Abstract
This study examines the alteration in Staphylococcus aureus gene expression following treatment with the type 2 fatty acid synthesis inhibitor AFN-1252. An Affymetrix array study showed that AFN-1252 rapidly increased the expression of fatty acid synthetic genes and repressed the expression of virulence genes controlled by the SaeRS 2-component regulator in exponentially growing cells. AFN-1252 did not alter virulence mRNA levels in a saeR deletion strain or in strain Newman expressing a constitutively active SaeS kinase. AFN-1252 caused a more pronounced increase in fabH mRNA levels in cells entering stationary phase, whereas the depression of virulence factor transcription was attenuated. The effect of AFN-1252 on gene expression in vivo was determined using a mouse subcutaneous granuloma infection model. AFN-1252 was therapeutically effective, and the exposure (area under the concentration-time curve from 0 to 48 h [AUC0–48]) of AFN-1252 in the pouch fluid was comparable to the plasma levels in orally dosed animals. The inhibition of fatty acid biosynthesis by AFN-1252 in the infected pouches was signified by the substantial and sustained increase in fabH mRNA levels in pouch-associated bacteria, whereas depression of virulence factor mRNA levels in the AFN-1252-treated pouch bacteria was not as evident as it was in exponentially growing cells in vitro. The trends in fabH and virulence factor gene expression in the animal were similar to those in slower-growing bacteria in vitro. These data indicate that the effects of AFN-1252 on virulence factor gene expression depend on the physiological state of the bacteria.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus is a leading cause of skin and soft tissue infections in the United States (1), and its prevalence has reached epidemic proportions in the world (2). The spread of multidrug-resistant organisms has spurred the development of new drugs to attack this infectious agent. One such compound is AFN-1252, a potent small-molecule inhibitor of staphylococcal type 2 fatty acid synthesis (FASII) that targets the enoyl-acyl carrier protein (ACP) reductase step (FabI) (3–5). The suitability of targeting bacterial fatty acid synthesis to combat Gram-positive bacteria was questioned by Brinster et al. (6), who concluded that the ability of these bacteria to incorporate exogenous fatty acids into membrane phospholipids would render inhibition of the pathway ineffective in the context of a host where such fatty acids are readily available. However, the molecular basis for the differential susceptibility of Gram-positive pathogens to FASII inhibitors has been uncovered, providing an understanding of why FASII inhibitors are effective against S. aureus even in the presence of an extracellular source of fatty acids (7). These data resolved the debate concerning the validity of using fatty acid synthesis inhibitors to treat S. aureus infections (8, 9) and are consistent with the examples of fatty acid synthesis inhibitors that show efficacy in S. aureus-infected animal models (5, 8, 10–14).
The production of extracellular mediators (virulence factors) plays an important role in the pathogenicity of S. aureus (15). The sae regulatory system is a central downstream effector in the regulatory network that controls the expression of major virulence genes (16). SaeRS is a 2-component regulator where a membrane-bound sensor (SaeS) activates a transcriptional regulator (SaeR) that influences virulence gene expression by direct interaction with target promoters (17–19). S. aureus strains with deletions in saeRS are less virulent than their wild-type counterparts, illustrating that SaeRS-regulated virulence factor expression is critical for pathogenesis in animal infection models (19–21). A number of antibiotic classes are known to modulate the expression of staphylococcal virulence factors under laboratory conditions (22–30). For example, the increased expression of virulence factors by the β-lactams is mediated by SaeRS in S. aureus (31). The impact of fatty acid synthesis inhibitors on virulence factor gene expression has not been studied in detail, although a report in 1984 that cerulenin treatment blunted alpha-toxin secretion (32) suggests that such inhibitors may also influence virulence factor production. Whether the laboratory studies correlating virulence gene expression and antibiotic treatment are recapitulated in vivo remains an open question.
There are also pathway-specific gene expression changes that are associated with specific antibiotic classes, and these expression profiles generate a characteristic signature for the specific branch of metabolism inhibited by the antibiotic (33, 34). With antibiotics targeting bacterial fatty acid synthesis, the upregulation of fab genes is a transcriptional signature that is not shared by other antibiotic classes (33, 34). In S. aureus, the expression of the fab gene set is controlled by the transcriptional regulator FapR (35), and the upregulation of fab gene expression following the inhibition of fatty acid synthesis is reflected by increased levels of mRNA encoding the Fab enzymes (36). Despite these correlations between patterns of virulence gene expression and specific pathway inhibitors in the laboratory, there is little understanding of whether these experiments translate in vivo. The goals of this study were to evaluate the efficacy and pharmacokinetic parameters of AFN-1252 in a mouse subcutaneous (s.c.) granuloma infection model, to determine if virulence and pathway-specific gene expression are altered by AFN-1252 treatment in vitro, and to test whether these effects of AFN-1252 are recapitulated in vivo. The mouse subcutaneous granuloma S. aureus infection model was selected to facilitate acquiring samples at specific treatment intervals for transcriptional analysis, pharmacokinetics, and efficacy.
MATERIALS AND METHODS
Bacterial strains.
S. aureus strain RN4220 was obtained from the American Type Culture Collection. Strain PDJ22 (ΔsaeR) was constructed by the insertion of a group II intron into the saeR gene of strain RN4220 at 42 bp after the initiation codon (37) using the primer design software provided by Sigma-Aldrich (Targetron system). The insertion was verified by PCR using primers outside the intron insertion site. The USA300 and Wood46 strains were obtained through the Network of Antimicrobial Resistance in S. aureus (NARSA) program supported under NIAID/NIH contract no. HHSN272200700055C, and the Newman strain was kindly provided by Mark Hart (NCTR, Jefferson, AR). For in vitro studies, S. aureus strains were grown in tryptone broth (TB) to mid-log phase (optical density at 600 nm [OD660], 0.45) or stationary phase (OD600, 2.2) and then split into 2 aliquots for treatment with solvent control (dimethyl sulfoxide [DMSO]) or treatment with AFN-1252 (0.05 μg/ml), cerulenin (500 μg/ml), or clarithromycin (1.5 μg/ml). For in vivo studies, S. aureus Wood46 was cultured overnight on tryptic soy agar (TSA), and the cells were suspended in tryptic soy broth and counted to generate an infecting inoculum of 8.0 log10 CFU/ml.
Affymetrix array analysis.
The effect of AFN-1252 on global abundance of gene transcripts was analyzed using the S. aureus Affymetrix array technology. Strain RN4220 was grown to an OD600 of 0.45 in tryptone broth. The cultures were then split into 2 aliquots and treated with 50 ng/ml AFN-1252 or an equal volume of DMSO (0.1% final concentration) for 15 min. RNA was extracted from the cells using the Ambion RNAqueous kit (Applied Biosystems) according to the manufacturer's instructions with the modification of using lysostaphin for 15 min at room temperature to lyse the cells. The quality of the RNA was checked with Agilent Technologies 2100 Bioanalyzer Laboratory-on-a-chip before proceeding with the microarray. Affymetrix protocols were used to synthesize, fragment, and terminally label the cDNA. Hybridization, washing, and scanning of cDNA to GeneChip S. aureus Genome Array were performed according to the manufacturer's instructions (Affymetrix). The arrays were analyzed with GeneChip operating software, and global scaling was used to normalize the data from different arrays. Spotfire DecisionSite 9.1.1 was used to analyze the data. Changes were reported if they were greater than ±1.4-fold with a P value of <0.05 using the Student t test to compare values. There were 6 data sets used for the analysis (3 treated with DMSO and 3 treated with AFN-1252).
Measurement of mRNA levels.
Quantitative reverse transcription-PCR (qRT-PCR) was used to determine the levels of transcript mRNAs. Bacterial strains were grown in tryptone broth at 37°C to mid-log phase (OD600, 0.45) for log-phase experiments or at the transition to stationary phase (OD600, 2.2). The cultures were treated with selected antibiotics or DMSO solvent as the control. Laboratory-grown cells were harvested by centrifugation and washed with 1 ml of RNAlater solution (Ambion). Total RNA was then immediately isolated from the bacterial cells using the RNAqueous kit (Ambion) per the manufacturer's instructions, except that treatment with lysostaphin was included to lyse the cells and LiCl was used to precipitate the RNA. The pelleted RNA was resuspended in nuclease-free water, and a 10-μg aliquot was treated with the Turbo DNA-free kit (Ambion) to remove any remaining genomic DNA. First-strand cDNA was generated using random primers and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR was performed using the ABI Prism 7700 Sequence Detection System, and samples were run in triplicate. Negative controls (distilled water) and RNA samples without the reverse transcription step were run in duplicate. Dissociation curves were generated after each real-time PCR run to check for nonspecific amplification. PCR mixtures were composed of SYBR green PCR Master Mix (Applied Biosystems), 150 nM (each) primer, and cDNA synthesized from either 10 ng or 15 ng of total RNA for in vitro and in vivo samples, respectively. The expression levels of various housekeeping genes were analyzed (38) under the different treatment conditions and compared to 16S RNA, and gmk was the least altered and was used as the calibrator to compare mRNA levels between strains and treatments. Template curves consisting of at least 7 points ranging from 50 ng to 50 pg of total RNA input were run for each primer set to verify that they produced linear responses and had the same relative efficiency in the PCR as in the gmk calibrator. The primers used are listed in Table S1 in the supplemental material. The values were compared using the threshold cycle (CT) method (39), and the amount of cDNA present (2−ΔCT) was reported relative to the gmk calibrator.
Mouse granuloma infection model.
Subcutaneous (s.c.) air pockets were aseptically formed on the dorsal aspect of anesthetized female, CD-1 mice (6 to 8 weeks of age) 5 days prior to infection. Air pockets were immediately injected with 0.4% croton oil (irritant), and 1 ml of sterile intravenous (i.v.) saline solution was injected into pouches 3 days later. Five-day-old pouches were infected with 7.1 to 7.5 log10 CFU of S. aureus Wood46, and animals were orally (p.o.) dosed with 100 mg AFN-1252/kg of body weight at 2, 26, and/or 50 h after infection. AFN-1252 was formulated as a 1% poloxamer suspension (Pluronic F-127; Sigma-Aldrich). Control mice were not treated. Pouch fluid was collected from animals at defined time points postdosing and transferred into tubes for RNA extraction, CFU enumeration, or determination of AFN-1252 concentration by liquid chromatography-mass spectrometry (LC-MS) analysis. Additionally, heart blood was collected at the same time points and processed for plasma separation and collection. Pouch fluid samples used for CFU counting were immediately 10-fold serially diluted in sterile 1× phosphate-buffered saline (PBS), plated onto tryptic soy agar (TSA) containing 5% activated charcoal, and incubated overnight at 37°C. Pouch fluid and plasma samples were stored at −80°C if they were not immediately analyzed or processed for analysis.
Bacterial RNA extraction from pouch fluid exudates.
Pouch fluid was transferred into 2-ml screw-cap tubes loaded with 1 ml of TRIzol (Invitrogen) and one-half volume of 0.1-mm zirconia-silica beads (sterile). Tubes were placed into the bead beater and processed for 1 min at maximum speed. Ice-cold chloroform was added to each tube and then cold centrifuged (4°C) for 15 min at 12,000 × g. A second treatment with TRIzol was done for each upper phase, followed by another chloroform extraction and centrifugation step. RNA was precipitated by adding ice-cold isopropanol to upper phases, and RNA was pelleted in each sample by cold centrifugation for 10 min at 15,000 × g. After RNA pellets were resuspended in 75% ethanol, they were cold centrifuged for 5 min at 7,500 × g and dried for 10 min at room temperature. RNase-free water was added to each RNA sample and then treated with Turbo DNase (Ambion) for 30 min at 37°C. Turbo DNase inactivating reagent was added to the samples, and they were centrifuged for 2 min at 10,000 × g. Upper phases were transferred into RNase-free tubes and stored at −80°C until analyzed by qRT-PCR as described above.
Quantification and pharmacokinetic analysis of AFN-1252 in plasma and pouch fluid.
Plasma and pouch fluid samples taken from study animals were placed into the wells of a V-bottomed 96-well plate and then mixed with the internal standard solution of deuterated AFN-1252 and methanol. The mixtures were vacuum filtered through an Isolute SLE+ extraction plate (Biotage), ethyl acetate was added to each well, and dissolved compound was eluted from the Isolute SLE+ plate under low vacuum for 5 min. Ethyl acetate was evaporated, and samples were reconstituted in a methanol-water-formic acid (500:500:1) solution. Samples were loaded into the autosampler of a Thermo Finnigan Surveyor high-pressure liquid chromatograph (HPLC) coupled to an LCQ Deca mass spectrometer, and three separate injections of each sample were used to calculate the concentration of AFN-1252 against the peak response ratio of the calibration curve. The AFN-1252 concentration for each time point was averaged between studies and entered into PK Solutions 2.0 software (Summit Research Services, Montrose, CO) to calculate the pharmacokinetic parameters for both pouch and plasma samples.
Microarray data accession number.
The complete, triplicate data set can be viewed and accessed under accession number GSE19400 in the NCBI GEO database (www.ncbi.nlm.nih.gov/geo).
RESULTS
Gene expression changes associated with FASII inhibition in S. aureus.
An Affymetrix array experiment was used to obtain a global view of the effect of the abrupt cessation of fatty acid synthesis at the enoyl-ACP reductase step (FabI) by AFN-1252 (7) on the pattern of cellular gene expression. The complete list of genes with significant expression changes (P < 0.05) is found in Table S2 in the supplemental material, and the complete, triplicate data set can be viewed and accessed under accession number GSE19400 in the NCBI GEO database (www.ncbi.nlm.nih.gov/geo). Previous work established the mechanism for the alteration in fab gene expression in response to FASII inhibitors. In Bacillus subtilis and S. aureus, the activation of FASII gene expression that occurs following the interruption of fatty acid synthesis is controlled by the FapR transcriptional regulator (35, 40–42). FapR is a repressor that binds to the fab gene promoters and represses transcription. FapR is released from its binding sites by either malonyl coenzyme A (malonyl-CoA) or malonyl-ACP (43). The inhibition of FASII leads to the accumulation of malonyl-CoA due to the continued production of this intermediate by acetyl-CoA carboxylase coupled with its impaired utilization due to FASII inhibition (41). Therefore, an increased expression of FASII genes that constitute the FapR regulon in S. aureus was anticipated and observed following AFN-1252 treatment (see Table S2). There may be an unknown stress response that contributes to the regulation of fabH, but stresses induced by pH (44), alcohol (45), or antibiotics that target other pathways (33, 34) do not alter fabH expression. The upregulation of the FapR-regulated genes in AFN-1252-treated cells was verified by the quantitative measurement of fabH (and plsX [data not shown]) gene expression following AFN-1252 treatment (Fig. 1). The fabH mRNA abundance increased approximately 3-fold within 15 min of AFN-1252 treatment. The same level of fabH transcriptional activation occurred after blocking FASII at the β-ketoacyl-ACP synthase step using cerulenin, but not with clarithromycin, a protein synthesis inhibitor (Fig. 1). Among the FASII genes controlled by this mechanism, fabH was the most highly activated by FASII inhibition in S. aureus and was selected as the marker for the genetic response of cells to FASII inhibition.
Fig 1.
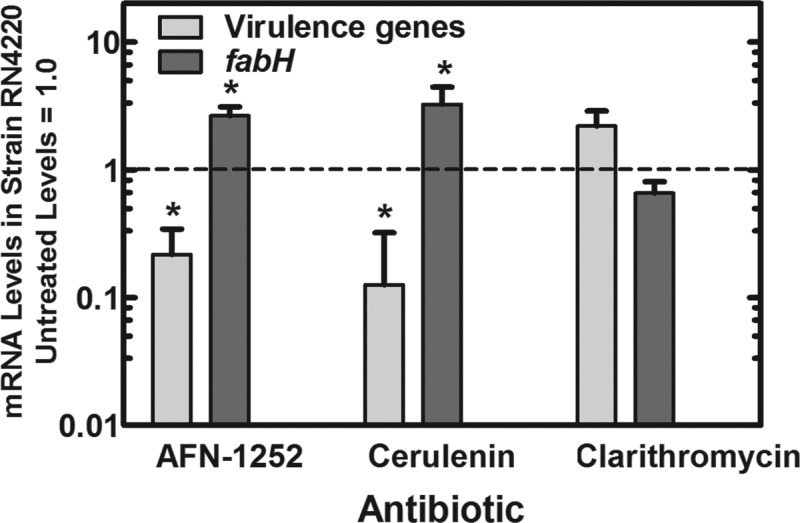
Depression of virulence factor mRNA levels in S. aureus treated with fatty acid synthesis inhibitors. Strain RN4220 was grown to mid-log phase in tryptone broth and treated with either AFN-1252 (50 ng/ml), cerulenin (500 μg/ml), or clarithromycin (1.5 μg/ml) for 15 min to arrest either lipid or protein synthesis. The MICs for these drugs in strain RN4220 determined in tryptone broth as described previously (7) were as follows: AFN-1252, 4 ng/ml; cerulenin, 62 μg/ml; and clarithromycin, 310 ng/ml. RNA was extracted, and the levels of four randomly selected virulence factor mRNAs (saeP, ehp, efb, and hlgC) were calculated and averaged, in addition to fabH, by quantitative RT-PCR. Significance between treated and untreated samples was calculated using the Student t test, and standard deviations were obtained from triplicate determinations derived from triplicate experiments. fabH mRNA was significantly increased in response to AFN-1252 and cerulenin (*, P < 0.05) but not significantly altered by clarithromycin. The average levels of virulence gene mRNAs were significantly decreased in response to AFN-1252 and cerulenin (*, P < 0.05). Clarithromycin-treated cells had a modest increase in virulence gene expression, but it was not significant.
A second result was the significant repression of a group of transcripts associated with S. aureus virulence. Specifically, the mRNA levels for α-, β-, and γ-hemolysins and two fibrinogen binding proteins (Ehp and Efb) decreased over 3-fold within 15 min of AFN-1252 treatment in the Affymetrix array experiment (Table 1). A unifying feature of the virulence gene expression pattern was that the virulence genes are positively regulated by the SaeRS two-component regulator (46–48). The SaeRS system also autoregulates its own expression (saeR and saeS genes) along with the saeP and saeQ genes, which are located adjacent to the saeRS operon (49). Therefore, the involvement of the SaeRS two-component system in the response to AFN-1252 was also directly indicated by the reduced expression of all four sae genes (Table 1). A set of four virulence factor genes that exhibited significant levels of downregulation by AFN-1252 treatment and represented the overall trend in virulence factor expression were randomly selected for quantitative analysis of SaeRS-regulated gene expression (hlgC, ehp, efb, and saeP) (Table 1). The inhibition of virulence gene expression was validated by combining the average levels of four transcripts (hlgC, ehp, efb, and saeP) that were coordinately downregulated following AFN-1252 treatment (Fig. 1). Similar reductions in virulence factor expression were associated with cerulenin treatment (Fig. 1). However, the average expression levels for these four virulence markers were elevated 2-fold in cells treated with the protein synthesis inhibitor clarithromycin. These data indicated that the attenuation of virulence factor gene expression was linked to the inhibition of fatty acid biosynthesis.
Table 1.
Virulence-related genes downregulated by AFN-1252
| Locus tag | Gene | Fold changea | P value | Function |
|---|---|---|---|---|
| SA0309 | geh | 1.54 | 0.002 | Lipase |
| SA0660 | saeS | 1.77 | 0.009 | Two-component sensor |
| SA0661 | saeR | 2.45 | 0.003 | Two-component regulator |
| SA0662 | saeQ | 4.94 | 0.000 | Hypothetical |
| SA0663 | saeP | 4.40 | 0.000 | Hypothetical |
| SA1000 | ehp | 3.63 | 0.001 | Fibrinogen binding protein |
| SA1003 | efb | 3.49 | 0.000 | Fibrinogen binding protein |
| SA1007 | 1.70 | 0.009 | Alpha-toxin | |
| SA1750 | 2.05 | 0.002 | Map-w | |
| SA1752 | 8.18 | 0.000 | β-Hemolysin | |
| SA1811 | hlb | 3.63 | 0.000 | β-Hemolysin |
| SA1812 | 2.72 | 0.005 | γ-Hemolysin | |
| SA2206 | sbi | 3.72 | 0.001 | IgG binding protein |
| SA2207 | hlgA | 4.21 | 0.000 | γ-Hemolysin |
| SA2208 | hlgB | 4.99 | 0.001 | γ-Hemolysin |
| SA2209 | hlgC | 4.78 | 0.000 | γ-Hemolysin |
| SA2461 | icaD | 1.54 | 0.007 | Intracellular adhesion |
| SA2462 | icaC | 1.61 | 0.003 | Intracellular adhesion |
Average fold decrease in mRNA abundance derived from the Affymetrix array experiment. Triplicate control and experimental samples were analyzed. Shown are S. aureus genes with significantly (P < 0.05) different expression levels (±1.4-fold) following a 15-min treatment with 50 ng/ml AFN-1252. See Table S2 in the supplemental material for a summary of all significantly altered genes and accession number GSE19400 in the NCBI GEO database for the complete data set.
AFN-1252 effect on virulence factor mRNA levels was mediated by SaeRS.
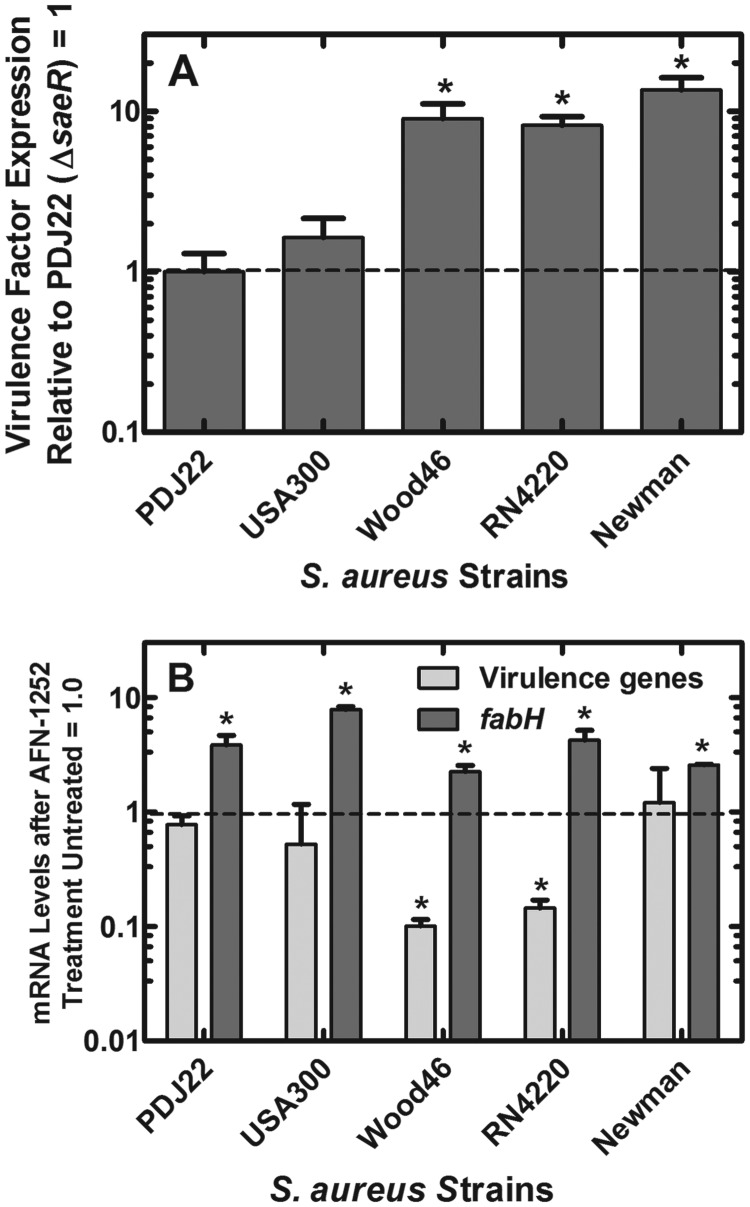
The idea that the effect of AFN-1252 was mediated by the sae system was tested by the construction of deletion strain PDJ22 (ΔsaeR) from strain RN4220, which would inactivate signaling through SaeRS. Experiments with this strain were corroborated by the analysis of additional S. aureus strains with known lesions in the SaeRS pathway (Fig. 2A). The average expression level of the virulence factors was lowest in strain PDJ22 (ΔsaeR), which lacked an intact SaeRS signaling pathway. The levels of virulence factor expression in a panel of strains were compared by normalizing their levels of expression to those in strain PDJ22, set to 1 (Fig. 2A). The USA300 isolate is wild type with respect to the regulatory system involving SaeRS and did not express high levels of virulence factors in vitro. This was in agreement with previous findings indicating that external stimuli, like serum or antimicrobial peptides, were required for virulence factor production in other isolates of USA300 (50, 51). Strains RN4220 (52) and Wood46 (53) expressed higher levels of SaeRS-directed virulence factor mRNA due to inactivating mutations in rsbU and sigB, respectively, which are negative regulators of SaeRS (54). Strain Newman has a point mutation in the sensory domain of SaeS that produces constitutively active signaling through SaeRS (16, 55), and this strain had the highest average levels of virulence factor transcripts, >10-fold higher than those in strain PDJ22 (Fig. 2A).
Fig 2.
Relative virulence factor production by S. aureus strains. (A) Basal level of virulence factor mRNA levels in a panel of S. aureus strains. The expression level in strain PDJ22 (ΔsaeR) was then set to 1, and the expression levels in the indicated strains were compared to the levels in the saeR-knockout strain. The differences between strains PDJ22 and USA300 were not significant. Strains Wood46, RN4220, and Newman exhibited significantly higher levels of virulence factor mRNA than did strains PDJ22 and USA300. *, P < 0.05. (B) Levels of virulence factor and fabH mRNA in strains treated with AFN-1252. In both experiments, the mRNA levels of four virulence-related genes (ehp, efb, hlgC, and saeP) and fabH were measured by qRT-PCR using gmk as the internal control, and triplicate determinations for each gene were averaged. Expression of fabH was significantly increased in all cases (*, P < 0.05). Virulence gene expression was only significantly decreased by 8- to 10-fold in strains Wood46 and RN4220 (*, P < 0.05). All strains had the same MIC for AFN-1252, 4 ng/ml (7). Significance between treated and untreated samples was calculated using the Student t test, and error bars indicate standard deviations.
The ability of AFN-1252 to impact virulence factor mRNA levels in this strain collection correlated with the activity of the SaeRS system (Fig. 2B). AFN-1252 did not alter virulence factor mRNA levels in strain PDJ22 (ΔsaeR), and there was also a minimal effect in the USA300 isolate, in which the intact SaeRS pathway appeared not to be actively inducing virulence factor expression in vitro. In contrast, AFN-1252 treatment significantly reduced virulence factor mRNA levels in strains RN4220 and Wood46. Importantly, the high levels of virulence factor mRNA in strain Newman were not impacted by AFN-1252 (Fig. 2B), which is presumably due to the constitutive and unregulated signaling through the SaeRS system. AFN-1252 treatment significantly upregulated the expression of fabH in all of the strains, showing that the pathway-specific action of the drug was a universal occurrence. These data suggested a link between the effects of AFN-1252 treatment on virulence factor transcription and SaeRS regulatory activity in S. aureus.
Therapeutic effect of AFN-1252 on S. aureus in the mouse granuloma model.
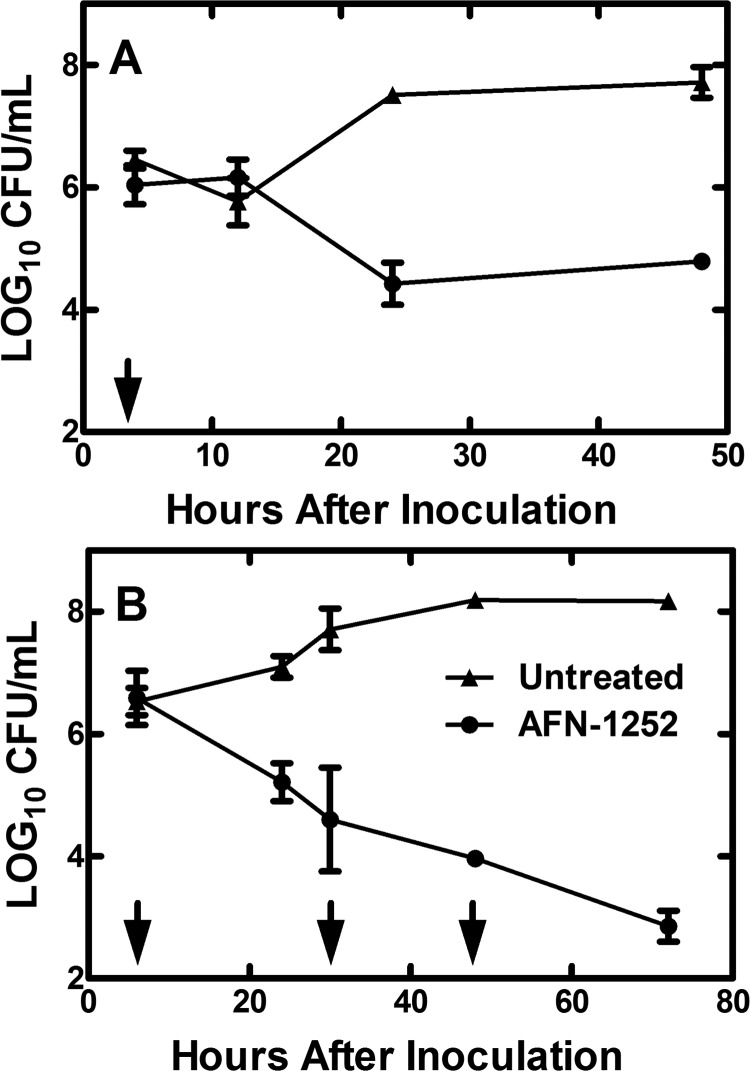
S. aureus Wood46, a clinical isolate known to produce virulence factors (53) (Fig. 2A), was selected for use in the mouse granuloma infection model. Selection of this strain was based on its prior use in animal models, as well as the in vitro effects of AFN-1252 on Wood46 gene transcription. The first experiments involved administering a single oral dose of AFN-1252 (100 mg/kg) 2 h after infecting the granuloma pouches and examining the effect of AFN-1252 on pouch-associated CFU within 48 h of dosing (Fig. 3A). Little change in the bacterial load was detected within the first 12 h of AFN-1252 treatment, but between 24 and 48 h, there was a 3.0-log10 reduction in the mean CFU/ml of pouch fluid collected from AFN-1252-treated animals compared to untreated animals (Fig. 3A). Wood46 counts in untreated pouch fluid increased to 7.5 log10 CFU/ml at 24 h, while bacterial counts in AFN-1252-treated pouches dropped to 4.4 log10 CFU/ml in 24 h. The pouch-associated CFU counts did not change between 24 and 48 h in the AFN-1252-treated animals. This response to a single dose of AFN-1252 was similar to the transient effect of a single oral dose of linezolid in the rat granuloma model, which initially showed a decrease in S. aureus CFU counts within 48 h of dosing that was followed by a rebound in bacterial growth (56). When 100 mg/kg of AFN-1252 was orally administered once a day for 3 days (+2, +26, and +50 h), AFN-1252 reduced 72-hour S. aureus CFU counts in the pouches of treated animals by 5 orders of magnitude compared to untreated controls, which was near the detection limit for this model (Fig. 3B). These data illustrate that orally administered AFN-1252 can reach therapeutic levels in the mouse granuloma pouch and effectively resolve the S. aureus infection associated with the pouch fluid.
Fig 3.
Effect of AFN-1252 treatment on S. aureus in the mouse granuloma model. Subcutaneous mouse granuloma pouches were infected with 3.2 × 107 CFU of strain Wood46. (A) An oral dose of 100 mg/kg of AFN-1252 was administered at 2 h postinfection (arrow). (B) Oral doses of 100 mg/kg of AFN-1252 were administered at 2, 26, and 50 h postinfection (arrows). At the indicated times in both panels, the pouch fluid was recovered and bacterial CFU/ml of pouch fluid were determined in triplicate. CFU in pouches from animals treated with AFN-1252 (●) were compared to that in untreated controls (▲). Error bars indicate standard deviations.
Pharmacokinetics of AFN-1252 in the mouse granuloma model infected with S. aureus.
AFN-1252 levels were measured in the plasma and pouch fluid of mice with S. aureus Wood46-infected granulomas over a 48-hour time course following a single oral dose of AFN-1252 at 100 mg/kg (Fig. 4). The maximum concentration of drug in serum (Cmax) of AFN-1252 was higher in the plasma (3.5 μg/ml) of infected animals than in pouch fluid (2.1 μg/ml), and the time to maximum concentration of drug in serum (Tmax) for each compartment was estimated to be approximately 1 h after treatment. Additionally, the elimination half-lives for AFN-1252 were similar for the pouch fluid (4.9 h) and plasma (4.4 h). The calculated exposure (area under the concentration-time curve from 0 to 48 h [AUC0–48]) of AFN-1252 in plasma was 22.5 μg · h/ml, while the AUC0–48 of AFN-1252 in granuloma pouch fluid was 19.9 μg · h/ml. Therefore, orally dosed AFN-1252 quickly (Tmax of 1.0 h) penetrated infected pouch fluid at an exposure level comparable to that for plasma (85% of the plasma AUC) and remained detectable in pouch fluid for 24 h (Fig. 4). The pharmacokinetic data supported the efficacy results of AFN-1252 in the model and validated the use of this animal model to measure the in vivo effects of AFN-1252 on S. aureus gene expression.
Fig 4.
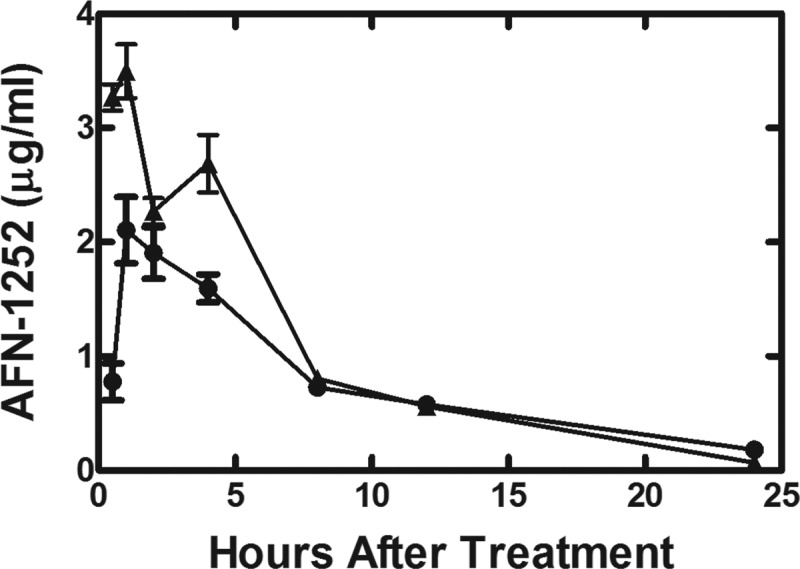
Levels of AFN-1252 in serum and pouch fluid following a single oral dose. S. aureus Wood46-infected animals received a single oral dose of AFN-1252 (100 mg/kg) 2 h after infection (time zero). At the indicated times, blood plasma and pouch fluid were sampled and AFN-1252 levels were determined in plasma (▲) and in the pouch exudate (●) by LC-MS. Mean values represent the plasma and pouch fluid levels of 3 to 9 samples harvested at the indicated times after AFN-1252 dosing. A total of 3 replicates were analyzed for each sample by LC-MS, and the limit of detection for the assay was 0.01 μg/ml. Error bars indicate standard deviations. AFN-1252 levels at 48 h were below detection.
S. aureus Wood46 virulence factor and fabH expression in AFN-1252-treated pouches.
We were able to examine S. aureus mRNA expression in the mouse granuloma model for up to 12 hours after treatment with AFN-1252, but the substantial decrease in the number of recovered bacteria in the treated pouches after 12 h (>99% reduction by 24 h) (Fig. 3A) precluded obtaining a sufficient sample for RNA analysis at later time points. Pouch samples used in mRNA expression experiments were taken over the 12 h that immediately followed the single dose of AFN-1252 (100 mg/kg), when there was little change in the bacterial load in both treated and untreated animals (Fig. 5A). During the first 12 h following oral administration of AFN-1252, there was a substantial and persistent 3.5- to 6.5-fold increase in the levels of fabH mRNA harvested from pouch-associated Wood46 cells at all time points (Fig. 5B), which was higher than the increase in fabH levels in the in vitro cultures of Wood46 after being exposed to AFN-1252 (∼2-fold). The highest levels of fabH mRNA were present in the 2-hour samples (6.5-fold), and elevation in fabH level persisted up to 12 h (∼3.5-fold). These data indicated the presence of AFN-1252 at levels in the pouch fluid that inhibit the FASII cycle for the entire 12 h following AFN-1252 dosing.
Fig 5.
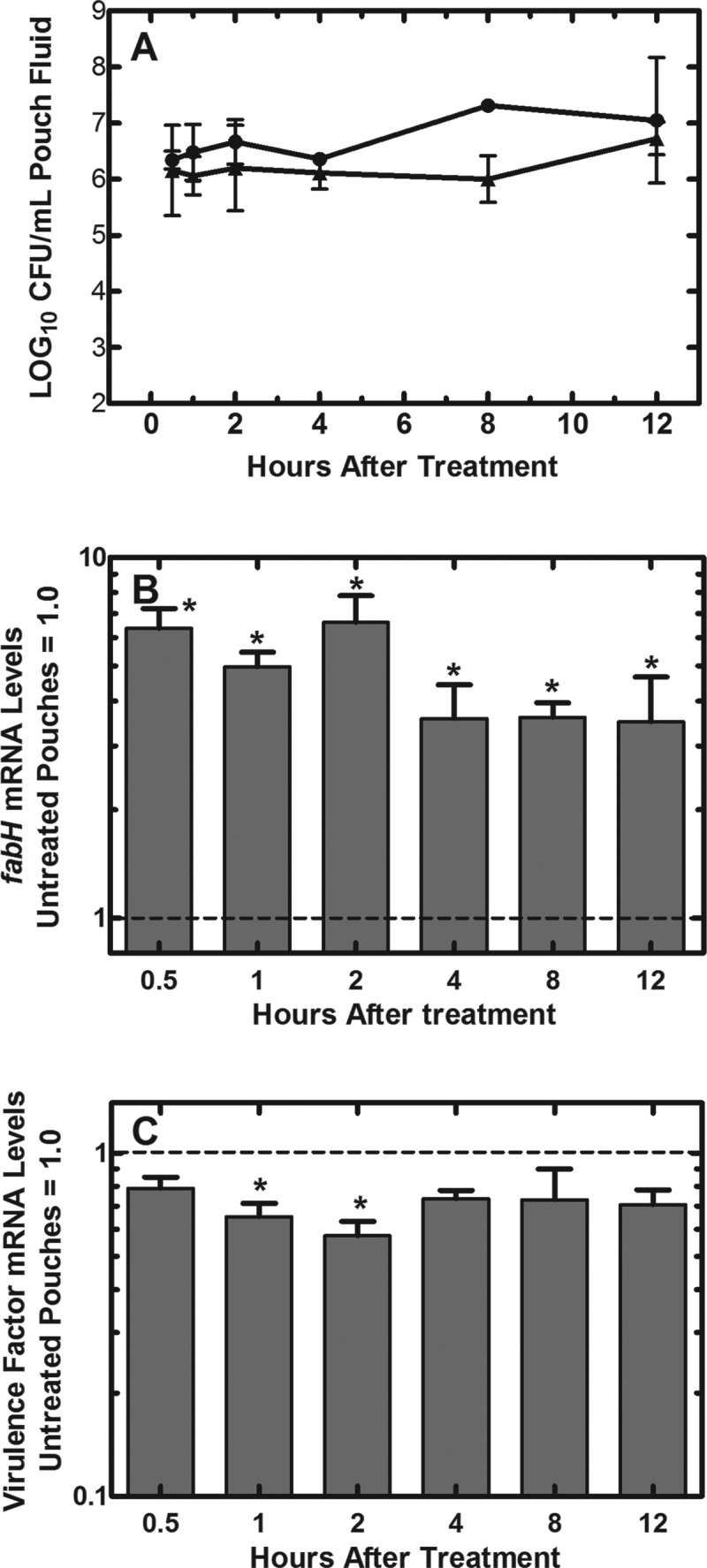
Virulence factor and fabH expression levels following treatment of infected pouches with AFN-1252. (A) Bacterial load in the pouches of AFN-1252-treated (▲) and untreated (●) pouches. (B) The levels of fabH mRNA in AFN-1252-treated samples relative to fabH expression levels in untreated samples. At all time points, fabH transcript levels were significantly elevated over controls (P < 0.05). (C) The average levels of 4 virulence factor mRNAs expressed in the AFN-1252-treated pouches relative to the expression levels in untreated controls. qRT-PCR was performed using gmk as the internal standard. The significance of differences between samples was determined by the Student t test. The modest reduction in virulence gene expression levels was significant only at 1 and 2 h (*, P < 0.05). Error bars indicate standard deviations. The exception is the 8-h untreated-pouch time point in panel A, which consisted of only a single determination.
The impact of AFN-1252 on S. aureus Wood46 virulence factor mRNA levels was not as strong in the mouse granuloma model as was observed on the laboratory bench (Fig. 5C). Two hours after AFN-1252 dosing, there was an average ∼2-fold reduction in virulence factor mRNA levels and mRNA levels continued to be suppressed for up to 12 h (1.6-fold). In comparison, there was a 3- to 4-fold decrease in average virulence factor mRNA levels following AFN-1252 treatment of S. aureus Wood46 cultures in the laboratory (Fig. 2B). One important difference in the two experiments was that the laboratory data were obtained in rapidly growing cells (Table 1; Fig. 1), whereas the CFU of strain Wood46 did not increase during the course of the experiment in the pouch model (Fig. 5A). The unchanged level of pouch-associated CFU during the first 12 h may reflect removal of S. aureus cells by the immune system coupled with their replacement by cell division, which has been previously shown with Escherichia coli in this model (57). However, it stands to reason that pouch-associated bacteria may be growing at a lower rate than in the log-phase laboratory model explored in Fig. 1. Therefore, we examined if the growth rate of S. aureus Wood46 influenced the AFN-1252 effect on gene expression in laboratory cultures (Fig. 6). The same gene panel used in vivo was also used to compare the effect of AFN-1252 on gene expression in the logarithmic growth phase to the effect in cells whose growth rate had decreased by 80% as they entered early stationary phase. Whereas AFN-1252 robustly suppressed virulence factor gene expression in log-phase Wood46 (OD600, 0.45), there was little alteration in the levels of virulence factor mRNA in Wood46 by AFN-1252 when the cells were harvested at an OD600 of 2.2 (Fig. 6). A comparison of the basal levels of virulence factor expression in the slower-growing cells to that in the log-phase cells showed that there was a 2-fold increase in the average levels of virulence factor mRNA in the denser cell cultures over those in the log-phase cells. The effect of AFN-1252 on fabH mRNA levels was higher in the slower-growing cultures than in the log-phase cells (Fig. 6). This finding reflected the higher average increase in fabH mRNA in the pouch model (Fig. 5B) over that in logarithmically growing cells (Fig. 6). However, the total cellular levels of fabH mRNA were lower in slower-growing stationary cells than in cells in log phase. Thus, the higher fold induction following AFN-1252 treatment was due to the lower basal levels of fabH mRNA in the slower-growing cell population. These experiments illustrated that the inverse relationships of fabH and virulence gene expression levels in exponential-phase lab cultures of S. aureus Wood46 after exposure to AFN-1252 were not reproduced in Wood46 cells harvested from infected granuloma pouches, which may be explained by the different physiological states of the cells in each environment. This divergence in genetic responses to AFN-1252 treatment may not only be the result of in vitro versus in vivo differences (immune response) but could also be the result of the growth rate and/or density of Wood46 in the pouch fluid. In the future, it may be of interest to evaluate additional strains in the animal model to further understand the interaction between AFN-1252 and the SaeRS virulence regulation pathway in S. aureus.
Fig 6.
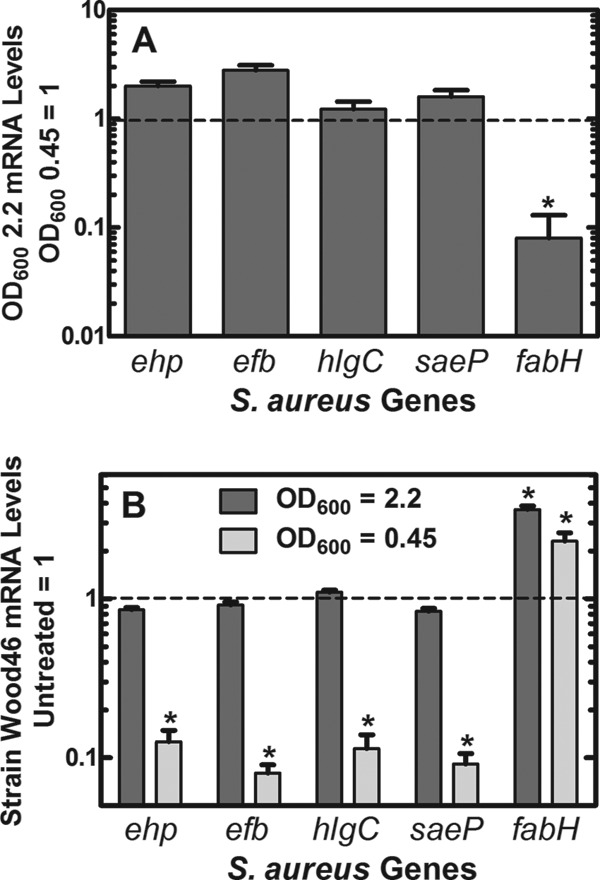
Growth rate dependence of AFN-1252 on virulence factor and fabH mRNA levels. Strain Wood46 was subjected to treatment with AFN-1252 (50 ng/ml) for 15 min either during the logarithmic phase of growth (OD600, 0.45) or after growth slowed as the cells began to enter stationary phase (OD600, 2.2). Transcript abundance in triplicate samples was measured in triplicate by qRT-PCR using gmk as the calibrator. (A) mRNA levels detected at an OD600 of 0.45 were set to 1.0 and compared to the same measurements at an OD600 of 2.2. fabH mRNA was significantly decreased in the samples at the OD600 of 2.2 (*, P < 0.05). mRNA levels for the 4 virulence factors were either unchanged (hlgC) or modestly elevated (∼2-fold) with ehp, efb, and saeP. (B) mRNA levels before AFN-1252 treatment were normalized to 1, and the changes in virulence factor and fabH mRNAs triggered by AFN-1252 are plotted. There was no significant difference between virulence gene expression in cells treated with AFN-1252 at an OD600 of 2.2, but all 4 virulence factors were decreased by about an order of magnitude in cells treated with AFN-1252 at an OD600 of 0.45 (*, P < 0.05). fabH mRNAs were significantly increased in response to AFN-1252 under both experimental conditions (*, P < 0.05). Significance between treated and untreated samples was calculated using the Student t test, and error bars indicate standard deviations.
DISCUSSION
Our data link the repression of virulence gene expression in S. aureus by AFN-1252 to the function of the SaeRS 2-component system. The suite of virulence factors downregulated by AFN-1252 is coordinately regulated by SaeRS, and virulence factor attenuation does not occur in cells lacking a functional SaeRS system or in cells with constitutive SaeR signaling. Most 2-component regulators have sensor components with a single transmembrane helix connecting the external ligand binding domain with an internal kinase domain (58). SaeRS belongs to a small subset of these regulators with the external ligand binding domain replaced by a series of membrane-spanning helices (59). These regulators are thought to sense perturbations in the membrane environment (18). One well-characterized member of this family is DesK from Bacillus subtilis (60). This regulator utilizes the transmembrane segment to sense the fluidity of the membrane and adjusts the transcription of an acyl desaturase to adjust the biophysical properties of the phospholipid bilayer. Similarly, SaeRS is thought to sense perturbations in the intramembrane environment, and a point mutation in the first transmembrane segment of SaeS leads to constitutive signaling through SaeRS (48). Membrane perturbations by antibacterial host factors (51), by biocides (55), or by altering membrane phospholipid composition (61) all affect SaeRS-dependent virulence factor production. Similarly, we propose that the abrupt cessation of phospholipid synthesis in AFN-1252-treated cells alters the membrane properties and signaling through SaeRS in S. aureus. This effect is most pronounced in rapidly growing cells that are actively forming new membrane phospholipid but is attenuated in slower-growing cultures where the rate of new phospholipid formation is reduced.
The importance of virulence factors in S. aureus pathogenesis has focused laboratory research on how antibiotics affect the expression of virulence genes. A survey of 31 antibiotics showed a strong induction of virulence factor expression with subinhibitory concentrations of β-lactams and an almost complete inhibition of expression by clindamycin (25). Linezolid is another protein synthesis inhibitor that reduces exoprotein production in S. aureus at subinhibitory concentrations (23, 30). As anticipated from the mechanism of action of linezolid, the primary effect of this functional class of antibiotics on virulence factor expression is mediated through translation rather than transcription (24). Our results extend this body of work to identify fatty acid synthesis inhibitors as a class of antibiotics that attenuate virulence factor transcription via their effect on the SaeRS system. However, the laboratory results do not exactly match the findings in the animal model. The pathway-specific transcriptional effect of AFN-1252 on FASII gene expression was preserved in the mouse granuloma model infected with strain Wood46, but the reduction in virulence gene transcription was attenuated. One potential reason for this difference is that the cells in the pouch are growing slower than are cells in laboratory culture. Although the pouch CFU did not change during the 12 h of our experiments, this level of viable bacteria may reflect the elimination of the bacteria by the host immune system coupled with their replacement by dividing bacteria (57). In support of the idea that slower-growing cells impact the effect of AFN-1252 on virulence factor expression, a comparison between rapidly and slow-growing cells in the laboratory recapitulated the reduced impact of AFN-1252 on virulence factor expression in slower-growing cells. However, this may not be the only factor that influences the expression of virulence genes in the pouch. The fact that the level of virulence factor expression is known to depend on the type of medium used for cell growth in laboratory experiments (62) suggests that host environment plays an important role. Factors are present in the animal that are absent in the laboratory that may activate the SaeRS system to counteract the effect of AFN-1252 (50, 51), or RNA turnover may be reduced by S. aureus stress response pathways that are activated in response to the host (63). Our evaluation of virulence factor mRNA levels in the mouse granuloma model illustrates that the extension of laboratory findings on virulence gene expression to in vivo infection models needs further research.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Frank for his expert technical assistance.
The research at St. Jude Children's Research Hospital was supported by National Institutes of Health grant GM034496 (C.O.R.), Cancer Center Support grant CA21765, and the American Lebanese Syrian Associated Charities. The research at the University of North Texas Health Science Center was supported by funds from Affinium Pharmaceuticals. Nachum Kaplan is an employee of Affinium Pharmaceuticals.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Published ahead of print 4 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02307-12.
REFERENCES
- 1. Como-Sabetti K, Harriman KH, Buck JM, Glennen A, Boxrud DJ, Lynfield R. 2009. Community-associated methicillin-resistant Staphylococcus aureus: trends in case and isolate characteristics from six years of prospective surveillance. Public Health Rep. 124:427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885 [DOI] [PubMed] [Google Scholar]
- 3. Karlowsky JA, Laing NM, Baudry T, Kaplan N, Vaughan D, Hoban DJ, Zhanel GG. 2007. In vitro activity of API-1252, a novel FabI inhibitor, against clinical isolates of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 51:1580–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karlowsky JA, Kaplan N, Hafkin B, Hoban DJ, Zhanel GG. 2009. AFN-1252, a FabI inhibitor, demonstrates a staphylococcal-specific spectrum of activity. Antimicrob. Agents Chemother. 53:3544–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaplan N, Albert M, Awrey D, Bardouniotis E, Berman J, Clarke T, Dorsey M, Hafkin B, Ramnauth J, Romanov V, Schmid MB, Thalakada R, Yethon J, Pauls HW. 2012. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrob. Agents Chemother. 56:5865–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86 [DOI] [PubMed] [Google Scholar]
- 7. Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. 2011. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U. S. A. 108:15378–15383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. 2010. Essentiality of FASII pathway for Staphylococcus aureus. Nature 463:E3 doi:10.1038/nature08667 [DOI] [PubMed] [Google Scholar]
- 9. Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2010. Reply to “Essentiality of FASII pathway for Staphylococcus aureus”. Nature 463:E4 doi:10.1038/nature08667 [Google Scholar]
- 10. Payne DJ, Miller WH, Berry V, Brosky J, Burgess WJ, Chen E, DeWolf JW, Jr, Fosberry AP, Greenwood R, Head MS, Heerding DA, Janson CA, Jaworski DD, Keller PM, Manley PJ, Moore TD, Newlander KA, Pearson S, Polizzi BJ, Qiu X, Rittenhouse SF, Slater-Radosti C, Salyers KL, Seefeld MA, Smyth MG, Takata DT, Uzinskas IN, Vaidya K, Wallis NG, Winram SB, Yuan CC, Huffman WF. 2002. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob. Agents Chemother. 46:3118–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361 [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, Gonzalez I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB. 2007. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. U. S. A. 104:7612–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freiberg C, Pohlmann J, Nell PG, Endermann R, Schuhmacher J, Newton B, Otteneder M, Lampe T, Habich D, Ziegelbauer K. 2006. Novel bacterial acetyl-coenzyme A carboxylase inhibitors with antibiotic efficacy in vivo. Antimicrob. Agents Chemother. 50:2707–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Escaich S, Prouvensier L, Saccomani M, Durant L, Oxoby M, Gerusz V, Moreau F, Vongsouthi V, Maher K, Morrissey I, Soulama-Mouze C. 2011. The MUT056399 inhibitor of FabI is a new antistaphylococcal compound. Antimicrob. Agents Chemother. 55:4692–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foster TJ. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948–958 [DOI] [PubMed] [Google Scholar]
- 16. Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giraudo AT, Cheung AL, Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53–58 [DOI] [PubMed] [Google Scholar]
- 18. Sun F, Li C, Jeong D, Sohn C, He C, Bae T. 2010. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 192:2111–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177 doi:10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J. Infect. Dis. 201:241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 199:1698–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith K, Gould KA, Ramage G, Gemmell CG, Hinds J, Lang S. 2010. Influence of tigecycline on expression of virulence factors in biofilm-associated cells of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gemmell CG, Ford CW. 2002. Virulence factor expression by Gram-positive cocci exposed to subinhibitory concentrations of linezolid. J. Antimicrob. Chemother. 50:665–672 [DOI] [PubMed] [Google Scholar]
- 24. Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 195:202–211 [DOI] [PubMed] [Google Scholar]
- 25. Ohlsen K, Ziebuhr W, Koller KP, Hell W, Wichelhaus TA, Hacker J. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 42:2817–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J. Antimicrob. Chemother. 66:979–984 [DOI] [PubMed] [Google Scholar]
- 27. Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 28. Herbert S, Barry P, Novick RP. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joo HS, Chan JL, Cheung GY, Otto M. 2010. Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:4942–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernardo K, Pakulat N, Fleer S, Schnaith A, Utermohlen O, Krut O, Müller S, Krönke M. 2004. Subinhibitory concentrations of linezolid reduce Staphylococcus aureus virulence factor expression. Antimicrob. Agents Chemother. 48:546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuroda H, Kuroda M, Cui L, Hiramatsu K. 2007. Subinhibitory concentrations of β-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268:98–105 [DOI] [PubMed] [Google Scholar]
- 32. Saleh FA, Freer JH. 1984. Inhibition of secretion of staphylococcal alpha toxin by cerulenin. J. Med. Microbiol. 18:205–216 [DOI] [PubMed] [Google Scholar]
- 33. Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, Freiberg C, Ziegelbauer K, Rock CO, Ivanov I, Loferer H. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freiberg C, Fischer HP, Brunner NA. 2005. Discovering the mechanism of action of novel antibacterial agents through transcriptional profiling of conditional mutants. Antimicrob. Agents Chemother. 49:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schujman GE, Paoletti L, Grossman AD, de Mendoza D. 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell 4:663–672 [DOI] [PubMed] [Google Scholar]
- 36. Wenzel M, Patra M, Albrecht D, Chen DY, Nicolaou KC, Metzler-Nolte N, Bandow JE. 2011. Proteomic signature of fatty acid biosynthesis inhibition available for in vivo mechanism-of-action studies. Antimicrob. Agents Chemother. 55:2590–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhong J, Karberg M, Lambowitz AM. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 31:1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Theis T, Skurray RA, Brown MH. 2007. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J. Microbiol. Methods 70:355–362 [DOI] [PubMed] [Google Scholar]
- 39. Winer J, Jung CK, Shackel I, Williams PM. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270:41–49 [DOI] [PubMed] [Google Scholar]
- 40. Schujman GE, Guerin M, Buschiazzo A, Schaeffer F, Llarrull LI, Reh G, Vila AJ, Alzari PM, de Mendoza D. 2006. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 25:4074–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schujman GE, Altabe S, de Mendoza D. 2008. A malonyl-CoA-dependent switch in the bacterial response to a dysfunction of lipid metabolism. Mol. Microbiol. 68:987–996 [DOI] [PubMed] [Google Scholar]
- 42. Albanesi D, Reh G, Guerin ME, Schaeffer F, Debarbouille M, Buschiazzo A, Schujman GE, de Mendoza D, Alzari PM. 2013. Structural basis for feed-forward transcriptional regulation of membrane lipid homeostasis in Staphylococcus aureus. PLoS Pathog. 9:e1003108 doi:10.1371/journal.ppat.1003108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martinez MA, Zaballa ME, Schaeffer F, Bellinzoni M, Albanesi D, Schujman GE, Vila AJ, Alzari PM, de Mendoza D. 2010. A novel role of malonyl-ACP in lipid homeostasis. Biochemistry 49:3161–3167 [DOI] [PubMed] [Google Scholar]
- 44. Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289–2303 [DOI] [PubMed] [Google Scholar]
- 45. Korem M, Gov Y, Rosenberg M. 2010. Global gene expression in Staphylococcus aureus following exposure to alcohol. Microb. Pathog. 48:74–84 [DOI] [PubMed] [Google Scholar]
- 46. Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74:4655–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steinhuber A, Goerke C, Bayer MG, Doring G, Wolz C. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adhikari RP, Novick RP. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959 [DOI] [PubMed] [Google Scholar]
- 50. Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, Shabb DW, Diep BA, Chambers HF, Otto M, DeLeo FR. 2011. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6:e18617 doi:10.1371/journal.pone.0018617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190:3419–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karlsson-Kanth A, Tegmark-Wisell K, Arvidson S, Oscarsson J. 2006. Natural human isolates of Staphylococcus aureus selected for high production of proteases and α-hemolysin are σB deficient. Int. J. Med. Microbiol. 296:229–236 [DOI] [PubMed] [Google Scholar]
- 54. Novick RP, Jiang D. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709–2717 [DOI] [PubMed] [Google Scholar]
- 55. Schafer D, Lam TT, Geiger T, Mainiero M, Engelmann S, Hussain M, Bosserhoff A, Frosch M, Bischoff M, Wolz C, Reidl J, Sinha B. 2009. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters response to biocide exposure. J. Bacteriol. 191:7306–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jabes D, Candiani G, Romano G, Brunati C, Riva S, Cavaleri M. 2004. Efficacy of dalbavancin against methicillin-resistant Staphylococcus aureus in the rat granuloma pouch infection model. Antimicrob. Agents Chemother. 48:1118–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Motley ST, Morrow BJ, Liu X, Dodge IL, Vitiello A, Ward CK, Shaw KJ. 2004. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell. Microbiol. 6:849–865 [DOI] [PubMed] [Google Scholar]
- 58. Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mascher T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133–144 [DOI] [PubMed] [Google Scholar]
- 60. Cybulski LE, Martin M, Mansilla MC, Fernandez A, de Mendoza D. 2010. Membrane thickness cue for cold sensing in a bacterium. Curr. Biol. 20:1539–1544 [DOI] [PubMed] [Google Scholar]
- 61. Sievers S, Ernst CM, Geiger T, Hecker M, Wolz C, Becher D, Peschel A. 2010. Changing the phospholipid composition of Staphylococcus aureus causes distinct changes in membrane proteome and membrane-sensory regulators. Proteomics 10:1685–1693 [DOI] [PubMed] [Google Scholar]
- 62. Ray B, Ballal A, Manna AC. 2009. Transcriptional variation of regulatory and virulence genes due to different media in Staphylococcus aureus. Microb. Pathog. 47:94–100 [DOI] [PubMed] [Google Scholar]
- 63. Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.