman BP, Berg D.J. Van den, Shen H, T.T., Baylin SB, Laird PW, Getz G, Noble M, Voet D, Saksena G, Gehlenborg N, DiCara D, Zhang J, Zhang H, Wu CJ, Liu SY, Shukla S, Lawrence MS, Zhou L, Sivachenko A, Lin P, Stojanov P, Jing R, Park RW, Nazaire MD, Robinson J, Thorvaldsdottir H, Mesirov J, Park PJ, Chin L, Thorsson V, Reynolds SM, Bernard B, Kreisberg R, Lin J, Iype L, Bressler R, Erkkila T, Gundapuneni M, Liu Y, Norberg A, Robinson T, Yang D, Zhang W, Shmulevich I, Ronde J.J. De, Schultz N, Cerami E, Ciriello G, Goldberg AP, Gross B, Jacobsen A, Gao J, Kaczkowski B, Sinha R, Aksoy BA, Antipin Y, Reva B, Shen R, Taylor BS, Chan TA, Ladanyi M, Sander C, Akbani R, Zhang N, Broom BM, Casasent T, Unruh A, Wakefield C, Hamilton SR, Cason RC, Baggerly KA, Weinstein JN, Haussler D, Benz CC, Stuart JM, Benz SC, Sanborn JZ, Vaske CJ, Zhu J, Szeto C, Scott GK, Yau C, Ng S, Goldstein T, Ellrott K, Collisson E, Cozen AE, Zerbino D, Wilks C, Craft B, Spellman P, Penny R, Shelton T, Hatfield M, Morris S, Yena P, Shelton C, Sherman M, Paulauskis J, Gastier-Foster JM, Bowen J, Ramirez NC, Black A, Pyatt R, Wise L, White P, Bertagnolli M, Brown J, Chan TA, Chu GC, Czerwinski C, Denstman F, Dhir R, Doerner A, Fuchs CS, Guillem JG, Iacocca M, Juhl H, Kaufman A, Kohl B, Le X. Van, Mariano MC, Medina EN, Meyers M, Nash GM, Paty PB, Petrelli N, Rabeno B, Richards WG, Solit D, Swanson P, Temple L, Tepper JE, Thorp R, Vakiani E, Weiser MR, Willis JE, Witkin G, Zeng Z, Zinner MJ, Zornig C, Jensen MA, Sfeir R, Kahn AB, Chu AL, Kothiyal P, Wang Z, Snyder EE, Pontius J, Pihl TD, Ayala B, Backus M, Walton J, Whitmore J, Baboud J, Berton DL, Nicholls MC, Srinivasan D, Raman R, Girshik S, Kigonya PA, Alonso S, Sanbhadti RN, Barletta SP, Greene JM, Pot DA, Shaw KRM, Dillon LAL, Buetow K, Davidsen T, Demchok JA, Eley G, Ferguson M, Fielding P, Schaefer C, Sheth M, Yang L, Guyer MS, Ozenberger BA, Palchik JD, Peterson J, Sofia HJ, Thomson E. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Henry K.T. Mc, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- [26].De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- [29].Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren M.J. van, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- [34].Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, Chen K, Walker J, McDonald S, Bose R, Ornitz D, Xiong D, You M, Dooling DJ, Watson M, Mardis ER, Wilson RK. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews. Genetics. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- [38].Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- [39].Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nature reviews. Genetics. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nature methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- [41].Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, Kalyana-Sundaram S, Huang C, Shankar S, Jing X, Iyer M, Hu M, Sam L, Grasso C, Maher CA, Palanisamy N, Mehra R, Kominsky HD, Siddiqui J, Yu J, Qin ZS, Chinnaiyan AM. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome research. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, Willis J, Moore JH, Tesar PJ, Laframboise T, Markowitz S, Lupien M, Scacheri PC. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews. Genetics. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- [45].Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- [46].Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- [48].Lieberman-Aiden E, Berkum N.L. van, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rickman DS, Soong TD, Moss B, Mosquera JM, Dlabal J, Terry S, MacDonald TY, Tripodi J, Bunting K, Najfeld V, Demichelis F, Melnick AM, Elemento O, Rubin MA. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci USA. 2012;109:9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, Wei CL, Ruan Y, Sung WK. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome biology. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome research. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Weinstein JN. Integromic analysis of the NCI-60 cancer cell lines. Breast Dis. 2004;19:11–22. doi: 10.3233/bd-2004-19103. [DOI] [PubMed] [Google Scholar]
- [54].Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, Pommier Y, Weinstein JN. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Le Cao KA, Gonzalez I, Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yli-Harja O, Ylipaa A, Nykter M, Zhang W. Cancer systems biology: signal processing for cancer research. Chin J Cancer. 2011;30:221–225. doi: 10.5732/cjc.011.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Guan YF, Li GR, Wang RJ, Yi YT, Yang L, Jiang D, Zhang XP, Peng Y. Application of next-generation sequencing in clinical oncology to advance personalized treatment of cancer. Chinese journal of cancer. 2012;31:463–470. doi: 10.5732/cjc.012.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- [62].Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- [63].Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- [64].Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D’Andrea K, Pushparajan A, Hayden JE, Brown KD, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Michor F, Iwasa Y, Nowak MA. Dynamics of cancer progression. Nat Rev Cancer. 2004;4:197–205. doi: 10.1038/nrc1295. [DOI] [PubMed] [Google Scholar]
- [66].Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- [68].Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]