Abstract
This study aimed to compare epithelial cells derived from human embryonic stem cells (hESCs) to human ameloblast-lineage cells (ALCs), as a way to determine their potential use as a cell source for ameloblast regeneration. Induced by various concentrations of bone morphogenetic protein 4 (BMP4), retinoic acid (RA) and lithium chloride (LiCl) for 7 days, hESCs adopted cobble-stone epithelial phenotype (hESC-derived epithelial cells (ES-ECs)) and expressed cytokeratin 14. Compared with ALCs and oral epithelial cells (OE), ES-ECs expressed amelogenesis-associated genes similar to ALCs. ES-ECs were compared with human fetal skin epithelium, human fetal oral buccal mucosal epithelial cells and human ALCs for their expression pattern of cytokeratins as well. ALCs had relatively high expression levels of cytokeratin 76, which was also found to be upregulated in ES-ECs. Based on the present study, with the similarity of gene expression with ALCs, ES-ECs are a promising potential cell source for regeneration, which are not available in erupted human teeth for regeneration of enamel.
Keywords: ameloblast, cytokeratin, dental epithelial cells, human embryonic stem cells, odontogenesis
Introduction
Regenerative medicine promises novel therapies for tissue regeneration, including the potential to replace missing teeth with bioengineered tissues. Tooth formation is guided by reciprocal signaling interactions between the cranial neural crest-derived mesenchymal cells (forming dentin and pulp) and ectoderm-derived dental epithelial cells (forming enamel). Classic tissue recombination experiments have been used to successfully regenerate mouse, rat and pig tooth organs,1,2,3,4,5,6 and demonstrate that both epithelial and mesenchymal components are required for tooth organ regeneration. Dental mesenchymal stem cells from either human exfoliated deciduous or adult dental pulp are available for dentin/pulp complex bioengineering,7,8,9,10 while a dental epithelial component does not exist in the erupted human tooth for enamel tissue regeneration. Therefore, identification of alternative cell sources for dental epithelial cell regeneration remains a challenge.
Mammalian dental epithelial cells are unique in that they are derived from oral ectoderm and are responsible for the formation of the highly mineralized enamel. The ectoderm-derived oral epithelium located at sites where teeth will form, proliferates and differentiates to form a dental epithelial band (also referred as dental placode), which is the first sign of tooth formation. In communication with adjacent dental mesenchyme, dental epithelial cells progress through multiple stages of differentiation in bud and cap stages of tooth morphogenesis. In the subsequent bell stage, cytodifferentiation of dental epithelial cells results in ameloblast-lineage cells (ALCs), including presecretory, secretory and maturation ameloblasts. Enamel matrix protein secretion and matrix mineralization is initiated by secretory ameloblasts. Secreted matrix proteins self-assemble to form the unique structure of mineralized enamel.
Previous studies have shown that mouse embryonic stem cells and c-kit-positive mouse bone marrow cells can be differentiated into ameloblast-like cells.4,11,12,13 These studies of stem cells from mouse suggest that human embryonic stem cells (hESCs) could be a viable cell source for regeneration of dental epithelial cells.
hESCs are derived from the inner cell mass of blastocyst-stage human embryos. hESCs can self-renew and give rise to all types of cell in the human body.14 When induced with bone morphogenetic protein 4 (BMP4) and α-retinoic acid (RA), hESCs differentiate toward cells with an epithelial morphology.15 Cytokeratin expression is a characteristic of epithelial cells, and the cytokeratin expression pattern of hESC-derived epithelial cells (ES-ECs) has not been characterized and compared with the cytokeratin expression patterns of other epithelia.
Cytokeratins (CKs) are the typical intermediate filament proteins of epithelia, which are known to be expressed in an epithelial type- and stage-specific manner. CKs are chemically very stable, long and unbranched filaments of ∼10 nm in diameter, which are important for maintaining mechanical stability and integrity of epithelial cells and tissues at both the single cell and epithelial sheet levels. The CK gene family consists of 54 distinct functional genes in humans. CKs are heterodimers by pairing one type I keratin and one type II keratin (1∶1) molecule.16
In this study, we characterized the expression patterns of CKs and genes associated with early enamel organ epithelial differentiation in ES-ECs, as compared to human fetal skin epithelium (SE), fetal oral buccal mucosal epithelial cells (OEs) and ALCs. These results will direct our further studies to explore the potential for using ES-ECs to regenerate ALCs.
Materials and methods
Maintenance of hESCs
The hESC line (WA09) was purchased from the WiCell Research Institute (Madison, WS, USA). The cells were maintained on a layer of mitosis inactivated mouse embryonic fibroblasts (MEFs) in unconditioned medium: Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 medium containing 20% Knockout serum replacement (Life Technologies, Carlsbad, CA, USA), non-essential amino acids (Millipore, Billerica, MA, USA), 1 mmol⋅L−1 L-glutamine, 0.1 mmol⋅L−1 beta-mercaptoethanol and 4 ng⋅mL−1 basic fibroblast growth factor, 50 mg⋅mL−1 penicillin and streptomycin (Sigma-Aldrich, St Louis, MO, USA).17
Epithelial induction of hESCs
As described by Metallo and co-workers,15 BMP4 and α-RA were used to induce differentiation of hESCs to an epithelial lineage. In brief, hESCs were grown for 2 days on Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) coated culture-ware in MEF-conditioned medium. The MEF-conditioned medium was then supplemented with varying concentrations of BMP4 (Life Technologies, Carlsbad, CA, USA) and RA (Sigma-Aldrich, St Louis, MO, USA). In parallel studies, lithium chloride (LiCl), a Wnt signaling activator,18 was added to the induction medium.
hESCs were induced for 7 days using the following conditions: Group A, 25 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA; Group B, 25 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA+100 µmol⋅L−1 LiCl; Group C, 25 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA+1 mmol⋅L−1 LiCl; Group D, 12.5 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA; Group E, 12.5 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA+100 µmol⋅L−1 LiCl; Group F, 12.5 ng⋅mL−1 BMP4+1 µmol⋅L−1 RA+1 mmol⋅L−1 LiCl. Upon induction, cell morphology was recorded daily using a phase contrast microscope. After 7 days' induction, cells were collected and total RNA was extracted using RNeasy Mini kit (Qiagen, Dusseldorf, Germany) according to manufacturer's instruction. SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA) was used to synthesize cDNA.19 In brief, 2 µg total RNA, 200 ng random primers, 1 µL 10 mmol⋅L−1 dNTP mix, 4 µL 5× first-strand buffer, 1 µL 0.1 mol⋅L−1 DTT and 1 µL SuperScript III reverse transcriptase were used to constitute a 20-μL reaction, which was incubated at 50 °C for 1 h.
Quantitative polymerase chain reaction (PCR) was carried out using the ABI 7500 system (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). Primers and TaqMan probes used for detecting endogenous control 18S and target genes were purchased from Applied Biosystems. TaqMan probes can hybridize to the target DNA sequence and emit fluorescence as PCR reaction goes on. The intensity of released fluorescence is proportional to the amount of the PCR amplicon. The real-time guantitative PCR (qPCR) conditions were as follows: 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. To quantify the relative expression levels of the target gene, the comparative CT (threshold cycle) method was used. The corresponding arithmetic formulas used are the following: ΔCT=CTtarget gene−CTGAPDH; and CTLinear=2−(ΔCTcondition1−ΔCTcondition2). CTLinear value represents the fold change in mRNA expression levels between the two conditions, assuming a doubling of the amplified product with each PCR cycle.
Characterization of amelogenesis-associated genes in differentiated cells
Conventional PCR was used to analyze the gene expression of Fgf8, a signaling molecule upregulated in placode stage of dental epithelial cells; Msx1, Msx2, Shh and Pitx2 have been identified to be important factors in early stages of amelogenesis. In addition, gene expression of amelogenin, the characteristic enamel matrix protein marker for secretory ameloblasts was determined. The gene of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. A cDNA library of human fetal tooth organs was used as a positive control for amelogenin amplification. PCR was performed at 95 °C for 5 min, 34 cycles of 95 °C for 30 s, 57 °C for 30 s and 72 °C for 1 min, followed by 72 °C for 5 min. Primer sequences for target genes are shown in Table 1. After amplification, equal volumes of PCR products were separated by electrophoresis on agarose gels.
Table 1. Sequences of primers used to amplify genes associated to odontogenesis.
| Genes | Sequences of primers | Size/bp |
|---|---|---|
| Amg | Forward GGCTGCACCACCAAATCATCC | 383 |
| Reverse CCCGCTTGGTCTTGTCTGTCG | ||
| Fgf8 | Forward GTTGCACTTGCTGGTCCTCT | 220 |
| Reverse GAGTTGGTAGGTCCGGATGA | ||
| Msx1 | Forward TCCTCAAGCTGCCAGAAGAT | 342 |
| Reverse TCTCCAGCTCTGCCTCTTGT | ||
| Shh | Forward CCAATTACAACCCCGACATC | 339 |
| Reverse CCGAGTTCTCTGCTTTCACC | ||
| Msx2 | Forward ACACAAGACCAATCGGAAGC | 222 |
| Reverse GCAGCCATTTTCAGCTTTTC | ||
| Pitx2 | Forward ACTTTACCAGCCAGCAGCTC | 369 |
| Reverse GTGGGGAAAACATGCTCTGT | ||
| GAPDH | Forward ACCACAGTCCATGCCATCAC | 452 |
| Reverse TCCACCACCCTGTTGCTGTA |
Characterization of CKs in differentiated cells
All fetal tissues were collected from 16- to 20-week-old human fetal cadavers under guidelines approved by Committee on Human Research at the University of California, San Francisco. Human fetal OEs and ALCs were cultured following previously published protocols.20,21 Briefly, dissected fetal oral buccal mucosal epithelia were minced and further dispersed by incubating with 2 mg⋅mL−1 collagenase/dispase (Roche, Basel, Switzerland) at 37 °C for 2 h. Dissected tooth organs were digested with 2 mg⋅mL−1 collagenase/dispase at 37 °C for 2 h. After washing, the tissue mass was further digested with 0.05% trypsin/ethylene diaminetetraacetic acid (EDTA) for 5 min at 37 °C. Epithelial cells were selectively grown in supplemented keratinocyte growth medium (KGM-2) (Lonza, Basel, Switzerland) with 0.05 mmol⋅L−1 calcium, 1% penicillin and streptomycin on BD Primaria Tissue Culture Dishes (BD Biosciences, Franklin Lakes, NJ, USA).
Total RNA was purified from cultured OEs, ALCs and dissected fetal facial SE by using RNeasy Mini RNA kit (Qiagen, Dusseldorf, Germany). cDNA was synthesized by using SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA), and served as templates to amplify CKs using the same PCR conditions as described above. Primers used to amplify CKs were listed in Table 2.
Table 2. Sequences of primers used to patterning cytokeratins.
| Genes | Sequences of primers | Size/bp |
|---|---|---|
| Krt2 | Forward TTAGTGTGGCTGGAGGAGGT | 485 |
| Reverse GCTGTCGATATACCCCTGGA | ||
| Krt76 | Forward GGTGGTCCTGGTGTATTTGG | 324 |
| Reverse GGATTCAAAACAAGGCTCCA | ||
| Krt1 | Forward AGGAGGTGGACGTGGTAGTG | 335 |
| Reverse AGGAGGCAAATTGGTTGTTG | ||
| Krt10 | Forward AGCATGGCAACTCACATCAG | 324 |
| Reverse TCATTTCCTCCTCGTGGTTC | ||
| Krt6 | Forward TCAGGTCACCGTCAACAAGA | 319 |
| Reverse CATGTTCCTCAGCTCCGAAT | ||
| Krt16 | Forward AGCCCATTTTGCAGATTGAC | 365 |
| Reverse GAACCAGGTCTCAGCGTCTC | ||
| Krt25 | Forward CCTGGTTCAACGAGAAGAGC | 484 |
| Reverse CTGCGTTGGTCTACCTCCTC | ||
| Krt71 | Forward CCACTCTCAGCTCCATCTCC | 379 |
| Reverse GGCCACTCTTGGTTGTTTGT | ||
| Krt14 | Forward CAGTTCACCTCCTCCAGCTC | 348 |
| Reverse GAGGTTCTGCATGGTCACCT | ||
| Krt19 | Forward GAATCGCAGCTTCTGAGACC | 345 |
| Reverse GCACCTTGTCCAGGTAGGAG | ||
| Krt7 | Forward CAGGAACTCATGAGCGTGAA | 346 |
| Reverse GGGTGGGAATCTTCTTGTGA | ||
| Krt5 | Forward CTTGTGGAGTGGGTGGCTAT | 440 |
| Reverse CCACTTGGTGTCCAGAACCT | ||
| Krt4 | Forward CTACAACCTCAGGGGGAACA | 401 |
| Reverse GCTCAAGGTTTTTGCTGGAG | ||
| Krt13 | Forward TGATTGGTTTCCCTTCCTCA | 400 |
| Reverse TGCAGAAAGGCAGGAAACTT |
Results
hESCs were induced toward an epithelial fate
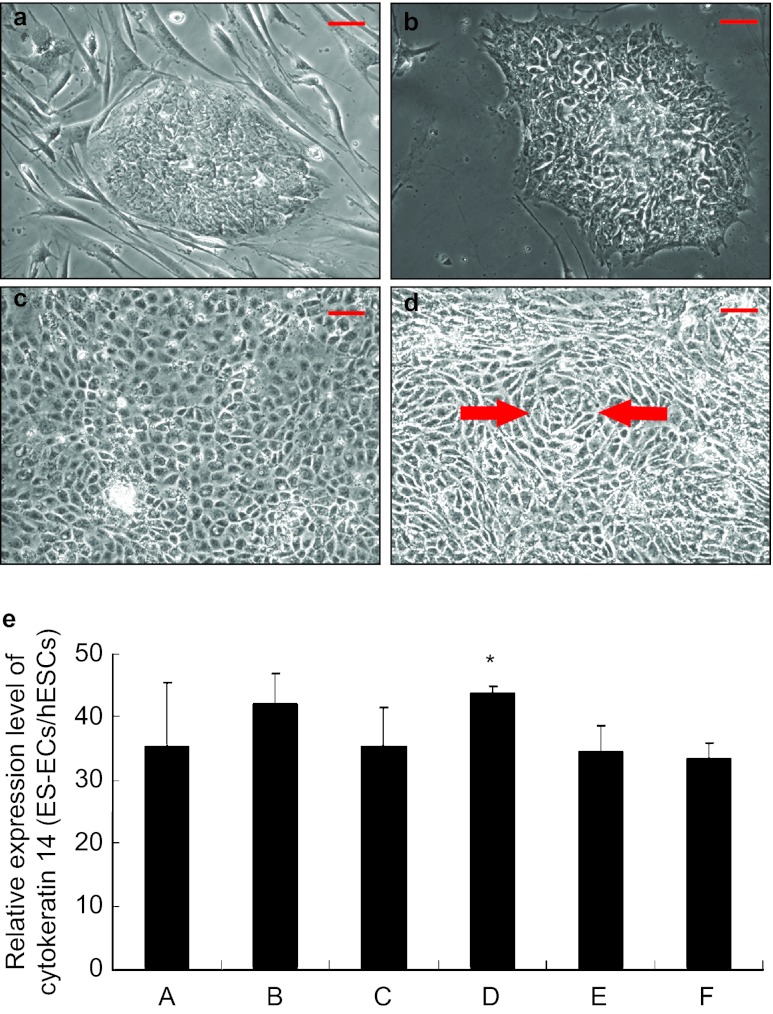
hESCs were maintained on MEF feeder cells and formed typical tightly packed embryonic stem cell colonies (Figure 1a). Prior to induction, hESCs were passaged to Matrigel-coated culture ware, and still formed the tightly packed colonies (Figure 1b). After culture with epithelial induction medium for 7 days, hESCs transformed into a cobble-stone epithelial phenotype (Figure 1c), and some cells formed concentric cell nests indicated by arrows in Figure 1d. Expression of CK14, a CK marker for epithelial cells, was upregulated with the induction with BMP4 and RA (Figure 1e). Expression of CK14 increased an average 44-fold in the cells induced with 12.5 ng⋅mL−1 BMP4 and 1 µmol⋅L−1 RA as compared to that of undifferentiated hESCs (column D in Figure 1e). Further increasing the concentration of BMP4 did not significantly enhance the efficiency of differentiation. Additional LiCl had no synergetic effects on upregulation of CK14 in the induced cells.
Figure 1.
hESCs adopted an epithelial phenotype after induction. (a) hESCs grown on mouse embryonic fibroblast feeder layer formed well-defined embryonic stem cell colonies containing epithelioid cells on the periphery and polygonal cells within the colony. (b) After being transferred to a feeder-free culture-ware, hESCs on Matrigel grew as monolayer. Colonies were well formed. Monolayer hESCs had dominant nuclei containing reticulated nucleoli with a high nucleus-to-cytosol ratio. (c) After culture with either 25 or 12.5 ng⋅mL−1 BMP4 and 1 µmol⋅L−1 RA for 7 days, hESCs adopted a cobble-stone like morphology. (d) A representative phase contrast microscopy image is shown of hESCs induced with 12.5 ng⋅mL−1 BMP4 and 1 µmol⋅L−1 RA and formed concentric cell nests indicated by red arrows. (e) CK14 expression was significantly upregulated upon the induction. Induction with 12.5 ng⋅mL−1 BMP4 and 1 µmol⋅L−1 RA resulted in an average 44-fold upregulation of CK14 as compared to that of control hESCs (column D). The columns represent mean values plus standard deviation of three independent experiments. Further increasing the concentration of BMP4 did not significantly enhance the efficiency of differentiation. Additional LiCl had no synergetic effects on the upregulation of CK14 in the induced cells. Column A, treated with 25 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA; Column B, treated with 25 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA + 100 μmol⋅L−1 LiCl; Column C, treated with 25 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA + 1 mmol⋅L−1 LiCl; Column D, treated with 12.5 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA; Column E, treated with 12.5 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA + 100 μmol⋅L−1 LiCl; Column F, treated with 12.5 ng⋅mL−1 BMP4 +1 mmol⋅L−1 RA + 1 mmol⋅L−1 LiCl. Scale bar: 50 µm. BMP4, bone morphogenetic protein 4; hESC, human embryonic stem cell; LiCl, lithium chloride; RA, retinoic acid.
ES-ECs showed upregulation of early stage of odontogenesis-associated genes Fgf8 and Msx1
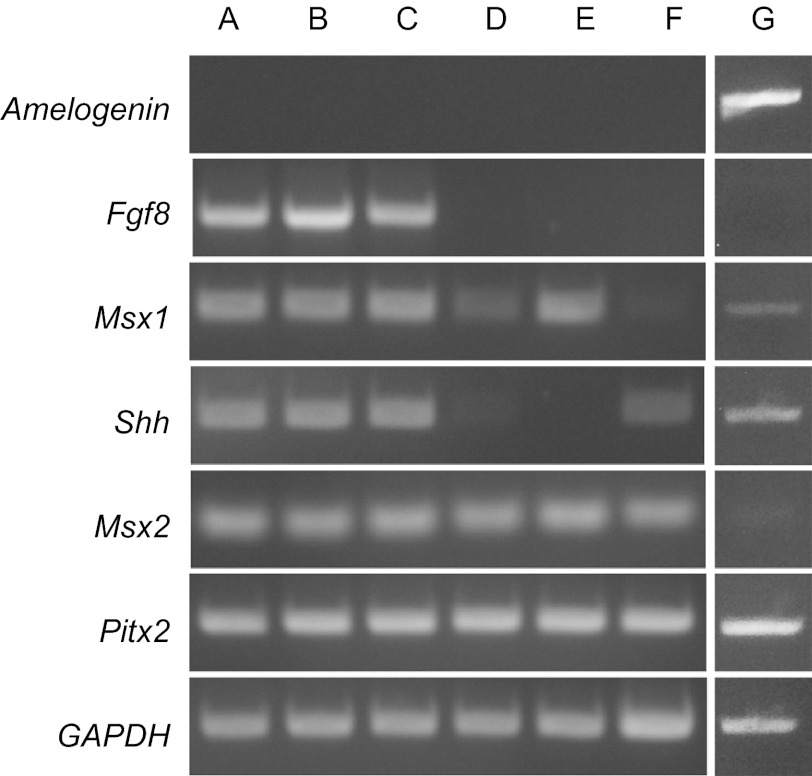
As shown in Figure 2, differentiated ES-ECs (lane A–C) expressed Fgf8 and Msx1 in addition to Shh, Msx2 and Pitx 2 that were expressed by non-induced hESCs (lane F). ALCs (lane E) and OEs (lane D) expressed Msx1, Msx2 and Pitx2, but no Fgf8 and Shh. None of inducers including BMP4, RA and LiCl was capable of inducing amelogenin expression in those induced epithelial cells.
Figure 2.
hESCs were induced to express genes associated with odontogenesis indicated by conventional PCR and agarose gel electrophoresis. hESCs were induced with 12.5 ng⋅mL−1 BMP4 and 1 µmol⋅L−1 RA (lane A), 12.5 ng⋅mL−1 BMP4, 1 µmol⋅L−1 RA and 100 µmol⋅L−1 LiCl (lane B), 12.5 ng⋅mL−1 BMP4, 1 µmol⋅L−1 RA and 1 mmol⋅L−1 LiCl (lane C). Oral epithelial cells (lane D), ALCs (lane E) and non-induced hESCs (lane F) were compared with ES-ECs in the expression of genes related to odontogenesis. Fgf8 and Msx1 were expressed in the induced ES-ECs (lane A–C), while no expression was detected in undifferentiated hESCs (lane F). Both ES-ECs (lane A–C) and non-induced hESCs (lane F) had detectable Shh expression. Pitx2 and Msx2 were detected in all cells. None of cultured cells had detectable amelogenin expression. Amelogenin expression was only detected in the positive control using human fetal tooth bud cDNA library as template (lane G). GAPDH was detected in all groups with equal intensity. BMP4, bone morphogenetic protein 4; ES-EC, hESC-derived epithelial cell; hESC, human embryonic stem cell; LiCl, lithium chloride; RA, retinoic acid.
Although there was no detectable expression of Fgf8 and Msx1 in hESCs (lane F), ES-ECs (lane A–C) expressed detectable Msx1, similar to that of ALCs (lane E). We did not find any expression of Shh in ALCs at mRNA level, though mRNA expression of this molecule was detected in all the induced ES-ECs. Pitx2 and Msx2 were detected in all groups (lane A–F). Gene expression of secretory ameloblast specific enamel matrix protein amelogenin was not detected in any of the cultured cells (lane A–F). cDNA library of human fetal tooth bud was used as a positive control for amelogenin expression. GAPDH used as an endogenous control was detected in all groups with the similar intensity.
Both ES-ECs and ALCs expressed CK76
The pattern of CK expression is shown in Figure 3a. CK71, CK5, CK14, CK7, CK19, CK13, CK2, CK1, CK10, CK6 and CK16 were the CKs that could be detected at mRNA level in all ALCs, OEs, SE and ES-ECs. CK25 was only detected in SE, and CK4 was mainly detected in ALCs and OEs. Expression of CK76 was more intense in ALCs, and was somewhat less intense in ES-ECs and OEs, but was not detected in SE and hESCs (Figure 3b).
Figure 3.
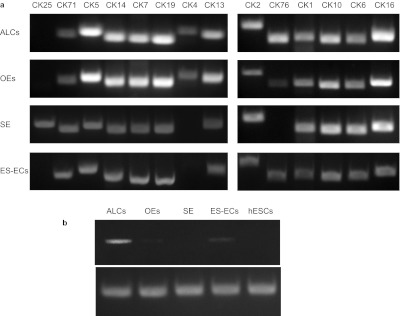
Conventional PCR analysis shows the expression pattern of CKs in human ALCs, fetal OEs, fetal SE and ES-ECs. (a) CK25 was only present in SE while absent in ALCs, OEs and ES-ECs. CK4 was present in both ALCs and OEs, while absent in SE and ES-ECs. (b) One representative DNA electrophoresis image shows that CK76 was present in ALCs, ES-ECs and OEs, while absent in SE and hESCs. ALC, ameloblast-lineage cell; BMP4, bone morphogenetic protein 4; CK, cytokeratin; ES-EC, hESC-derived epithelial cell; hESC, human embryonic stem cell; OE, oral epithelial cell; SE, skin epithelium.
Discussion
The focus of this study was to determine whether the epithelial cells derived from hESCs could be used as a source of epithelial progenitor cells for ameloblasts. In this study, we demonstrated that BMP4 at 12.5 ng⋅mL−1 and RA at 1 µmol⋅L−1 are sufficient to differentiate hESCs into cells with an epithelial morphology and upregulated CK14. Increased concentrations of BMP4 to 25 ng⋅mL−1 did not further enhance the efficiency of induction. BMP4 has been characterized as an inducer of epidermal differentiation.22 The effect of RA on epithelial lineage specification dependents on the context of bone morphogenetic protein signaling.15
We conducted further studies to differentiate ES-ECs towards amelogenin-producing cells by using LiCl. LiCl is known to be an activator of Wnt signaling.23 Wnt signaling plays a critical role during tooth development,24,25,26,27,28,29 as shown by the lack of Lef1, a downstream effector of Wnt signaling pathway, which results in developmental defects in all epithelial appendages including tooth.30 However, supplementation of LiCl in the inducing medium did not result in the expression of amelogenin in ES-ECs.
The cultured ALCs and ES-ECs did not have any detectable amelogenin expression, indicating that neither cell type had reached a stage of differentiation characteristics of secretory ameloblasts. Conditioned medium collected from human ALCs could not induce ES-ECs to express amelogenin. We have found that coculture of ALCs with either Matrigel or dental pulp stem cells can result in upregulated amelogenin expression.31 Therefore, further differentiation of ES-ECs to secretory ameloblasts likely requires a complex of signaling molecules derived from dental mesenchyme or extracellular matrix.
In addition to Msx2 and Pitx2, ES-ECs expressed odontogenic associated genes including Fgf8, Msx1 and Shh. Fgf8 and Msx1 have been identified to be critical for the initial stage of tooth formation. Transgenic mice carrying a homozygous Msx1 germline mutation display a phenotype of complete absence of the incisor.32 Inactivation of Fgf8 can severely disturb mandibular and maxillary development with no molar teeth formed, while the incisor remains.33 Expression of Fgf8 and Msx1 in the ES-ECs is a promising finding toward identifying a cell source for ameloblast regeneration. Shh is either temporally expressed early at the epithelial thickening stage or later at the enamel knot of tooth formation. Shh did not present in human ALCs, which are derived from bell stage of tooth organs. In contrast, ES-ECs do express Shh, which further supports the possibility that ES-ECs are similar to early differentiating enamel organ epithelial cells.
We sought to characterize these cells utilizing CKs as markers specific for different types of epithelial cells. For example, CK25 is among the group of CKs that are highly specific for the inner root sheath (IRS) of the hair follicle.34 The keratinocytes of all three IRS compartments including the Henle layer, the Huxley layer and the IRS cuticle35,36 synthesize CK25.37 Our studies indicated that CK25 was certainly specific to human fetal skin epithelia, which contain hair follicles, while no CK25 expression was detected in hESCs, human fetal OEs, ALCs and ES-ECs, although skin epithelia, mucosal epithelia and ALCs are all derived from ectoderm.
The paired type II keratin CK4 and the type I keratin CK13 indicates a mucosal path of differentiation for the non-keratinizing internal stratified squamous epithelia. A previous study reported that CK4 is present in the entire suprabasal compartment of mucosal stratified squamous epithelia, while it is completely absent in the epidermis.38 In this study, expression of CK4 in both human oral buccal mucosal epithelial cells and ALCs which originate from the local thickened oral epithelial cells,39 suggests the same origin of these two epithelial sources. No CK4 was detected in human fetal facial SE, which confirmed that this CK is specific for mucosa and non-keratinized epithelia.
It is interesting that except for SE, all ALCs, OEs and ES-ECs expressed CK76, which is known to be expressed in suprabasal cells of oral masticatory epithelia. Given the fact that OEs and ALCs are derived from the same progenitor cell source, the upregulation of CK76 in ES-ECs suggests that ES-ECs have increased homology to ALCs and OEs, but do not commit to the fate of SE. Combined with the fact that ES-ECs expressed Fgf8, Msx1 and Shh, we anticipate that ES-ECs hold the potential to be differentiated into an ameloblast phenotype.
Conclusion
In this study, we have identified the effective concentrations of BMP4 and RA to drive differentiation of hESCs toward an epithelial lineage phenotype. We found that this protocol could increase the expression of Fgf8, Shh and Msx1 in ES-ECs. Expression of Msx1 in ES-ECs was similar to the levels found in ALCs. ES-ECs had upregulated expression of CK76, a CK that is differentially expressed in ALCs as compared to SE. These studies suggest that these ES-ECs are useful as an alternative cell source to regenerate enamel-forming cells as availability of dental epithelial cells in humans is significantly constrained. Studies to identify factors including the human dental mesenchyme that can promote the further differentiation of ES-ECs will further facilitate the use of hESCs and ES-ECs for tooth tissue regeneration.
Acknowledgments
We appreciate Dr Sean P Palecek to share the epithelial induction protocol with us. This study was supported by NIH/NIDCR grants R03 DE019507-02 to Yan Zhang, R21 DE018633 to Pamela K DenBesten and 2011SCU11999-3/NSFC81200760to Li-Wei Zheng.
References
- Kollar EJ, Baird GR. The influence of the dental papilla on the development of tooth shape in embryonic mouse tooth germs. J Embryol Exp Morphol. 1969;21 1:131–148. [PubMed] [Google Scholar]
- Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32 2:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Duailibi MT, Duailibi SE, Young CS, et al. Bioengineered teeth from cultured rat tooth bud cells. J Dent Res. 2004;83 7:523–528. doi: 10.1177/154405910408300703. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Modino SA, Miletich I, et al. Stem-cell-based tissue engineering of murine teeth. J Dent Res. 2004;83 7:518–522. doi: 10.1177/154405910408300702. [DOI] [PubMed] [Google Scholar]
- Komine A, Suenaga M, Nakao K, et al. Tooth regeneration from newly established cell lines from a molar tooth germ epithelium. Biochem Biophys Res Commun. 2007;355 3:758–763. doi: 10.1016/j.bbrc.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Nakao K, Morita R, Saji Y, et al. The development of a bioengineered organ germ method. Nat Methods. 2007;4 3:227–230. doi: 10.1038/nmeth1012. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, et al. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97 25:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batouli S, Miura M, Brahim J, et al. Comparison of stem-cell-mediated osteogenesis and dentinogenesis. J Dent Res. 2003;82 12:976–981. doi: 10.1177/154405910308201208. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Sonoyama W, Ono M, et al. Simvastatin induces the odontogenic differentiation of human dental pulp stem cells in vitro and in vivo. . J Endod. 2009;35 3:367–372. doi: 10.1016/j.joen.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Yang X, van der Kraan PM, Bian Z, et al. Mineralized tissue formation by BMP2-transfected pulp stem cells. J Dent Res. 2009;88 11:1020–1025. doi: 10.1177/0022034509346258. [DOI] [PubMed] [Google Scholar]
- Hu B, Nadiri A, Kuchler-Bopp S, et al. Tissue engineering of tooth crown, root, and periodontium. Tissue Eng. 2006;12 8:2069–2075. doi: 10.1089/ten.2006.12.2069. [DOI] [PubMed] [Google Scholar]
- Ning F, Guo Y, Tang J, et al. Differentiation of mouse embryonic stem cells into dental epithelial-like cells induced by ameloblasts serum-free conditioned medium. Biochem Biophys Res Commun. 2010;394 2:342–347. doi: 10.1016/j.bbrc.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Hu B, Unda F, Bopp-Kuchler S, et al. Bone marrow cells can give rise to ameloblast-like cells. J Dent Res. 2006;85 5:416–421. doi: 10.1177/154405910608500504. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282 5391:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Ji L, de Pablo JJ, et al. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 2008;26 2:372–380. doi: 10.1634/stemcells.2007-0501. [DOI] [PubMed] [Google Scholar]
- Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129 6:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimanskaya I, Hipp J, Rezai KA, et al. Derivation and comparative assessment of retinal pigment epithelium from human embryonic stem cells using transcriptomics. Cloning Stem Cells. 2004;6 3:217–245. doi: 10.1089/clo.2004.6.217. [DOI] [PubMed] [Google Scholar]
- Hedgepeth CM, Conrad LJ, Zhang J, et al. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev Biol. 1997;185 1:82–91. doi: 10.1006/dbio.1997.8552. [DOI] [PubMed] [Google Scholar]
- Kotewicz ML, D'Alessio JM, Driftmier KM, et al. Cloning and overexpression of Moloney murine leukemia virus reverse transcriptase in Escherichia coli. . Gene. 1985;35 3:249–258. doi: 10.1016/0378-1119(85)90003-4. [DOI] [PubMed] [Google Scholar]
- Coppe C, Zhang Y, Den Besten PK. Characterization of primary dental pulp cells in vitro. . Pediatr Dent. 2009;31 7:467–471. [PubMed] [Google Scholar]
- Yan Q, Zhang Y, Li W, et al. Differentiation of human ameloblast-lineage cells in vitro. Eur J Oral Sci 2006114Suppl 1154–158.discussion 64–65, 380–381. [DOI] [PubMed] [Google Scholar]
- Coraux C, Hilmi C, Rouleau M, et al. Reconstituted skin from murine embryonic stem cells. Curr Biol. 2003;13 10:849–853. doi: 10.1016/s0960-9822(03)00296-3. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93 16:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92 1:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Sarkar L, Cobourne M, Naylor S, et al. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc Natl Acad Sci U S A. 2000 25;97 9:4520–4524. doi: 10.1073/pnas.97.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mech Dev. 1999;85 1/2:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67 2:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128 8:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- He P, Zhang Y, Kim SO, et al. Ameloblast differentiation in the human developing tooth: effects of extracellular matrices. Matrix Biol. 2010;29 5:411–419. doi: 10.1016/j.matbio.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6 4:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, et al. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13 23:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arin MJ, Mueller FB. Keratins and their associated skin disorders. Eur J Dermatol. 2007;17 2:123–129. doi: 10.1684/ejd.2007.0123. [DOI] [PubMed] [Google Scholar]
- Baharvand H, Hajheidari M, Ashtiani SK, et al. Proteomic signature of human embryonic stem cells. Proteomics. 2006;6 12:3544–3549. doi: 10.1002/pmic.200500844. [DOI] [PubMed] [Google Scholar]
- Lynch MH, O'Guin WM, Hardy C, et al. Acidic and basic hair/nail (‘hard') keratins: their colocalization in upper cortical and cuticle cells of the human hair follicle and their relationship to ‘soft' keratins. J Cell Biol. 1986;103 (6 Pt 2): :2593–2606. doi: 10.1083/jcb.103.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J, Langbein L, Rogers MA, et al. Hair follicle-specific keratins and their diseases. Exp Cell Res. 2007;313 10:2010–2020. doi: 10.1016/j.yexcr.2007.02.032. [DOI] [PubMed] [Google Scholar]
- van Muijen GN, Ruiter DJ, Franke WW, et al. Cell type heterogeneity of cytokeratin expression in complex epithelia and carcinomas as demonstrated by monoclonal antibodies specific for cytokeratins nos. 4 and 13. Exp Cell Res. 1986;162 1:97–113. doi: 10.1016/0014-4827(86)90429-5. [DOI] [PubMed] [Google Scholar]
- Zeichner-David M, Diekwisch T, Fincham A, et al. Control of ameloblast differentiation. Int J Dev Biol. 1995;39 1:69–92. [PubMed] [Google Scholar]