Abstract
Daclatasvir (DCV; BMS-790052) is a hepatitis C virus (HCV) NS5A replication complex inhibitor (RCI) with picomolar to low nanomolar potency and broad genotypic coverage in vitro. Viral RNA declines have been observed in the clinic for both alpha interferon-ribavirin (IFN-α–RBV) and IFN-RBV-free regimens that include DCV. Follow-up specimens (up to 6 months) from selected subjects treated with DCV in 14-day monotherapy studies were analyzed for genotype and phenotype. Variants were detected by clonal sequencing in specimens from baseline and were readily detected by population sequencing following viral RNA breakthrough and posttreatment. The major amino acid substitutions generating resistance in vivo were at residues M28, Q30, L31, and Y93 for genotype 1a (GT-1a) and L31 and Y93 for GT-1b, similar to the resistance substitutions observed with the in vitro replicon system. The primary difference in the resistance patterns observed in vitro and in vivo was the increased complexity of linked variant combinations observed in clinical specimens. Changes in the percentage of individual variants were observed during follow-up; however, the overall percentage of variants in the total population persisted up to 6 months. Our results suggest that during the 14-day monotherapy, most wild-type virus was eradicated by DCV. After the end of DCV treatment, viral fitness, rather than DCV resistance, probably determines which viral variants emerge as dominant in populations.
INTRODUCTION
Daclatasvir (DCV; BMS-790052) is a hepatitis C virus (HCV) NS5A replication complex inhibitor (RCI) with picomolar to low nanomolar potency and broad genotypic coverage in vitro. In the replicon system, 50% (i.e., half-maximal) effective concentrations (EC50s) of DCV are 50 and 9 pM against genotype 1a (GT-1a) and GT-1b, respectively (1, 2). Its in vitro potency translated into anti-HCV activity in the clinic. Initial viral RNA declines with high sustained virologic response (SVR) have been achieved for both interferon-ribavirin (IFN-RBV) and IFN-RBV-free regimens in combination therapies (1, 3–8).
In a 14-day multiple-ascending-dose (MAD) monotherapy study, chronically infected patients, treated with DCV at 1, 10, 30, 60, and 100 mg QD (once daily) or 30 mg BID (twice daily) for 14 days (4 subjects per cohort), generally experienced rapid and marked viral load declines (3, 4). Although viral breakthrough (VBT) was observed for both GT-1a- and -1b-infected patients, RNA declined below the level of detection (<10 IU/ml) in several GT-1b-infected patients, and viral RNA remained detectable in the majority of GT-1a-infected patients (3, 4).
Genome variants of HCV NS5A that emerged in viral specimens collected during and after treatment with DCV in vivo (clinical cases) and in vitro (replicons) are similar (1, 2, 4, 9). To date, all amino acid substitutions observed in vitro that are associated with resistance to DCV and its analogs synthesized by us mapped to the N-terminal region of NS5A (1, 2, 9, 10). For GT-1b, the major resistance substitutions observed were at residues 31 and 93 of NS5A, while for GT-1a, the major resistance substitutions observed also included residues 28 and 30 (1, 2, 9). The major variants detected in infected patients treated with DCV in the 14-day MAD monotherapy study were similar. However, additional variants, especially variants with linkage of two or more resistance-associated amino acid substitutions, were also detected in vivo (4, 11).
The primary objective of this study was to determine if the variants that emerged during monotherapy with DCV persisted after treatment ended (up to 6 months posttreatment) or if the population of variants decayed, with a return to wild-type (WT) virus. This information could inform future decisions about retreatment of patients that experience viral breakthrough or viral rebound. This report expands and extends our previous study of resistance variants that emerged during the MAD study of DCV (4). Genotypic analysis of viral variants determined by both population and clonal sequencing and phenotypic analysis using transient-replicon-replication assays are discussed.
MATERIALS AND METHODS
Compounds.
NS5A replication complex inhibitor daclatasvir (DCV; BMS-790052), NS3 protease inhibitor asunaprevir (ASV; BMS-650032), and NS5B polymerase inhibitor BMS-791325 have been described previously (1, 4, 9).
Genotypic analysis of clinical specimens.
Genotypic analysis of clinical specimens has been previously described (4). Basically, 2 amplicons from each sample were obtained by PCR using 2 different primer sets. The percentages of amino acid substitutions present in each sample were derived from visual inspection of the population cDNA sequence chromatograms and are the average of estimates from the two amplicons. For the cloning sequence analysis, amplicons from selected time points were cloned and NS5A sequence from individual cDNA clones was obtained as described previously (2). To determine the relative sensitivity of detecting sequence variants, reconstitution experiments were performed with DNA mixtures containing both wild-type and resistant (Y93H) variants. Mixtures of wild-type and Y93H variant DNA at ratios of 100:0, 95:5, 90:10, 80:20, and 60:40 were sequenced. The experiment revealed that the variant could be readily detected at 20% of the wild-type population (results not shown) (4).
In vitro analysis of replicon variants.
Amino acid substitutions were introduced into HCV reference replicons (genotype 1b, Con1; genotype 1a, H77c) as previously described (2). Inhibitor sensitivities and replicative ability (fitness) were assessed in transient-replication assays, as previously described (2).
Study design and clinical specimens.
To examine safety, pharmacokinetics, and antiviral effect across the potential clinical dose range, DCV was dosed as monotherapy in a double-blind, placebo-controlled, sequential-panel, MAD study with patients chronically infected with HCV genotype 1. Six dose regimens were evaluated (1 mg once daily, 10 mg once daily, 30 mg once or twice daily, 60 mg once daily, and 100 mg once daily). Five patients in each panel were randomized to receive a 14-day course of orally administered BMS-790052 or placebo in a ratio of 4:1; thus, a total of 24 patients received DCV (3, 4). The study was approved by the institutional review boards in all study centers and conducted in accordance with good clinical practice and ethical principles that have their origin in the Declaration of Helsinki (3, 4, 6). Informed written consent was obtained from all patients. HCV RNA levels were determined using the Roche Cobas TaqMan HCV test, version 2.0 (Roche, Pleasanton, CA; lower limit of quantification, 25 IU/ml; lower limit of detection, 10 IU/ml). Viral breakthrough was defined as an HCV RNA increase by at least 0.5 log10 following HCV RNA nadir while receiving DCV. Serum specimens were collected for genotypic analysis at baseline (BL, or time zero [T0]) and days 1 (4, 8, and 12 h after the first dose [shown throughout as T4, T8, etc.]), 2, 4, 7, 14, 15, 16, 17, 21, 28, 42 (or 45), 98 (or 92 or 116), and 182 (or 177) as indicated in Tables 3 to 14. Following amplification of the NS5A coding region, a genotypic analysis was performed by population sequencing to determine the emergence of viral variants following the administration of multiple doses of DCV and the follow-up until 182 days. The complete study design and resistance analysis methodology have been described elsewhere (3, 4).
Table 3.
Variant persistence in HCV patients after treatment QD with 1 mg of DCV by population sequence
| Patient | Genotype | Time | RNA (IU/ml) | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M/L28 | Q/R30 | L31 | Y93 | |||||
| A | 1a | T0 | 1.1E+06 | WT | WT | WT | WT | |
| T12 | 2.1E+03 | WT | WT | WT | WT | |||
| Day 14 | 3.9E+05 | T (∼45%, ∼45%) | H (∼10%, ∼10%), R (∼30%, ∼10%) | M (0, ∼30%) | C (0, ∼20%) | |||
| Day 15 | 7.9E+05 | T (∼35%) | H (∼2.5%), R (∼40%) | M (∼10%) | C (∼15%) | |||
| Day 21 | 6.3E+05 | T (∼30%) | H (∼7.5%), R (∼40%) | M (∼20%) | C (∼7.5%) | |||
| Day 28 | 2.0E+06 | T (∼25%) | H (∼5%), R (∼40%) | M (∼12.5%) | C (∼5%) | |||
| Day 42 | 3.2E+06 | T (∼27.5%) | H (∼5%), R (∼40%) | M (∼20%) | ||||
| B | 1a | T0 | 3.2E+05 | WT | WT | WT | WT | H58P (∼5%) |
| Day 14 | 7.9E+04 | T (∼10%) | R (∼5%) | M (∼50%) | H58P (∼55%) | |||
| Day 15 | 1.0E+05 | T (∼10%) | R (∼10%) | M (∼50%) | H58P (∼20%) | |||
| Day 21 | 3.2E+05 | T (∼10%) | R (∼7.5%) | M (∼35%) | H58P (∼5%) | |||
| Day 28 | 1.3E+06 | T (∼7.5%) | R (∼5%) | M (∼37.5%) | T21I (∼5%) | |||
| Day 42 | 3.2E+06 | T (∼5%) | H (∼10%), R (∼5%) | M (∼60%) | T21I (∼7.5%) | |||
| Day 98 | 1.6E+06 | T (<2.5%) | H (∼10%) | M (∼77.5%) | T21I (∼25%) | |||
| Day 182 | 1.3E+05 | T (<5%) | H (∼7.5%) | M (∼90%) | T21I (∼50%), H58P (∼52.5%) | |||
| C | 1b | T0 | 5.4E+04 | WT | WT | WT | WT | |
| Day 4 | <25 | ND | ND | ND | ND | |||
| Day 7 | Undetectable | ND | ND | ND | ND | |||
| Day 15 | Undetectable | ND | ND | ND | ND | |||
| Day 21 | Undetectable | ND | ND | ND | ND | |||
| Day 28 | Undetectable | ND | ND | ND | ND | |||
| Day 42b | 1.0E+02 | WT | WT | WT | WT | |||
| Day 98 | 1.0E+04 | WT | WT | WT | WT | |||
| D | 1b | T0 | 5,765 | WT | WT | WT | WT | |
| Day 7 | <25 | ND | ND | ND | ND | |||
| Day 14 | Undetectable | ND | ND | ND | ND | |||
| Day 15 | Undetectable | ND | ND | ND | ND | |||
| Day 21 | Undetectable | ND | ND | ND | ND | |||
| Day 28 | Undetectable | ND | ND | ND | ND | |||
Frequencies are quantitative estimates of variants based on chromatograms from population sequencing (estimated from the average of two PCR products). “WT” indicates consensus with the WT. ND, not determined (HCV RNA < 1,000 IU/ml). Data for T0 to day 14 have been reported previously (3, 4).
Data derived from one PCR product.
Table 14.
Number of clones for specimens from patient W (GT-1b infected), treated BID with 30 mg of DCV
| Mutation(s) | No. of clones at the indicated time pointa |
|||||||
|---|---|---|---|---|---|---|---|---|
| T0-1 | T0-2 | D4-1 | D4-2 | D42-1 | D42-2 | D182-1 | D182-2 | |
| None (WT) | 94 | 93 | 91 | 96 | ||||
| R30Q | 1 | |||||||
| L31I | 15 | |||||||
| L31V | 4 | |||||||
| P32del | 37 | 47 | 5 | |||||
| Y93C | 2 | 1 | 4 | |||||
| L31F-P32del | 1 | |||||||
| L31I-Y93H | 18 | 11 | 45 | 34 | ||||
| L31I-Y93N | 1 | |||||||
| L31I-Y93R | 4 | |||||||
| L31V-Y93H | 15 | 2 | 4 | |||||
| P32del-Y93H | 25 | 18 | ||||||
| Total clones | 96 | 95 | 95 | 96 | 95 | 95 | 48 | 48 |
Two different primer sets (-1 and -2) were used for the same specimen derived from each time point indicated.
RESULTS
The resistance profile and replication level (fitness) of genotype 1a and 1b variants observed in the 14-day monotherapy study of daclatasvir (DCV; BMS-790052) were characterized in vitro by introducing substitutions into wild-type (WT) replicons (2, 4). Transient-replicon-replication assays were performed to determine if the phenotypes observed are consistent with the impact of the specific substitutions identified. Tables 1 and 2 list variants identified in specimens obtained from subjects treated with DCV in the MAD study during treatment and follow-up. Since replication ability (fitness), EC50, and fold resistance for many variants have been reported previously, values for only some key mutants are included in Tables 1 and 2 (1, 2, 4, 9). Values for previously described variants (Tables 1 and 2) have been updated to reflect additional test occasions.
Table 1.
Profile of resistance to DCV in the in vitro replicon system: 1a replicona
| Mutation | Replication level (%) |
EC50 (ng/ml)b |
Fold resistance | ||
|---|---|---|---|---|---|
| Avg | SD | Avg | SD | ||
| None (WT) | 100 | 0.0044 | 0.0028 | 1 | |
| T21I | 169 | 9 | 0.0034 | 0.0003 | 0.8 |
| M28I | 200 | 38 | 0.0055 | 0.0003 | 1.2 |
| Q30E | 130 | 56 | 110.9 | 66 | 25,205 |
| L31M | 55 | 15 | 1.5 | 0.5 | 341 |
| L31V | 117 | 29 | 14.9 | 4.4 | 3,386 |
| V37A | 13 | 2 | 0.0037 | 0.0006 | 0.8 |
| V37M | 117 | 5 | 0.004 | 0.0009 | 0.9 |
| E62D | 89 | 39 | 0.0089 | 0.0044 | 2 |
| Y93C | 11 | 7 | 8.2 | 3 | 1,864 |
| Y93H | 18 | 11 | 23.9 | 7 | 5,432 |
| Y93N | 13 | 8 | 208.9 | 47.9 | 47,477 |
| T21I-L31M | 82 | 5 | 2.39 | 0.018 | 544 |
| M28A-Q30R | 45 | 11 | 1,262 | 273 | 287,283 |
| M28T-Q30K | 15 | 2 | >1,481 | >336,591 | |
| Q30H-H58D | 28 | 2 | >1,481 | >337,329 | |
| Q30R-L31M | 54 | 15 | 868 | 679 | 197,698 |
| Q30R-E62D | 44 | 29 | 113 | 13 | 25,682 |
| L31M-H58D | 41 | 4 | 294.5 | 10.5 | 67,055 |
| L31V-V37A | 10 | 0.1 | 47.1 | 6.6 | 10,717 |
| L31V-H58P | 100 | 0 | 54 | 6 | 12,312 |
| H58P-Y93H | 22 | 0.7 | 5.3 | 0.06 | 1,206 |
The GT-1a replicon is H77C with cell culture replication-enhancing mutations P1496L and S2204I. GT-1a Q30E, L31M, L31V, E62D, Y93C, Y93H, Y93N, and Q30R-E62D variants have been previously reported (1, 2, 4, 9, 11); additional GT-1a variants with M28A, M28T, M28V, Q30D, Q30H, Q30K, Q30R, H58D, H58P, M28T-Q30H, M28T-Q30R, M28V-Q30R, Q30H-Y93H, and Q30R-H58D substitutions have been previously reported (1, 2, 4, 9) but are not included in this table. SDs are for ≥3 replicates.
1 ng/ml = 1.35 nM. An EC50 of <10 nM indicates low resistance, an EC50 of 10 to 100 nM indicates moderate resistance, and an EC50 of >100 nM indicates high resistance.
Table 2.
Profile of resistance to DCV in the in vitro replicon system: 1b replicona
| Mutation | Replication level (%) |
EC50 (ng/ml)b |
Fold resistance | ||
|---|---|---|---|---|---|
| Avg | SD | Avg | SD | ||
| None (WT) | 100 | 0.0019 | 0.0002 | 1 | |
| L31I | 54 | 12 | 0.0027 | 0.0011 | 1.4 |
| P32del | 29.1 | 3.2 | >741 | >390,000 | |
| L31I-Y93H | 43 | 11 | 4.8 | 2.5 | 2,526 |
| P32del-Y93H | No replication | ND | |||
The GT-1b replicon is Con1 with cell culture replication-enhancing mutation S2204I. Additional GT-1b R30Q, L31M, L31V, F37L, Q54H, Q54N, Y93C, Y93H, L31M-Y93H, L31V-Y93H, Q54H-Y93H, and L31V-Q54H-Y93H variants have been previously reported (1, 2, 4, 9) and are not included in the table. SDs are for ≥3 replicates.
1 ng/ml = 1.35 nM. An EC50 of <10 nM indicates low resistance, an EC50 of 10 to 100 nM indicates moderate resistance, and an EC50 of >100 nM indicates high resistance.
Both population sequencing and clonal sequencing (with ≥10% variants in the clonal analysis) were used to identify variants. Due to the large amount of data generated, only representative examples from clonal analysis are presented here to support the general conclusions. A few variants with a low frequency of appearance (most are associated with double and triple substitutions) were not analyzed for phenotype because high levels of resistance are expected, based on the level of resistance of each individual mutant. Since phenotypic analysis of variants has been performed multiple times (n ≥ 3), standard deviations (SD) are available (Tables 1 and 2) to indicate the range of EC50s and replication levels (fitness) for WT and variant replicons.
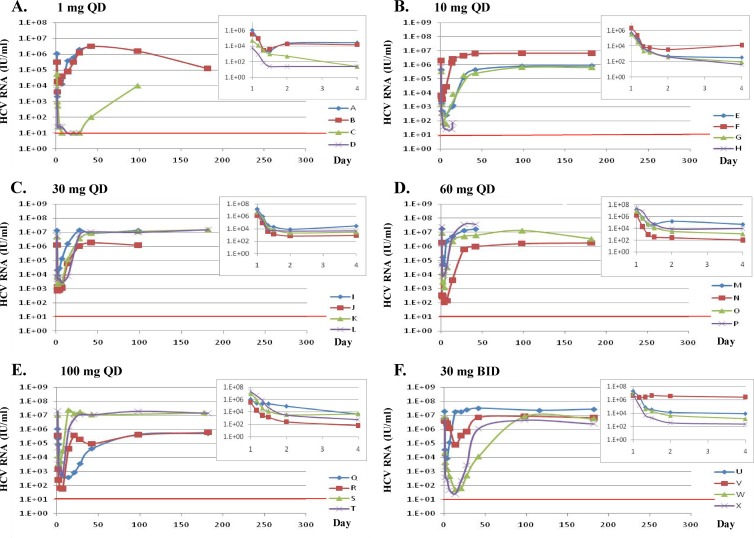
Figure 1 shows the changes in HCV RNA for each dosing cohort during the 14-day monotherapy and follow-up (up to 6 months) periods. The inset in each panel shows the initial changes in HCV RNA during the first 4 days of treatment (3, 4).
Fig 1.
Individual change from baseline in log10 HCV RNA by dose and HCV genotype during (14-day monotherapy) or following (until 180 days) DCV treatment. HCV RNA was set to 25 or 10 IU/ml if the observed value was less than the lower limit of quantification (25 IU/ml) or lower limit of detection (10 IU/ml), respectively, when deriving changes from baseline. Insets display HCV RNA change within 4 days of treatment for each cohort. The red line indicates the HCV RNA detection limit (10 IU/ml). (A) Patients A and B, infected with GT-1a, and patients C and D, infected with GT-1b; (B) patients E and F, infected with GT-1a, and patients G and H, infected with GT-1b; (C) patients I, J, K, and L, infected with GT-1a; (D) patients M, N, O, and P, infected with GT-1a; (E) patients Q, R, and S, infected with GT-1a, and patient T, infected with GT-1b; (F) patients U and V, infected with GT-1a, and patients W and X, infected with GT-1b. Data for T0 to day 14 have been reported previously (3, 4).
One-milligram cohort.
Patients infected with GT-1a and -1b experienced mean declines in HCV RNA of ∼1.5 log10 and ∼2.9 log10, respectively, on day 4 (Fig. 1A). Known resistant variants were not detected in any of the baseline (BL; time zero [T0]) specimens by population sequencing (Table 3). At the end of treatment (day 14), resistance-associated substitutions were detected at multiple residues in GT-1a-infected patients, primarily at residues 28, 30, 31, and 93 as previously reported (1, 2, 9). Variants with M28T, Q30H/R, L31M, and Y93C substitutions showed relatively low or moderate levels of resistance to DCV in vitro (EC50, <10 ng/ml, or <13.5 nM [Table 1]) (2, 4, 9). We regard an EC50 of <10 nM to indicate low resistance, 10 to 100 nM to indicate moderate resistance, and >100 nM to indicate high resistance (Tables 1 and 2). GT-1a variants with T21I and H58P amino acid substitutions were also observed (Table 3, patient B). Neither of these amino acid substitutions substantially altered DCV potency in vitro. Furthermore, the resistance of a variant with T21I in combination with L31M was similar to that of an L31M variant, suggesting that T21I did not contribute to DCV resistance (Table 1). In patient B, the relative percentages of T21I and H58P substitutions increased during follow-up (see day 182 results in Table 3), suggesting that these polymorphisms may have contributed to viral fitness. For both of the GT-1a-infected patients in this cohort (patients A and B), minor changes in the relative percentages of viral variants with substitutions at residues 28, 30, 31, and/or 93 were observed from the end of dosing through the follow-up period by population sequence analysis.
In two GT-1b-infected patients with low BL levels of virus, HCV RNA became undetectable at the end of the 14-day treatment (Table 3). Although viral rebound or relapse was observed at days 42 and 98 in patient C, only WT virus was observed by population sequencing. In GT-1b-infected patient D, HCV RNA was still undetectable at day 28, 14 days after treatment ended (Table 3). No further follow-up samples were obtained from this patient.
To validate the quantitative estimates (percent resistance variants) determined by population sequencing, and to explore linkage of resistant substitutions, clonal analysis was performed for selected specimens (T0, T12, day 14, and day 42) from patient A. Although no resistance variants were detected at BL by population sequencing (Table 3), clonal analysis revealed approximately 3% resistant variants (Table 4).
Table 4.
Number of clones for specimens from patient A (GT-1a infected), treated QD with 1 mg of DCV
| Mutation(s) | No. of clones at the indicated time point |
||||
|---|---|---|---|---|---|
| T0 | T12 | D14-1a | D14-2a | D42 | |
| None (WT) | 93 | 87 | 3 | ||
| M28T | 1 | 49 | 49 | 27 | |
| Q30H | 24 | 5 | 14 | ||
| Q30K | 2 | ||||
| Q30R | 2 | 31 | 15 | 33 | |
| L31M | 12 | 9 | |||
| H58D | 4 | ||||
| Y93C | 13 | 2 | |||
| Y93N | 1 | ||||
| M28T-Y93C | 2 | ||||
| Q30R-L31P | 1 | ||||
| L31M-Y93C | 1 | ||||
| Total | 96 | 88 | 104 | 100 | 92 |
Two different primer sets (-1 and -2) were used for the same specimen derived from day 14.
These variants (Q30R variant, 2%, and Y93N variant, 1%) were not observed at T12, when there was a 99.8% decline in viral RNA from BL, suggesting that they were at least partially suppressed by 1 mg of DCV or that they had poor fitness. Another variant of note, the Q30K variant, was detected as a very minor species in the posttreatment specimen (day 42) but displayed a relatively high level of DCV resistance, with an EC50 of 108 ng/ml, or 146 nM (2, 4, 9).
Overall, the clonal analysis of patient A viral variants yielded several important observations. First, resistant variants were rapidly enriched from ∼3% at BL to ∼97 to 100% at day 14. Second, while the percentages of individual resistant variants varied slightly over time, the combined total percentage of resistant variants remained constant from the end of treatment (day 14) through 28 days after treatment (day 42), suggesting that the resistant variants enriched or selected by 1 mg of DCV were relatively fit. Third, a low percentage of linked resistance substitutions were detected at day 14 (L31M-Y93C and M28T-Y93C) and day 42 (Q30R-L31P). Since linked variants were not detected among the 96 clones analyzed at BL, it is unclear if very low levels of variants with linked resistance-associated mutations were present at baseline or if these double amino acid substitution variants were selected during the 14-day treatment with 1 mg of DCV.
Ten-milligram cohort.
Patients infected with GT-1a and -1b experienced mean declines in HCV RNA of ∼2.7 log10 and ∼ 3.9 log10, respectively, on day 4 (Fig. 1B). No known resistant variants were detected in the BL or T4 specimens by population sequencing (Table 5). At day 14, resistant variants were observed in all GT-1a-infected and one of two GT-1b-infected patients. Viral RNA in another GT-1b-infected patient, patient H, was too low to evaluate (76 IU/ml) at day 15. Two notable observations were made in patient E: (i) the GT-1a Y93H variant became undetectable during the follow-up period (from day 28 to day 182), and (ii) resistant variants persisted up to ∼182 days after treatment.
Table 5.
Variant persistence in HCV patients after treatment QD with 10 mg of DCV by population sequence
| Patient | Genotype | Time | RNA (IU/ml) | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M/L28 | Q/R30 | L31 | Y93 | |||||
| E | 1a | T0 | 4.2E+05 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |
| T4 | 1.0E+05 | WT | WT | WT | WT | H58P (∼100%) | ||
| Day 14b | 1.0E+03 | H (∼100%) | H58P (∼100%) | |||||
| Day 15 | 1.3E+03 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |||
| Day 21 | 9.9E+03 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |||
| Day 28b | 1.2E+05 | H (∼30%) | M (∼50%), V (∼20%) | H58P (∼100%) | ||||
| Day 42 | 4.4E+05 | H (∼15%) | M (∼60%), V (∼20%) | H58P (∼100%) | ||||
| Day 98b | 7.9E+05 | M (∼95%), V (∼5%) | H58P (∼100%) | |||||
| Day 182 | 8.3E+05 | M (∼100%) | H58P (∼100%) | |||||
| F | 1a | T0 | 2.0E+06 | WT | WT | WT | WT | |
| T24 | 3.4E+03 | H (∼30%), R (∼40%) | H (∼10%) | |||||
| Day 14 | 1.3E+06 | E (∼5%), H (∼15%), R (∼20%) | M (∼5%), V (∼50%) | H (∼10%) | ||||
| Day 15 | 2.6E+06 | H (∼5%), R (∼15%) | M (∼5%), V (∼57.5%) | H (∼15%) | ||||
| Day 21 | 7.4E+06 | H (∼5%), R (∼10%) | V (∼60%) | H (∼5%) | ||||
| Day 28 | 4.3E+06 | H (∼12.5%), R (∼10%) | V (∼70%) | H (∼2.5%) | ||||
| Day 42 | 6.1E+06 | H (∼10%), R (∼7.5%) | M (∼2.5%), V (∼70%) | |||||
| Day 98 | 6.6E+06 | H (∼10%), R (∼7.5%) | M (∼5%), V (∼75%) | |||||
| Day 182 | 6.8E+06 | H (∼15%), R (∼2.5%) | M (∼5%), V (∼80%) | |||||
| G | 1b | T0 | 3.2E+05 | WT | WT | WT | WT | |
| Day 14 | 2.9E+03 | M (∼50%), V (∼50%) | H (∼100%) | |||||
| Day 15 | 8.1E+03 | M (∼60%), V (∼40%) | H (∼100%) | |||||
| Day 21 | 1.9E+05 | M (∼50%), V (∼50%) | H (∼100%) | |||||
| Day 28 | 1.7E+05 | M (∼50%), V (∼50%) | H (∼100%) | |||||
| Day 42 | 2.5E+05 | M (∼50%), V (∼50%) | H (∼100%) | |||||
| Day 98 | 6.6E+05 | M (∼50%), V (∼50%) | H (∼100%) | |||||
| Day 182 | 6.4E+05 | M (∼60%), V (∼40%) | H (∼100%) | |||||
| H | 1b | T0 | 6.8E+05 | WT | WT | WT | WT | Q54N (∼50%), Q54H (∼45%) |
| Day 14 | <25 | ND | ND | ND | ND | |||
| Day 15 | 76 | ND | ND | ND | ND | |||
Frequencies are quantitative estimates of variants based on chromatograms from population sequencing (estimated from the average of two PCR products). “WT” indicates consensus with the WT. ND, not determined (HCV RNA < 1,000 IU/ml). Data for T0 to day 14 have been reported previously (3, 4).
Data derived from one PCR product.
Clonal analysis was performed for selected specimens (T0 [BL], T24, day 14, day 15, and day 182) from patient F (Table 6). Although no resistant variants were detected at BL by population sequencing of BL samples from this patient, clonal analysis revealed ∼6% resistant variants (∼3% M28T, ∼1% Q30R, and ∼2% Y93H variants). An M28V amino acid substitution was also detected in BL samples from this subject, but this substitution did not confer resistance to DCV in vitro (4). At the end of the treatment (day 14), ∼99% of the detectable HCV population was made up of resistant variants. During the follow-up period (up to day 182), the total percentage of variants did not change (∼99%), demonstrating persistence of the resistant variants. Consistent with the population sequencing data, the most dominant variant in patient F at day 182 was the L31V variant (78%, or 119 out of 152 clones), with a moderate resistance to DCV in vitro (EC50, 14.9 ng/ml, or ∼20 nM [Table 1]) (2, 4, 9). A low percentage of variants (∼3 to 6%) with linked substitutions were observed by clonal analysis during the follow-up period.
Table 6.
Number of clones for specimens from patient F (GT-1a infected), treated QD with 10 mg of DCV
| Mutation(s) | No. of clones at the indicated time point |
||||
|---|---|---|---|---|---|
| T0 | T24 | D14 | D15 | D182 | |
| None (WT) | 87 | 16 | 1 | 5 | 2 |
| M28T | 3 | ||||
| M28V | 2 | ||||
| Q30E | 10 | 2 | |||
| Q30H | 23 | 13 | 5 | 13 | |
| Q30K | 4 | 11 | |||
| Q30R | 1 | 28 | 19 | 19 | 6 |
| L31M | 4 | 2 | 5 | ||
| L31V | 1 | 44 | 81 | 119 | |
| H58D | 4 | 3 | |||
| Y93C | 2 | ||||
| Y93H | 2 | 9 | 5 | 19 | |
| Y93N | 1 | ||||
| M28T-Y93C | 1 | ||||
| Q30H-L31P | 1 | ||||
| Q30H-L31V | 2 | ||||
| Q30R-L31M | 1 | 1 | |||
| Q30R-H58D | 2 | ||||
| Q30R-Y93H | 1 | 2 | |||
| Q30R-Y93N | 1 | ||||
| L31V-H58L | 1 | ||||
| L31V-H58P | 1 | ||||
| L31V-Y93H | 6 | ||||
| Q30H-L31P-Y93H | 1 | ||||
| Total clones | 95 | 86 | 109 | 150 | 152 |
Thirty-milligram cohort.
All patients in the 30-mg cohort were infected with GT-1a and experienced mean declines in HCV RNA of ∼3.1 log10 on day 4 (Fig. 1C). Although no known resistant variants were detected at BL by population sequencing, variants were readily detectable at day 7 or day 14 (Table 7). The most dominant amino acid substitution in this cohort at day 14 was Q30E, and viral variants with Q30E persisted through day 98 or day 182. A Q30E amino acid substitution conferred a relatively high level of resistance to DCV in vitro (EC50, 110.9 ng/ml or 150 nM [Table 1]) (2, 4, 9).
Table 7.
Variant persistence in HCV patients after treatment QD with 30 mg of DCV by population sequence
| Patient | Genotype | Time | RNA (IU/ml) | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M28 | Q30 | L31 | Y93 | |||||
| I | 1a | T0 | 1.4E+07 | WT | WT | WT | WT | |
| Day 7 | 1.3E+05 | T (∼40%) | E (∼17.5%), H (∼5%), R (∼10%) | M (∼2.5%), V (∼10%) | C (∼10%), H (∼2.5%), N (∼2.5%) | |||
| Day 14 | 1.6E+06 | T (∼5%) | E (∼70%), R (<5%) | V (∼15%) | H (∼2.5%), N (∼5%) | |||
| Day 15 | 1.3E+06 | T (∼5%) | E (∼60%), R (∼5%) | V (∼15%) | C (∼2.5%), N (∼5%) | |||
| Day 21 | 4.6E+06 | T (∼5%) | E (∼60%), R (∼5%) | V (∼15%) | C (∼5%), N (∼5%) | |||
| Day 28 | 1.5E+07 | T (∼5%) | E (∼70%), R (∼2.5%) | M (∼2.5%), V (∼12.5%) | C (∼2.5%), N (∼2.5%) | |||
| Day 42 | 8.8E+06 | T (∼5%) | E (∼70%), R (∼5%) | V (∼15%) | ||||
| Day 98 | 1.4E+07 | T (∼10%) | E (∼80%) | V (∼10%) | ||||
| J | 1a | T0 | 1.3E+06 | WT | WT | WT | WT | |
| Day 14 | 6.5E+04 | T (∼22.5%) | E (∼65%), K (∼5%) | V (∼5%) | ||||
| Day 15 | 1.4E+05 | T (∼10%) | E (∼55%), K (∼5%), R (∼5%) | V (∼15%) | ||||
| Day 21 | 6.3E+05 | T (∼15%) | E (∼72.5%), H (∼15%) | V (∼5%) | ||||
| Day 28 | 1.1E+06 | T (∼7.5%) | E (∼60%), R (∼5%) | V (∼10%) | ||||
| Day 42 | 1.9E+06 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |||
| Day 98 | 1.2E+06 | T (∼15%) | E (∼65%), H (∼2.5%), R (∼5%) | V (∼7.5%) | ||||
| K | 1a | T0 | 4.8E+06 | WT | WT | WT | WT | |
| Day 14 | 1.2E+05 | E (∼57.5%) | ||||||
| Day 15 | 1.1E+05 | T (∼7.5%) | E (∼50%), H (∼15%), K (∼17.5%), R (∼5%) | |||||
| Day 21 | 2.3E+06 | T (∼5%) | E (∼62.5%), H (∼7.5%), R (∼10%) | |||||
| Day 28 | 3.6E+06 | T (∼5%) | E (∼60%), K (∼5%), R (∼10%) | V (∼7.5%), I (∼5%) | ||||
| Day 42 | 8.7E+06 | T (∼10%) | E (∼57.5%), H (∼15%), K (∼2.5%), R (∼10%) | |||||
| Day 98 | 1.2E+07 | T (∼10%) | E (∼50%), H (∼17.5%), R (∼15%) | M (∼10%) | ||||
| L | 1a | T0 | 5.6E+06 | WT | WT | WT | WT | V37A (∼7.5%), V37 M (∼10%) |
| Day 7 | 2.6E+03 | T (∼30%) | E (∼15%), H (∼5%), R (∼12.5%) | V (∼5%) | C (∼5%) | V37A (∼20%) | ||
| Day 14 | 7.5E+03 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |||
| Day 15 | 1.3E+04 | T (∼15%) | E (∼40%), R (∼5%) | V (∼40%) | C (∼10%) | V37A (∼25%), V37 M (∼15%) | ||
| Day 28 | 6.7E+06 | T (∼10%) | E (∼25%), R (∼2.5%) | V (∼50%) | V37A (∼70%) | |||
| Day 42 | 1.1E+07 | T (∼15%) | E (∼20%), R (∼10%) | V (∼50%) | V37A (∼82.5%) | |||
| Day 98 | 1.0E+07 | T (∼30%) | E (∼10%), H (∼2.5%), R (∼5%) | V (∼55%) | V37A (∼90%) | |||
| Day 182 | 1.5E+07 | T (∼25%) | E (∼5%), H (∼2.5%), R (∼7.5%) | V (∼55%) | V37A (∼82.5%) | |||
Clonal analysis was performed on specimens derived from patient I for selected time points (T0 [BL], day 7, and day 14). Similar to the findings for patient A in the 1-mg cohort and patient F in the 10-mg cohort, no resistant variants were detected at BL by population sequencing, but variants were observed by clonal analysis (Table 8). At day 7, the dominant amino acid substitution was M28T (∼40%, or 38 of 94 clones), with a low level of resistance to DCV (3 ng/ml, or 4.1 nM) (2, 4, 9). However, at day 14, Q30E was dominant and was found in ∼65% of the clones.
Table 8.
Number of clones for specimens from patient I (GT-1a infected), treated QD with 30 mg of DCV
| Mutation(s) | No. of clones at the indicated time point |
||
|---|---|---|---|
| T0 | D7 | D14 | |
| None (WT) | 72 | 3 | 1 |
| M28T | 38 | 3 | |
| M28V | 3 | ||
| Q30D | 1 | ||
| Q30E | 15 | 62 | |
| Q30H | 7 | 2 | |
| Q30K | 1 | ||
| Q30R | 5 | ||
| L31M | 7 | ||
| L31V | 5 | 16 | |
| H58D | 1 | ||
| Y93C | 7 | 1 | |
| Y93N | 6 | ||
| M28T-Q30R | 2 | 2 | |
| M28T-Y93C | 1 | ||
| Q30E-Y93H | 2 | ||
| Q30H-Y93H | 1 | ||
| Q30R-L31M | 1 | ||
| Total clones | 75 | 94 | 96 |
Sixty-milligram cohort.
All patients in the 60-mg cohort were infected with GT-1a and experienced mean declines in HCV RNA of ∼3.6 log10 on day 4 (Fig. 1D). The M28V amino acid substitution, which does not confer resistance to DCV, was present in ∼40% of the BL population in patient M (Table 9). A Q30R substitution, which confers a low level of resistance to DCV, was also observed at BL in this patient. Although variants with Q30E were still dominant in 2 patients at day 14 and day 182 (patients N and O), other variants were dominant in the other GT-1a-infected patients. In patient P, the only amino acid substitution observed was Q30R, which conferred a relatively low level of resistance (EC50 = 5.4 ng/ml, or 7.3 nM) (2, 4, 9). The fact that Q30R variants were able to survive at the 60-mg treatment was most likely due to the linkage with E62D (11). When tested in vitro, E62D by itself did not confer resistance to DCV, but a replicon with Q30R in combination with E62D was highly resistant (EC50 = 113 ng/ml, or 153 nM [Table 1]) (11). For patient M, ≥90% of the variants had Q30H and Y93H substitutions from day 14 to day 42, suggesting the linkage of these substitutions (Table 9). A GT-1a Q30H-Y93H variant was highly resistant to DCV (EC50 = 409.8 ng/ml, or 553 nM) (2, 4, 9).
Table 9.
Variant persistence in HCV patients after treatment QD with 60 mg of DCV by population sequence
| Patient | Genotype | Time | RNA (IU/ml) | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M28 | Q30 | L31 | Y93 | |||||
| M | 1a | T0 | 1.7E+07 | V (∼40%) | R (∼10%) | |||
| Day 14 | 4.9E+06 | A (∼5%), T (∼5%), V (∼5%) | H (∼95%), R (∼5%) | H (∼95%) | ||||
| Day 15 | 7.1E+06 | A (∼5%), T (∼5%), V (∼5%) | H (∼95%), R (∼5%) | H (∼95%) | ||||
| Day 21 | 1.0E+07 | A (∼5%), T (∼5%), V (∼5%) | H (∼95%), R (∼5%) | H (∼95%) | ||||
| Day 28 | 1.3E+07 | A (∼5%), T (∼5%), V (∼5%) | H (∼95%), R (∼5%) | H (∼95%) | ||||
| Day 42 | 1.6E+07 | A (∼5%), T (∼5%), V (∼5%) | H (∼90%), R (∼10%) | H (∼90%) | ||||
| N | 1a | T0 | 1.8E+06 | V (∼2.5%) | ||||
| Day 14 | 3.9E+03 | A (∼7.5%), T (∼42.5%), V (∼10%) | E (∼45%), K (∼25%), R (∼15%) | N (∼10%) | ||||
| Day 15 | 5.8E+03 | T (∼20%) | E (∼45%), H (∼10%), R (∼2.5%) | M (∼5%) | N (∼40%) | |||
| Day 21 | 5.8E+05 | T (∼20%) | E (∼50%), H (∼5%), R (∼10%) | M (∼5%) | N (∼30%) | |||
| Day 28 | 6.0E+05 | T (∼20%) | E (∼60%), H (∼5%), R (∼20%) | M (∼2.5%) | N (∼15%) | |||
| Day 42 | 9.6E+05 | T (∼20%) | E (∼70%), H (∼5%), R (∼20%) | M (∼5%) | ||||
| Day 98 | 1.6E+06 | T (∼45%) | E (∼50%), R (∼40%) | M (∼5%) | ||||
| Day 182 | 1.8E+06 | T (∼40%) | E (∼60%), R (∼40%) | |||||
| O | 1a | T0 | 8.9E+06 | WT | WT | WT | WT | |
| Day 14 | 2.2E+06 | T (∼ 5%) | E (∼62.5%), R (∼5%) | N (∼5%) | ||||
| Day 15 | 2.1E+06 | T (∼ 5%) | E (∼75%), R (∼10%) | M (∼5%) | N (∼10%) | |||
| Day 21 | 4.0E+06 | T (∼ 5%) | E (∼67.5%), R (∼10%) | M (∼5%) | N (∼5%) | |||
| Day 28 | 5.1E+06 | T (∼ 5%) | E (∼70%), R (∼10%) | M (∼5%) | ||||
| Day 42 | 5.9E+06 | T (∼5%) | E (∼70%), R (∼10%) | M (∼5%) | ||||
| Day 98 | 1.3E+07 | T (∼10%) | E (∼60%), R (∼15%) | M (∼10%) | ||||
| Day 182 | 3.4E+06 | T (∼15%) | E (∼50%), R (∼20%) | M (∼15%) | ||||
| P | 1a | T0 | 2.4E+07 | WT | WT | WT | WT | E62D (∼100%) |
| Day 14 | 4.4E+06 | R (∼100%) | E62D (∼100%) | |||||
| Day 15 | 7.9E+06 | R (∼100%) | E62D (∼100%) | |||||
| Day 21 | 2.3E+07 | R (∼100%) | E62D (∼100%) | |||||
| Day 28 | 3.5E+07 | R (∼100%) | E62D (∼100%) | |||||
| Day 42 | 3.6E+07 | R (∼100%) | E62D (∼100%) | |||||
Clonal analysis was performed on specimens derived from T0 (BL), day 14, day 15, day 21, and day 182 from patient N. At BL, ∼90% of the HCV population was WT with respect to DCV sensitivity, but at the end of treatment (day 14) and day 182 (∼6 months after treatment), ∼95 to 99% were of the virus population harbored resistance-associated amino acid substitutions (Table 10). In addition to the Q30E amino acid substitution, another amino acid substitution, Y93N, that is associated with high-level DCV resistance (EC50 = 208.9 ng/ml, or 282 nM [Table 1]) (2, 4, 9) was apparent. A distinct difference from previous cohorts was also observed: the percentage of variants with double and triple linked amino acid substitutions was dramatically increased, suggesting the emergence of high-level resistance under higher selective pressure (Table 10). For example, linked M28T-Q30R amino acid substitutions, which confer a high level of DCV resistance in vitro (EC50 = 264 ng/ml, or 356 nM) (9), were also commonly observed in viral variants from day 14 through day 182. As with previous patients, the percentages of variants with linked amino acid substitutions recovered from patient N were similar from day 14 through day 182 (∼43% versus ∼45%).
Table 10.
Number of clones for specimens from patient N (GT-1a infected), treated QD with 60 mg of DCV
| Mutation(s) | No. of clones at the indicated time point |
|||||
|---|---|---|---|---|---|---|
| T0 | D14-1a | D14-2a | D15 | D21 | D182 | |
| None (WT) | 46 | 4 | 7 | 6 | 1 | |
| M28G | 2 | 5 | 2 | |||
| M28V | 4 | |||||
| Q30E | 36 | 26 | 57 | 51 | 59 | |
| Q30G | 1 | |||||
| Q30H | 4 | 6 | 2 | |||
| Q30R | 1 | 1 | ||||
| L31R | 1 | |||||
| L31V | 1 | |||||
| Y93N | 21 | 29 | 29 | 17 | ||
| M28A-Q30R | 14 | 2 | ||||
| M28T-Q30H | 27 | 16 | 11 | |||
| M28T-Q30K | 27 | |||||
| M28T-Q30R | 1 | 16 | 3 | 11 | 33 | |
| M28T-L31M | 6 | 6 | 8 | |||
| M28V-Q30E | 1 | |||||
| M28V-Y93N | 1 | 1 | ||||
| Q30E-Y93C | 1 | |||||
| Q30E-Y93N | 4 | 1 | 2 | |||
| Q30E-Y93T | 1 | |||||
| Q30H-Y93H | 1 | |||||
| Q30R-L31M | 4 | 9 | 9 | |||
| L31P-Y93N | 1 | |||||
| H58R-Y93N | 1 | |||||
| H58T-Y93T | 1 | 1 | ||||
| M28A-Q30R-Y93N | 3 | |||||
| M28T-Q30H-Y93N | 3 | 1 | ||||
| M28T-Q30H-Y93T | 1 | |||||
| M28T-Q30R-Y93N | 3 | |||||
| M28T-L31M-Y93N | 1 | 1 | ||||
| Q30E-H58P-Y93L | 1 | |||||
| Q30E-H58T-Y93T | 1 | 1 | ||||
| Q30R-L31M-Y93N | 1 | 1 | ||||
| M28T-Q30H-H58P-Y93L | 1 | |||||
| M28T-L31M-H58T-Y93T | 1 | |||||
| M28T-Q30R-L31T-H58P-Y93L | 1 | |||||
| Total clones | 51 | 64 | 93 | 150 | 151 | 151 |
Two different primer sets (-1 and -2) were used for the same specimen derived from day 14.
One-hundred-milligram cohort.
Patients infected with GT-1a and -1b experienced mean declines in HCV RNA of ∼3.3 log10 and ∼ 4.5 log10, respectively, on day 4 (Fig. 1E). Population sequencing revealed no resistant variants at BL for 3 patients infected with GT-1a (Table 11). Although multiple amino acid substitutions were observed during the 14-day monotherapy study, Q30E was the dominant substitution in two patients at day 182. In patient T, infected with GT-1b, 100% of the HCV at BL carried linked Q54H-Y93H amino acid substitutions, based on population sequencing. GT-1b Q54H did not confer resistance to DCV in vitro (2, 4), while Q54H-Y93H, similar to Y93H by itself, conferred a low level of resistance (2, 4). At the end of treatment (day 14) and during the entire 6-month follow-up period, all of the HCV detected had L31V-Q54H-Y93H linked amino acid substitutions. The combination of L31V-Q54H-Y93H amino acid substitutions conferred a moderate level of resistance in in vitro transient-replication assays (EC50 = 36.1 ng/ml, or ∼49 nM) (4).
Table 11.
Variant persistence in HCV patients after treatment QD with 100 mg of DCV by population sequence
| Patient | Genotype | Time | RNA (IU/ml) | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M/L28 | Q/R30 | L31 | Y93 | |||||
| Q | 1a | T0 | 1.1E+06 | WT | WT | WT | WT | |
| Day 14 | 3.6E+02 | T (∼50%) | E (∼50%), H (∼20%) | C (∼35%) | ||||
| Day 15 | 3.4E+02 | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | Failed to generate PCR products | |||
| Day 21b | 8.0E+02 | T (∼75%) | D (∼20%), H (∼80%) | |||||
| Day 28 | 3.5E+03 | E (∼50%) | N (∼50%) | |||||
| Day 42 | 4.1E+04 | T (∼25%) | D (∼10%), E (∼45%), H (∼10%) | C (∼10%) | ||||
| Day 98 | 4.4E+05 | T (∼20%) | D (∼7.5%). E (∼55%), H (∼5%) | |||||
| Day 182 | 5.1E+05 | T (∼5%) | E (∼80%) | |||||
| R | 1a | T0 | 3.5E+05 | WT | WT | WT | WT | |
| T12 | 1.4E+03 | H (∼60%) | H (∼25%) | |||||
| Day 14 | 4.2E+04 | T (∼50%) | E (∼10%), H (∼37.5%) | |||||
| Day 15 | 1.2E+05 | T (∼60%) | E (∼17.5%), H (∼50%), R (∼10%) | |||||
| Day 21 | 3.7E+05 | T (∼67.5%) | E (∼17.5%), H (∼70%), R (∼12.5%) | |||||
| Day 28 | 1.9E+05 | T (∼75%) | E (∼20%), H (∼65%), R (∼15%) | |||||
| Day 42 | 9.4E+04 | T (∼82.5%) | E (∼7.5%), H (∼82.5%), R (∼10%) | |||||
| Day 98 | 4.1E+05 | T (∼100%) | H (∼95%) | |||||
| Day 182 | 6.1E+05 | E (∼100%) | ||||||
| S | 1a | T0 | 1.0E+07 | WT | WT | WT | WT | |
| Day 14 | 2.4E+07 | R (∼100%) | H58D (∼100%) | |||||
| Day 15 | 7.6E+06 | T (<5%) | R (∼92.5%) | M (∼5%) | H58D (∼92.5%) | |||
| Day 21 | 1.3E+07 | T (<5%) | H (∼5%), R (∼85%) | M (∼5%) | H58D (∼85%) | |||
| Day 28 | 1.8E+07 | T (<5%) | H (∼2.5%), R (∼85%) | M (∼5%) | H58D (∼85%) | |||
| Day 42 | 1.1E+07 | T (∼5%) | R (∼92.5%) | M (<5%) | H58D (∼92.5%) | |||
| Day 92 | 2.0E+07 | R (∼95%) | M (∼5%) | H58D (∼95%) | ||||
| Day 177 | 1.5E+07 | T (∼2.5%) | R (∼95%) | M (∼5%) | H58D (∼92.5%) | |||
| T | 1b | T0 | 2.0E+07 | H (∼100%) | Q54H (∼100%) | |||
| Day 14 | 8.4E+05 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 15 | 4.6E+06 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 21 | 5.7E+06 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 28 | 1.1E+07 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 45 | 1.1E+07 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 98 | 2.0E+07 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
| Day 182 | 1.4E+07 | V (∼100%) | H (∼100%) | Q54H (∼100%) | ||||
Clonal analysis was performed for patient R specimens derived from T12 and day 14 (Table 12). Resistant variants comprised ∼87% of the HCV population (13 of 15 clones) at T12, suggesting that WT (DCV-sensitive) HCV at BL was rapidly eliminated. The amino acid substitutions detected at T12 (Q30H and Y93C) conferred relatively low levels of resistance (2, 4, 9). At day 14, all clones had at least one known resistance substitution, the majority of which conferred high levels of resistance, including Q30E, Q30K, and linked substitutions. M28T-Q30H were the most commonly observed linked substitutions (8 of 21 clones), and overall, about one-half (11 of 21 clones) of the clones had linked amino acid substitutions (Table 12). Replicons with linked M28T-Q30H substitutions were highly resistant to DCV (EC50 = 461 ng/ml, or 622 nM) (2). Since the plasma exposure of BMS-790052 in patient R ranged from 115 to 305 ng/ml (155 to 412 nM) at T24, day 4 (T72), day 7, and day 14 (3), the resistance conferred by M28T-Q30H likely explains the viral breakthrough during the treatment.
Table 12.
Number of clones for specimens from patient R (GT-1a infected), treated QD with 100 mg of DCV
| Mutation(s) | No. of clones at the indicated time point |
|
|---|---|---|
| T12 | D14 | |
| None (WT) | 2 | |
| Q30E | 5 | |
| Q30H | 12 | 2 |
| Q30K | 2 | |
| Q30R | 1 | |
| Y93C | 1 | |
| M28T-Q30H | 8 | |
| M28T-Q30R | 2 | |
| L31V-Y93F | 1 | |
| Total clones | 15 | 21 |
Thirty-milligrams-BID cohort.
Patients infected with GT-1a and -1b experienced a mean decline in HCV RNA of ∼1.8 log10 and ∼ 4.1 log10, respectively, on day 4 (Fig. 1F). No resistant variants were detected at BL for 1 of 2 patients infected with GT-1a (Table 13). Patient U had a complex resistance pattern, containing GT-1a substitutions at all major residues on day 182 (M28, Q30, L31, and Y93). In addition, the percentage of the low-level-resistance substitution H58D (EC50 = 2.2 ng/ml, or 3 nM) (4, 9) increased from ∼7.5% on day 14 to ∼40% on day 182. GT-1a-infected patient V had variants Q30H and Q30H-Y93H at BL. This patient experienced the lowest viral RNA decline in this cohort (Fig. 1F).
Table 13.
Variant persistence in HCV patients after treatment BID with 30 mg of DCV by population sequence
| Patient | Genotype | Time | RNA IU/ml | Substitution(s) (% frequency)a |
Additional mutation (% frequency) | |||
|---|---|---|---|---|---|---|---|---|
| M/L28 | Q/R30 | L31 | Y93 | |||||
| U | 1a | T0 | 2.0E+07 | WT | WT | WT | WT | |
| Day 14 | 1.8E+07 | T (∼7.5%) | E (∼70%), R (∼10%) | H58D (∼7.5%) | ||||
| Day 15 | 1.5E+07 | T (∼4%) | E (∼45%), H (∼5%), R (∼10%) | M (∼17.5%) | H58D (∼10%) | |||
| Day 21 | 1.8E+07 | T (∼2.5%) | E (∼80%), R (∼2.5%) | M (∼10%) | H58D (∼2.5%) | |||
| Day 28 | 2.5E+07 | T (∼12.5%) | E (∼45%), R (∼10%) | M (∼7.5%) | H58D (∼5%) | |||
| Day 42 | 3.2E+07 | T (∼10%) | E (∼62.5%), R (∼5%) | M (∼7.5%) | H58D (∼12.5%) | |||
| Day 116b | 2.3E+07 | T (∼5%) | E (∼25%), R (∼25%) | M (∼15%) | H (∼5%) | H58D (∼30%) | ||
| Day 182 | 2.7E+07 | T (∼7.5%), V (∼12.5%) | E (∼10%), H (∼10%), R (∼37.5%) | M (∼10%) | H (∼10%) | H58D (∼40%) | ||
| V | 1a | T0 | 4.3E+06 | H (∼100%) | H (∼45%) | |||
| Day 14 | 7.7E+04 | H (∼100%) | H (∼60%) | |||||
| Day 15 | 1.5E+05 | H (∼100%) | H (∼55%) | |||||
| Day 21 | 3.5E+05 | H (∼100%) | H (∼15%) | |||||
| Day 28 | 7.0E+05 | H (∼100%) | H (∼10%) | |||||
| Day 42 | 7.2E+06 | H (∼100%) | H (∼10%) | |||||
| Day 98 | 8.8E+06 | H (∼100%) | H (∼35%) | |||||
| Day 182 | 6.9E+06 | H (∼100%) | H (∼32.5%) | |||||
| W | 1b | T0 | 7.5E+06 | WT | WT | WT | WT | |
| Day 4 | 1.6E+03 | WT | WT | WT | WT | |||
| Day 14 | 5.0E+01 | ND | ND | ND | ND | |||
| Day 15 | 3.8E+01 | ND | ND | ND | ND | |||
| Day 21 | 6.2E+01 | ND | ND | ND | ND | |||
| Day 28 | 5.1E+02 | ND | ND | ND | ND | |||
| Day 42 | 1.1E+04 | I (∼27.5%), V (∼12.5%) | H (∼45%) | P32del (∼55%) | ||||
| Day 98 | 8.6E+06 | I (∼85%), V (∼10%) | H (∼95%) | F37L (∼10%), P32del (∼15%) | ||||
| Day 182 | 5.9E+06 | I (∼62.5%), V (∼12.5%) | H (∼72.5%) | F37L (∼2.5%), P32del (∼25%) | ||||
| X | 1b | T0 | 6.6E+06 | WT | WT | WT | WT | |
| Day 14 | <25 | ND | ND | ND | ND | |||
| Day 15 | <25 | ND | ND | ND | ND | |||
| Day 21 | 2.1E+02 | ND | ND | ND | ND | |||
| Day 28 | 2.8E+03 | I (∼72.5%), V (∼32.5%) | H (∼100%) | |||||
| Day 42 | 1.1E+06 | M (∼45%), V (∼55%) | H (∼100%) | |||||
| Day 98 | 4.6E+06 | I (∼70%), V (∼30%) | H (∼100%) | |||||
| Day 182 | 2.5E+06 | I (∼100%) | H (∼100%) | |||||
Frequencies are quantitative estimates of variants based on chromatograms from population sequencing (estimated from the average of two PCR products). “WT” indicates consensus with the WT. ND, not determined (HCV RNA < 1,000 IU/ml). Data for T0 to day 14 have been reported previously (3, 4).
Data derived from one PCR product.
No resistant variants were detected at BL for 2 patients infected with GT-1b (Table 13), and viral RNA became ≤50 IU/ml at the end of treatment. In both patients, viral rebound occurred during the follow-up period, and we were able to recover NS5A cDNA on days 42 and 28, respectively. In patient W, viral variants with a deletion of residue 32 (P32del) were observed. P32del was a novel mutation which has not previously been identified for GT-1b either in in vitro replicon or in the clinic. Because a deletion rather than a substitution occurred, it would suggest a high selective pressure by DCV in this GT-1b-infected patient. In the GT-1b replicon, P32del conferred a very high level of resistance to DCV (EC50 > 741 ng/ml, or >1,000 nM [Table 2]).
Clonal analysis was performed for patient W specimens from T0 (BL), day 4, day 42, and day 182 (Table 14). Consistent with the results derived from population sequencing, >95% of virus at BL and day 4 were wild type with respect to known DCV resistance-associated amino acid substitution. However, at day 42, all rebounding viruses had resistance mutations, the most notable being an in-frame deletion of NS5A codon 32 (P32del) that was present in up to 49.5% of the virus population. Variants with P32del linked to other substitutions, such as P32del-Y93H and L31F-P32del, were also observed. L31I alone did not confer resistance to DCV; Y93H conferred a low level of resistance, and L31I-Y93H conferred low resistance to DCV (EC50 = 4.8 ng/ml, or ∼6.5 nM [Table 2]).
Cross-resistance to other directly acting antiviral agents (DAAs).
During the course of NS5A resistance studies, an NS3 protease inhibitor(s) and/or NS5B polymerase inhibitors were always used as controls for cross-resistance. As expected, these inhibitors were equally potent against WT and NS5A-resistant replicon variants (data not shown) (4, 9).
DISCUSSION
Phase I and II clinical studies with DCV have revealed a profile that is effective and generally well tolerated, with marked and rapid initial declines in HCV RNA levels combined. In a 14-day monotherapy study of DCV, the initial HCV RNA decline observed was faster than that observed with any other DAAs reported to date. For example, patient N (GT-1a infected), in the 60-mg-QD cohort, experienced an ∼2-log10 viral decline 4 h after the first dose (3, 4). This impressive initial impact on the virus suggests that DCV may provide significant advantages, including shorter treatment duration, when included in DAA combination therapies. Since the initial decline in HCV RNA is predicted to include DCV-sensitive variants that contain preexisting mutations conferring resistance to other DAAs, DCV may enhance the overall effectiveness of other DAAs. This effect should lead to higher SVR rates and/or shorten the duration of treatment necessary to achieve SVR. Recent clinical results with DCV plus ASV in patients infected with GT-1b and DCV plus GS-7977 in patients infected with GT-1, -2, and -3 demonstrate the effectiveness of DCV in oral-only combination therapies (5, 12).
Despite the robust initial anti-HCV effect of DCV monotherapy and the fact that HCV RNA became undetectable in several GT-1b patients, viral RNA was detectable in all GT-1a patients, and viral breakthrough (VBT) was observed. To characterize the resistance profile of DCV, specimens from HCV-infected subjects treated with DCV in the 14-day monotherapy study were examined by population sequencing and clonal analysis. Resistant variants present at BL, as well as during and after DCV treatment, were examined. Our analysis indicated that population sequencing generated a relatively accurate estimate of variants in the population. For example, the complete analysis of patient A (GT-1a infected, 1-mg cohort [Tables 3 and 4]) included the clonal analysis of amplicons generated from the day 14 specimen by 2 sets of primers (D14-1 from A2A2 and D14-2 from A3A3). Population sequencing (using the amplicon D14-2 derived from primer set A3A3) revealed Y93C to be present at ∼20% in the day 14 specimen; clonal analysis gave a value of 13% (Table 4). Population sequencing obtained from the same specimen with the A2A2 primer set did not detect any Y93C, consistent with the clonal analysis using the same amplicon, D14-1 (Table 4).
Genotypic analysis of specimens derived from GT-1b-infected patients indicated that at low doses, relapse or rebound HCV was WT and the population remained WT during the follow-up period (patient C, 1-mg cohort [Table 3]). At higher doses, most, if not all, relapse or rebound virus was resistant (patients W and X, 30-mg-BID cohort [Table 13]). This suggests that during the 14-day monotherapy, DCV completely eradicated WT virus. Impressive inhibition of low levels of resistant variants detected at BL further supports this hypothesis. For example, patient T (100-mg cohort) had GT-1b HCV RNA of 2.0E+07 IU/ml at BL with the Y93H variant present at ∼100%. An ∼4.5 log10 HCV RNA decline was observed for patient T at day 7 (2, 3). The rebound virus was ∼100% variant with linked substitutions (L31V-Y93H), and it displayed a much higher level of resistance than the variant with Y93H alone (2, 4, 9). Since the Y93H substitution was observed at ∼100% at BL, it is likely the L31V-Y93H variant was also present at a very low level at BL and was enriched or selected as the Y93H variant was eradicated.
A quite different picture emerged for patients infected with GT-1a, even though viral titers were similar for patients infected with GT-1b and -1a. GT-1a variants with low levels of resistance to DCV were initially suppressed, but the rate of rebound was faster and the frequency of VBT was higher. This is not surprising, since (i) DCV is less potent on GT-1a than GT-1b in vitro (EC50 = 50 versus 9 pM, respectively), (ii) GT-1a variants with a single amino acid substitution show higher levels of resistance to DCV than GT-1b variants, and (iii) although both GT-1a and -1b variants with linked substitutions generally display high levels of resistance, such variants were more frequently detected in the specimens derived from patients infected with GT-1a during and after DCV treatment (1, 2, 4, 9). Almost all HCV variants detected in GT-1a-infected patients in all cohorts at the end of the 14-day monotherapy were resistant (Tables 3 to 13) (4).
Most variants with substitutions at residues 28, 30, and 31 were relatively stable during the follow-up period (up to 6 months), while variants with substitutions at residue 93 were much less stable, unless the residue 93 substitution was linked with a substitution(s) at other residues. This interpretation is consistent with the observations shown in Table 1 and data previously reported (2, 4, 9). Y93C/H/N variants had low replication capabilities (11 to 18% of WT level) compared to other variants with substitutions at residues 28, 30, and 31 (2, 4, 9). This would suggest that after 14 days of DCV monotherapy, the dominant species continued to be selected based on fitness and not resistance to DCV. One distinct difference in the GT-1a resistance patterns observed in vitro (2, 9) and in the clinic (4) is that the variants with linked substitutions are more complex in clinical specimens. Although variants that were >20% of the total population could be detected by population sequencing (4), additional linked substitutions were identified and confirmed by clonal analysis. The complexity of the substitution patterns became more apparent with the analysis of specimens from cohorts that received higher doses of DCV. One example is the analysis of specimens from patient N (60-mg cohort [Table 10]). It is not surprising that resistance patterns are more complex in treated patients than those selected in vitro in the replicon system, because HCV sequences derived from clinical specimens are more heterogeneous than replicons (13–15). It is logical that variants with linked substitutions have higher levels of resistance to DCV, but how do they persist with lower replication abilities, as shown in Table 1 and by data previously reported (2, 4, 9)? One likely explanation is that the persistent variants were fit in vivo, due to a selected compensatory mutation(s). No attempt was made to identify compensatory mutations, since our primary objective was to determine the cause of VBT during DCV monotherapy. Although persistence of NS5A variants was detected up to 6 months, more data for longer follow-up are necessary to determine the true duration or persistence of NS5A resistance.
The most common GT-1 resistance amino acid substitution observed in this 14-day DCV monotherapy study was Q30E. Likewise, Q30E was a predominant mutation identified in in vitro studies with GT-1a replicons (9). A replicon variant with this amino acid substitution displayed a relatively high level of resistance to DCV (EC50 = 110.9 ng/ml, or 150 nM [Table 1]) and replicated better than the wild-type replicon (2, 4, 9). Moreover, viral variants with Q30E substitutions persisted for at least 6 months following 14 days of DCV monotherapy (Tables 7 to 13). Despite these findings, a Q30E amino acid substitution was not detected in 1,497 GT-1a sequences in the European HCV database (16), suggesting that the Q30E variant should not be of particular concern when DCV is used in DAA combination therapies. As expected, all of the DCV-resistant replicon variants that we have examined, including a GT-1a Q30E variant, remained fully sensitive to inhibitors targeting NS3 protease and NS5B polymerase (4, 9). These results suggest that any preexisting DCV resistant viral variants would be suppressed by other DAAs in combination therapy.
Overall, through the 14-day DCV monotherapy, most wild-type virus was likely eradicated. After the end of DCV treatment, viral fitness, rather than DCV resistance, probably determines which viral variants emerge as dominant species to persist. This information may provide guidance for retreatment of patients who experience viral breakthrough or relapse through DCV-containing regimens.
ACKNOWLEDGMENTS
We thank Mark Cockett, Nick Meanwell, and Makonen Belema for valuable discussions and critical reading of the manuscript. We thank Xin Huang, Bernadette Kienzle, and Charles Tilford for assistance of DNA sequencing analysis.
This study was supported by Bristol-Myers Squibb (BMS).
The authors all are currently BMS employees.
Footnotes
Published ahead of print 12 February 2013
REFERENCES
- 1. Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun O'Boyle J-HII DR, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical efficacy. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nettles RE, Gao M, Bifano M, Chung E, Persson A, Marbury T, Goldwater R, DeMicco MP, Rodriguez-Torres M, Vutikullird A, Fuentes E, Lawitz E, Lopez-Talavera JC, Grasela DM. 2011. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology 54:1956–1965 [DOI] [PubMed] [Google Scholar]
- 4. Fridell RA, Wang C, Sun JH, O'Boyle DR, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935 [DOI] [PubMed] [Google Scholar]
- 5. Chayama K, Takahashi S, Toyota J, Karino Y, Ikeda K, Ishikawa H, Watanabe H, McPhee F, Hughes E, Kumada H. 2012. Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology 55:742–748 [DOI] [PubMed] [Google Scholar]
- 6. Lok AS, Gardiner DF, Lawitz E, Martorell C, Everson GT, Ghalib R, Reindollar R, Rustgi V, McPhee F, Wind-Rotolo M, Persson A, Zhu K, Dimitrova DI, Eley T, Guo T, Grasela DM, Pasquinelli C. 2012. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366:216–224 [DOI] [PubMed] [Google Scholar]
- 7. Pol S, Ghalib RH, Rustgi VK, Martorell C, Everson GT, Tatum HA, Hezode C, Lim JK, Bronowicki Abrams J-PGA, Brau N, Morris DW, Thuluvath PJ, Reindollar RW, Yin PD, Diva U, Hindes R, McPhee F, Hernandez D, Wind-Rotolo M, Hughes EA, Schnittman S. 2012. Daclatasvir for previously untreated chronic hepatitis C genotype-1 infection: a randomised, parallel-group, double-blind, placebo-controlled, dose-finding, phase 2a trial. Lancet Infect. Dis. 12:671–677 [DOI] [PubMed] [Google Scholar]
- 8. Lok A, Gardiner D, Lawitz E, Martorell C, Everson G, Ghalib R, Reindollar R, Rustgi V, McPhee F, Wind-Rotolo M, Persson A, Zhu K, Dimitrova DI, Eley T, Guo T, Grasela DM, Pasquinelli C. 2011. Quadruple therapy with BMS-790052, BMS-650032 and Peg-IFN/RBV for 24 weeks results in 100% SVR12 in HCV genotype 1 null responders. J. Hepatol. 54(Suppl 1):S536 (Abstract.) [Google Scholar]
- 9. Wang C, Huang H, Valera L, Sun JH, O'Boyle DR, II, Nower PT, Jia L, Qiu D, Huang X, Altaf A, Gao M, Fridell RA. 2012. Hepatitis C virus RNA elimination and development of resistance in replicon cells treated with BMS-790052. Antimicrob. Agents Chemother. 56:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lemm JA, O'Boyle DR, II, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. 2010. Identification of NS5A inhibitors. J. Virol. 84:482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun JH, O'Boyle DR, II, Zhang Y, Wang C, Nower P, Valera L, Roberts S, Nettles RE, Fridell RA, Gao M. 2012. Impact of a baseline polymorphism on the emergence of resistance to the hepatitis C virus nonstructural protein 5a replication complex inhibitor, BMS-790052. Hepatology 55:1692–1699 [DOI] [PubMed] [Google Scholar]
- 12. Sulkowski M, Gardiner D, Lawitz E, Hinestrosa F, Nelson D, Thuluvath P, Rodriguez-Torres M, Lok A, Schwartz H, Reddy KR, Eley T, Wind-Rotolo M, Huang Gao S-PM, McPhee F, Hindes R, Symonds B, Pasquinelli C, Grasela DM. 2012. Potent viral suppression with all-oral combination of daclatasvir (NS5A inhibitor) and GS-7977 (NS5B inhibitor), +/− ribavirin, in treatment-naïve patients with chronic HCV Gt1, 2 or 3. J. Hepatol. 56(Suppl 2):S560 (Abstract.) [Google Scholar]
- 13. Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188 [DOI] [PubMed] [Google Scholar]
- 14. Guedj J, Rong L, Dahari H, Perelson AS. 2010. A perspective on modelling hepatitis C virus infection. J. Viral Hepat. 17:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 83:5760–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Mercier PL, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky J-M, Rice CM, Trepo C, Penin F, Deleage G. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 35:D363–D366 doi:10.1093/nar/gkl970 [DOI] [PMC free article] [PubMed] [Google Scholar]