Abstract
We characterized carbapenem resistance mechanisms among 12 Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae (referred to here as KPC K. pneumoniae) clinical isolates and evaluated their effects on the activity of 2- and 3-drug combinations of colistin, doripenem, and ertapenem. All isolates were resistant to ertapenem and doripenem; 75% (9/12) were resistant to colistin. Isolates belonged to the ST258 clonal group and harbored blaKPC-2, blaSHV-12, and blaTEM-1. As determined by time-kill assays, doripenem (8 μg/ml) and ertapenem (2 μg/ml) were inactive against 92% (11/12) and 100% (12/12) of isolates, respectively. Colistin (2.5 μg/ml) exerted some activity (range, 0.39 to 2.5 log10) against 78% (7/9) of colistin-resistant isolates. Colistin-ertapenem, colistin-doripenem, and colistin-doripenem-ertapenem exhibited synergy against 42% (5/12), 50% (6/12), and 67% (8/12) of isolates, respectively. Expression of ompK35 and ompK36 porins correlated with each other (R2 = 0.80). Levels of porin expression did not correlate with colistin-doripenem or colistin-ertapenem synergy. However, synergy with colistin-doripenem-ertapenem was more likely against isolates with high porin expression than those with low expression (100% [8/8] versus 0% [0/4]; P = 0.002). Moreover, bactericidal activity (area under the bacterial killing curve) against isolates with high porin expression was greater for colistin-doripenem-ertapenem than colistin-doripenem or colistin-ertapenem (P ≤ 0.049). In conclusion, colistin-carbapenem combinations may provide optimal activity against KPC K. pneumoniae, including colistin-resistant isolates. Screening for porin expression may identify isolates that are most likely to respond to a triple combination of colistin-doripenem-ertapenem. In the future, molecular characterization of KPC K. pneumoniae isolates may be a practical tool for identifying effective combination regimens.
INTRODUCTION
Infections with Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae (referred to here as KPC K. pneumoniae) are widely encountered in the United States and are increasingly being reported worldwide (1–9). Mortality rates due to KPC K. pneumoniae infections are as high as 50% (1, 10–13), and optimal therapy is not well defined (14). Previous observational studies have shown that combinations of two or three antimicrobial agents may achieve better clinical outcomes than monotherapy (12, 14, 15), especially when the combination includes a carbapenem (16). In keeping with this clinical experience, we and others have demonstrated that the combination of colistin and doripenem is bactericidal and synergistic against KPC K. pneumoniae isolates in vitro (17–19). Doripenem was chosen to represent the carbapenems because it is more stable against hydrolysis by KPC than other agents in the same class (20). Despite these data, a subset of patients with KPC K. pneumoniae bacteremia does not respond to this combination clinically (21). The treatment of KPC K. pneumoniae infections may be further complicated by the presence of extended-spectrum β-lactamases (ESBLs) and porin mutations that impact carbapenem responses (3, 8, 9, 22), as well as the emergence of colistin resistance (18, 23, 24).
In a recent study, a double carbapenem regimen of doripenem-ertapenem rapidly reduced bacterial counts of a KPC K. pneumoniae isolate in vitro and in vivo (25), even though the isolate was resistant to both agents. In light of these findings and our previous doripenem-colistin data, we investigated the novel combination of colistin-doripenem-ertapenem against 12 colistin-susceptible and -resistant KPC K. pneumoniae isolates in vitro. At the same time, we determined whether the efficacy of colistin-carbapenem combinations was impacted by the presence of specific ESBL genes or levels of porin expression by the isolates.
(Preliminary data were presented at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 9 to 12 September 2012.)
MATERIALS AND METHODS
Isolates and phenotypic and genotypic characterization.
Twelve KPC K. pneumoniae clinical isolates obtained from unique patients at the University of Pittsburgh Medical Center between March 2010 and June 2011 underwent testing. The isolates were stored at −80°C and passaged at least twice at 37°C before experimentation. Stock solutions of antimicrobial agents were prepared in our laboratory in sterile water, aliquoted, and stored at −70°C. MICs were determined by standard broth microdilution and disk diffusion methods (26). Breakpoints for resistance were defined according to the Clinical and Laboratory Standards Institute (27). All isolates underwent multilocus sequence typing (MLST), and full-length blaKPC genes were characterized by PCR and DNA sequencing (28–32).
Time-kill studies.
Time-kill studies were performed using a final volume of 25 ml of cation-adjusted Mueller-Hinton broth and an initial inoculum of ∼1 × 106 CFU/ml (33). The antimicrobial agent concentrations were chosen based on achievable serum levels from published pharmacokinetic data (34, 35): colistin, 2.5 μg/ml; doripenem, 8 μg/ml; and ertapenem, 2 μg/ml. As detailed in Results, colistin-susceptible isolates were also tested against colistin (1 μg/ml). Flasks were incubated with shaking at 37°C, and bacterial colonies were enumerated at 0, 4, 8, 12, and 24 h. The degree of killing was defined as the difference between the log10 combination of the starting inoculum and the log10 concentration of KPC K. pneumoniae after 24 h of incubation with the agent(s). Bactericidal activity was defined as ≥3 log10 killing. Synergy was defined as a difference in the log10 CFU/ml of isolates incubated with colistin and the log10 CFU/ml of isolates incubated with combination agents of ≥2 at 24 h. Antagonism was defined as ≥1-log10-greater growth of isolates in response to combination agents than colistin alone. The area under the bacterial killing curve (AUBC) for time-kill graphs was determined using GraphPad Prism 6 software (GraphPad, San Diego, CA).
Analysis of porin expression and levels.
DNase-treated RNA from individual isolates was obtained from late-exponential-phase cultures using a RiboPure bacterial kit (Ambion). cDNA was made using qScript cDNA mix (Quanta Biosciences). Quantitative reverse transcription PCR (qRT-PCR) for porin genes ompK35 and ompK36 was then performed in a TaqMan gene expression assay (ABI) using the one-step SYBR green kit (SYBR green FastMix ROX; Quanta Biosciences). The primers used in these experiments were 5′GCAATATTCTGGCAGTGGTGATC3′ for the ompK35 forward primer, 5′ACCATTTTTCCATAGAAGTCCAGT3′ for the ompK35 reverse primer, 5′TTAAAGTACTGTCCCTCCTGG3′ for the ompK36 forward primer, and 5′TCAGAGAAGTAGTGCAGACCGTCA3′ for the ompK36 reverse primer. Amplification was done in a 20-μl final volume using the following protocol: denaturation at 95°C for 10 min, followed by 40 cycles consisting of 15 s at 95°C for denaturation, 11 s at 54°C for annealing, and 22 s at 72°C for elongation. The expression levels were expressed in cycle threshold (CT) units, which is the first cycle at which the signal is detected above a preset threshold. Relative quantities of mRNA from each gene of interest were determined by the comparative threshold CT. Expression of each gene was normalized to that of the housekeeping gene rpoB; the primers used were 5′AAGGCGAATCCAGCTTGTTCAGC3′ for the rpoB forward primer and 5′TGACGTTGCATGTTCGCACCCATCA3′ for the rpoB reverse primer. Data were expressed as the relative expression of ompK35 and ompK36 compared to that in a control isolate that is known to express both porins (K. pneumoniae ATCC 13883) (36). Each experiment was performed in triplicate, and the data are presented as mean CT values from three experiments. The presence of OmpK36 porin was assessed by SDS-PAGE as previously described (37).
Statistical analysis.
Comparisons between two groups of antimicrobial agents were made by Fisher's exact test for categorical variables and Mann-Whitney test or unpaired t test for continuous variables. Correlations between pairs of variables were assessed using the Spearman rank test. Receiver operating characteristic (ROC) analysis was conducted to determine optimal cutoffs for porin expression to predict synergy among 2- or 3-drug combinations. Significance was defined as a P value of ≤0.05 (two-tailed).
RESULTS
Phenotypic and genotypic characterization of KPC K. pneumoniae.
All isolates were resistant in vitro to amikacin, aztreonam, ciprofloxacin, piperacillin-tazobactam, and carbapenems. Specifically, the median MIC of doripenem was 128 μg/ml (range, 32 to 512 μg/ml), and that of ertapenem was >256 μg/ml (range, 128 to >256 μg/ml) (Table 1). Ninety-two percent (11/12), 17% (2/12), and 58% (7/12) of isolates were resistant to cefepime, doxycycline, and gentamicin, respectively. Seventy-five percent (9/12) of isolates were resistant to colistin, with MICs ranging from 4 to 64 μg/ml; the MICs against the 3 susceptible isolates were 2 μg/ml.
Table 1.
Bactericidal activity of single drugs and 2- and 3-drug combinations against KPC K. pneumoniae isolates at 24 h
| Isolate | MIC (μg/ml) |
Porin expressiona |
Killing activity (log10 CFU/ml)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colistin | Doripenem | Ertapenem | ompK36 | ompK35 | Colistin | Doripenem | Ertapenem | Colistin-ertapenem | Colistin-doripenem | Doripenem-ertapenem | Colistin-doripenem-ertapenem | |
| 44 | 64 | 32 | 256 | 0.59 | 0.17 | 1.36 | −3.64 | −3.61 | 0.02 | 2.75 | −3.84 | 2.82 |
| 94 | 4 | 256 | >256 | 0.1 | 0.25 | 2.29 | −3.77 | −3.72 | 0.62 | 3.96 | −3.68 | 3.82 |
| 121 | 4 | 256 | >256 | 0.13 | 0.37 | 1.46 | −3.84 | −3.67 | 4.23 | 3.24 | −3.73 | 3.1 |
| 404 | 2 | 64 | >256 | 0.04 | 0.3 | 1.42 | −3.57 | −3.72 | 0.6 | 3.99 | −3.67 | 3.49 |
| 25 | 16 | 32 | 128 | 2.35 | 0.57 | −1.85 | 3.17 | −3.78 | 2.76 | 3.48 | 2.65 | 4.2 |
| 83 | 32 | 64 | >256 | 5.3 | 1.1 | 2.5 | −3.48 | −3.59 | 2.06 | 4.27 | −2.81 | 4.65 |
| 89 | 64 | 128 | >256 | 4.12 | 1.06 | 2.29 | −3.19 | −3.41 | 0.58 | 4.39 | −3.32 | 4.38 |
| 155 | 2 | 512 | >256 | 4.23 | 1.1 | −0.05 | −3.9 | −4.13 | 2.88 | 2.68 | −3.76 | 1.97 |
| 168 | 32 | 128 | >256 | 3.02 | 0.52 | −3.8 | −3.13 | −3.72 | 0.72 | 0.77 | −3.29 | 4.45 |
| 175 | 2 | 64 | >256 | 1 | 0.47 | −0.05 | −3.66 | −3.67 | 0.17 | 1.26 | −3.52 | 4.56 |
| 178 | 16 | 256 | >256 | 1.23 | 0.78 | 1.11 | −3.79 | −3.65 | 0.99 | 1.23 | −3.69 | 3.27 |
| 220 | 16 | 256 | >256 | 1.69 | 0.57 | 0.39 | −3.27 | −3.43 | 3.17 | 2.62 | −3.29 | 3.23 |
Relative porin expression normalized to rpoB gene expression and compared to K. pneumoniae ATCC 13883.
Degree of killing was defined as the difference between the log10 of starting inoculum and the log10 concentration of KPC K. pneumoniae after 24 h of incubation with drug(s). Negative numbers denote growth of isolates compared with starting inoculum. Positive numbers denote suppression of growth of isolates compared with starting inoculum. Bactericidal activity was defined as degrees of killing of ≥3-log10 CFU/ml. Colistin was tested at concentrations of 2.5 and 1 μg/ml against colistin-resistant (MIC > 2 μg/ml) and -susceptible (MIC = 2 μg/ml) isolates, respectively. Values that reflect bactericidal activity are in bold.
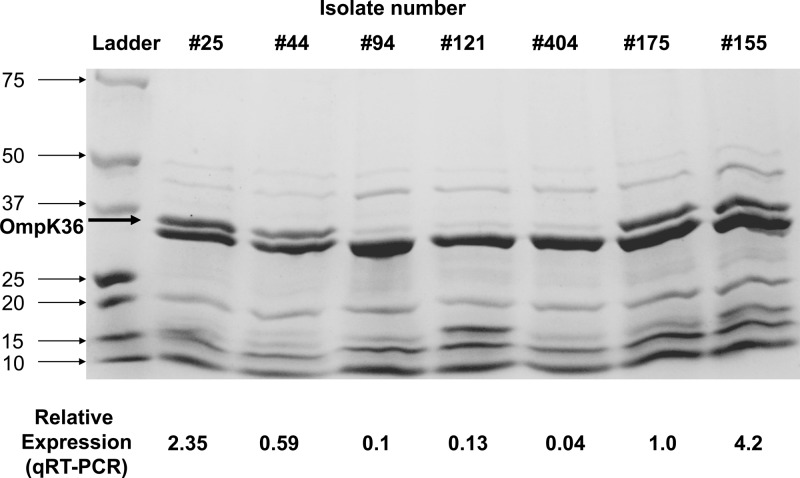
MLST showed that all isolates belonged to the epidemic K. pneumoniae ST258 clone. Genotyping β-lactamase resistance by both PCR and DNA sequencing revealed that all isolates harbored blaKPC-2, blaSHV-12, and blaTEM-1 but were negative for blaCTX-M, blaIMP, blaNDM, blaVIM, blaOXA-48, and blaAmpCs (blaACT-1, blaACC, blaBIL-1, blaCMY, blaDHA, blaFOX, blaLAT, blaMIR-1, and blaMOX) (28–32). Median ompK35 and ompK36 expression relative to the control K. pneumoniae strain ATCC 13883 was 0.55 (range, 0.17 to 1.1) and 1.46 (range, 0.1 to 5.3), respectively (Table 1). Expression of ompK35 and that of ompK36 was directly correlated with each other (R2 = 0.80). The levels of OmpK36 detected in KPC K. pneumoniae isolates were variable and correlated with qRT-PCR findings (Fig. 1). There was no evident OmpK35 production, consistent with our qRT-PCR data and previous studies (38).
Fig 1.
Outer membrane protein analysis of representative KPC K. pneumoniae isolates by 12% SDS-PAGE. The isolate number and relative expression of ompK36 by real-time RT-PCR are provided for each lane. The lane marked “Ladder” contains size markers (sizes are in kDa).
Bactericidal activity.
As expected, ertapenem (2 μg/ml) had no activity against any of the isolates during time-kill studies (33) (Fig. 2). Doripenem (8 μg/ml) exerted no activity against 92% (11/12) of isolates; it was bactericidal against the remaining isolate (isolate 25) (Table 1). Colistin (2.5 μg/ml) exerted some activity against 78% (7/9) of colistin-resistant KPC K. pneumoniae isolates (range, 0.39 to 2.5 log10 killing) but did not reach bactericidal levels. Colistin had no activity against the remaining resistant isolates (22%; 2/9). Colistin (2.5 μg/ml) was fully bactericidal against the 3 colistin-susceptible isolates at 12 and 24 h. For this reason, the colistin concentration was reduced to 1 μg/ml (0.5× MIC) to assess the interaction with carbapenems in subsequent combination time-kill studies of colistin-susceptible isolates.
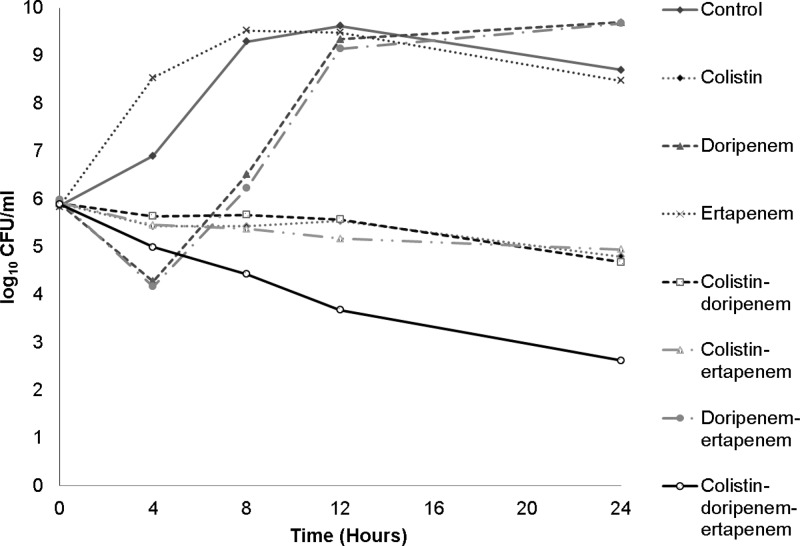
Fig 2.
Time-kill curve of single drugs and 2- and 3-drug combinations against a representative KPC K. pneumoniae isolate. Doripenem and colistin MICs against this isolate (isolate 178) were 256 μg/ml and 16 μg/ml, respectively. Times on the x axis are incubation times.
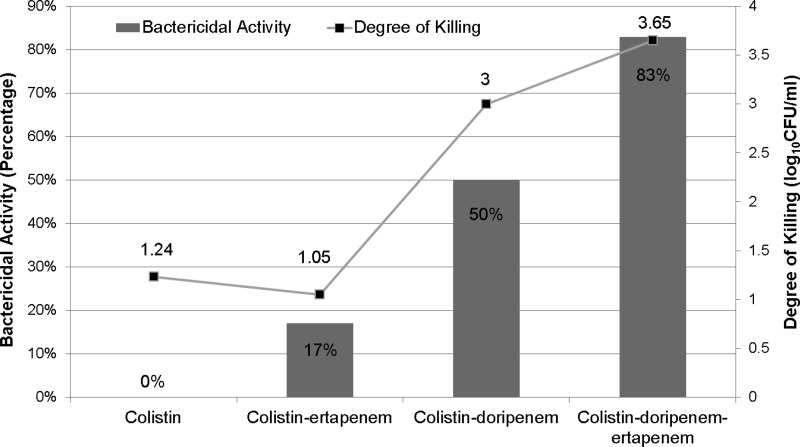
In combination time-kill experiments, colistin-ertapenem was bactericidal against only 17% (2/12) of isolates (versus colistin alone [0%]; P = 0.47) (Table 1; Fig. 3). Median killing of any isolate at 24 h did not improve when ertapenem was added to colistin (P = 0.31). The colistin-doripenem combination, however, was bactericidal against 50% (6/12) of isolates (P = 0.01 versus colistin alone). Median killing was increased from 1.24 log10 with colistin alone to 3.00 log10 with colistin-doripenem (P = 0.002).
Fig 3.
Bactericidal activity and degree of killing by single drugs and 2- and 3-drug combinations against 12 KPC K. pneumoniae isolates. The line graph represents the median degree of killing, and the bar graph represents the percentage of bactericidal activity of each drug regimen against the 12 KPC K. pneumoniae isolates. There was a stepwise improvement in bactericidal activity from colistin only and colistin-ertapenem to colistin-doripenem to colistin-doripenem-ertapenem. Bars (left y axis) show bactericidal activity, and the line (right y axis) shows the degree of killing by single drugs and 2- and 3-drug combinations.
The triple combination of colistin-doripenem-ertapenem was bactericidal against 83% (10/12) of isolates (P = 0.19 versus colistin-doripenem). Median killing also improved to 3.65-log10 with the triple combination compared to colistin-doripenem (P = 0.09).
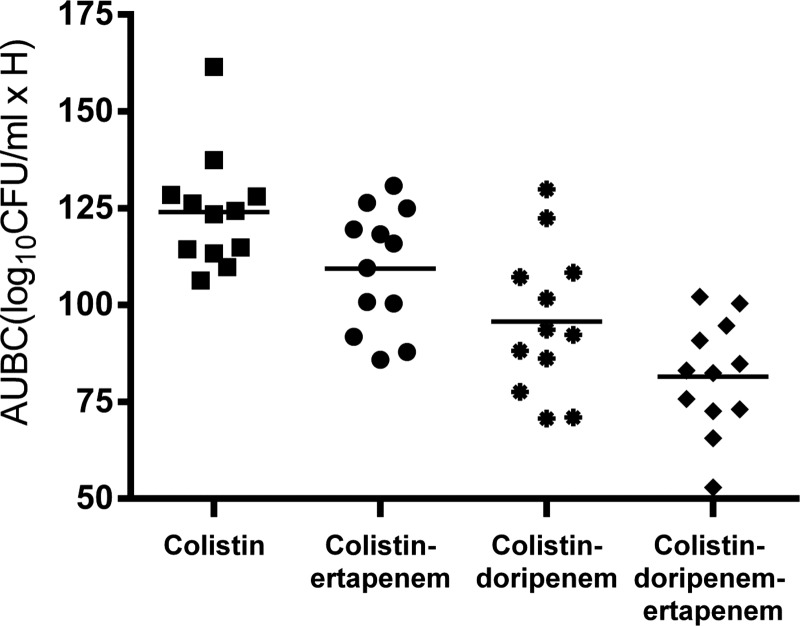
To further evaluate the effects of colistin and carbapenems in combination, we calculated the area under the bacterial killing curve (AUBC) for the time-kill graphs (Table 2). The median AUBCs for colistin-ertapenem and colistin-doripenem were significantly lower than those of colistin alone (P = 0.03 and P = 0.0005, respectively) (Fig. 4). The median AUBC of the triple combination was significantly lower than the AUBCs of colistin-ertapenem (P = 0.0002) and colistin-doripenem (P = 0.05).
Table 2.
Area under the bacterial killing curve (AUBC) of 2- or 3-drug combinations against 12 KPC K. pneumoniae isolates
| Drug(s) | Median AUBC (log10 CFU/ml · h) (interquartile range)a |
P value |
|
|---|---|---|---|
| Vs colistinb | Vs colistin-doripenemc | ||
| Colistin | 123.9 (114.1–128.1) | NA | 0.0005 |
| Colistin-ertapenem | 112.8 (98.3–120.9) | 0.03 | 0.07 |
| Colistin-doripenem | 93.0 (84.1–107.5) | 0.0005 | NA |
| Colistin-doripenem-ertapenem | 82.8 (73.0–91.8) | <0.0001 | 0.05 |
Calculated using GraphPad Prism 6 software. A more effective drug regimen yields a lower AUBC.
The AUBCs of colistin-ertapenem, colistin-doripenem, and colistin-doripenem-ertapenem were significantly lower than the AUBC of colistin alone.
The AUBC of colistin-doripenem-ertapenem was significantly lower than the AUBC of colistin-doripenem. NA, not applicable.
Fig 4.
Area under the bacterial killing curves (AUBC) of single drugs and 2- and 3-drug combinations against 12 KPC K. pneumoniae isolates. The median AUBC was significantly lower for colistin-doripenem-ertapenem than colistin with a carbapenem (colistin-ertapenem, P = 0.0002; colistin-doripenem, P = 0.05) and colistin only (P < 0.0001).
Synergistic activity.
Colistin-ertapenem, colistin-doripenem, and colistin-doripenem-ertapenem achieved synergy at 24 h against 42% (5/12), 50% (6/12), and 67% (8/12) of isolates, respectively. Antagonism was not observed with colistin-doripenem or colistin-doripenem-ertapenem but did occur with colistin-ertapenem against 25% (3/12) of isolates.
Associations between antimicrobial activity and porin expression by isolates.
There was no correlation between doripenem MIC and ompK35 or ompK36 expression (P = 0.29 and P = 0.97, respectively). There was also no correlation between porin expression and killing by doripenem (P = 0.78 and 0.98 for ompK35 and ompK36, respectively) or ertapenem (P = 0.19 and 0.40 for ompK35 and ompK36, respectively).
Using ROC, we identified optimal ompK35 and ompK36 expression cutoffs of 0.45 and 1.0, which identified isolates against which the triple combination of colistin-doripenem-ertapenem was synergistic. At these cutoffs, all isolates with high ompK35 expression had high ompK36 expression, and isolates with low ompK35 expression had low ompK36 expression. From this point forward, we refer to high ompK35/ompK36 expression as “high porin expression” and low ompK35/ompK36 expression as “low porin expression.” Synergy with colistin-doripenem-ertapenem was achieved against 100% (8/8) of isolates with high porin expression but none (0/4) of the isolates with low porin expression (P = 0.002). Synergy between colistin and doripenem was achieved against 63% (5/8) and 25% (1/4) of isolates with high and low porin expression, respectively (P = 0.5).
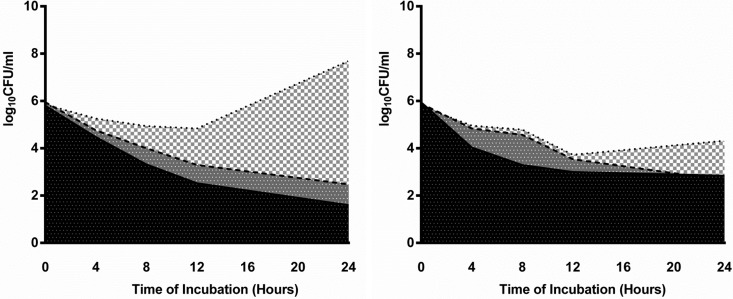
We also evaluated associations between porin expression and AUBCs (Fig. 5). For isolates with high porin expression, the triple combination had lower median AUBC than colistin-doripenem (74.45 versus 104.45; P = 0.049). However, for isolates with low porin expression, median AUBCs of the triple combination and colistin-doripenem were not significantly different (83.95 versus 89.25; P = 0.49). Colistin-doripenem had a lower median AUBC than colistin alone, regardless of the level of porin expression (P = 0.03 and 0.01).
Fig 5.
AUBCs of representative KPC K. pneumoniae isolates with high porin expression (isolate 25 [left]) and low porin expression (isolate 121 [right]). The dotted line, dashed line, and solid line represent time-kill curves of colistin, colistin-doripenem, and colistin-doripenem-ertapenem, respectively. Black, AUBC of colistin-doripenem-ertapenem; dark gray, difference in AUBC between colistin-doripenem and colistin-doripenem-ertapenem; light gray, difference in AUBC between colistin and colistin-doripenem.
DISCUSSION
There are three particularly notable findings from this study. First, the data corroborate previous observations by our group and others that the colistin-doripenem combination is more effective than colistin-ertapenem or any of the agents alone against KPC K. pneumoniae isolates, exhibiting greater killing and more often achieving bactericidal activity and synergy (17–19). In fact, the beneficial effects of colistin-doripenem were evident with isolates that were highly resistant to both agents, as determined by MICs. Second, the addition of ertapenem to colistin-doripenem further enhanced killing, bactericidal activity, and synergy, despite the fact that the colistin-ertapenem combination itself was no more effective than colistin alone. Third, the enhanced activity of colistin-doripenem-ertapenem was observed exclusively with KPC K. pneumoniae isolates with high levels of ompK35 or ompK36 expression. In contrast, the benefits of colistin-doripenem over colistin alone were independent of porin expression. Our findings are consistent with a recent clinical observation that treatment with colistin-carbapenem combinations led to favorable survival among patients with KPC K. pneumoniae bacteremia (15). Moreover, they suggest that the triple combination of colistin-doripenem-ertapenem may have a role in the treatment of at least some KPC K. pneumoniae infections, and screening for porin expression may identify isolates that are most susceptible to this combination. Indeed, the study offers a model by which molecular characterization of difficult-to-treat microbial isolates can be used to identify effective antimicrobial regimens.
To our knowledge, this is the first study to evaluate the activity of antimicrobial combinations against KPC K. pneumoniae in the context of molecular mechanisms of resistance. In this regard, our most important finding is that “one-size-fits-all” approaches to identifying optimal antimicrobial regimens against resistant pathogens are not likely to be effective, even for isolates from a single center that share similar genetic backgrounds. Indeed, all isolates in the study belonged to the ST258 international clone and carried blaKPC-2 carbapenemase, blaSHV-12 ESBL, and blaTEM-1 β-lactamase genes. Nevertheless, the range of porin expression and responses to antimicrobial agents were quite heterogeneous and could not be predicted solely based on MICs of individual agents. Of course, the molecular characteristics of isolates from other centers may differ from ours. As such, the efficacy of colistin-doripenem with or without ertapenem in our experience may not be representative of isolates from other centers or for clonal groups that carry different KPC and β-lactamase genes. Therefore, we must acknowledge that our findings need to be confirmed by follow-up studies elsewhere.
Our data highlight how molecular characterization of antimicrobial-resistant isolates can provide insights into the possible mechanisms and extent of antimicrobial synergy. Colistin exerts its bactericidal effects by permeabilizing the outer membrane of Gram-negative bacteria (39–41). Porins are outer membrane channels that allow molecules, including β-lactams, to diffuse into their periplasmic active sites (42). Absence or reduced expression of ompK35 and ompK36 porins, which typically results from mutations within promoter regions or coding sequences, can contribute to carbapenem resistance (36, 42–46). The lack of correlation between the levels of porin expression by our isolates and carbapenem MICs suggests that this mechanism may be less relevant in the face of high-level resistance due to KPC as well as other β-lactamases and mechanisms. The synergy we found between colistin and doripenem is consistent with a model in which membrane permeabilization by the former agent facilitates increased access of the latter to its penicillin-binding protein targets, allowing it to overcome its hydrolysis by KPC (47–49). In the time-kill studies, colistin (2.5 μg/ml) was active against the majority of colistin-resistant KPC K. pneumoniae isolates, indicating that subinhibitory concentrations still induced some degree of membrane permeabilization. The fact that synergy between colistin and doripenem was independent of the level of porin expression suggests that doripenem's inhibitory effects were maximized in the presence of colistin; either high expression of porin channels did not appreciably increase doripenem access, or further access did not enhance doripenem's effects. Along these lines, there are two possible explanations for the correlation between high levels of porin expression and additional synergy with the addition of ertapenem to colistin-doripenem. First, the enhanced access of both carbapenems through porins may allow ertapenem to act as a suicide substrate that binds to KPC and permits doripenem to be freely available at the penicillin-binding site (25). Alternatively, the effects may not be specific to either agent but rather stem from increased cumulative carbapenem concentrations that saturate KPC.
In contrast to a previous study showing that the doripenem-ertapenem combination rapidly reduced KPC K. pneumoniae bacterial counts in in vitro chemostat and mouse thigh infection models, we did not observe any synergy with this combination (25). The discrepancies between our findings and the earlier data offer further evidence that results from particular centers or with particular isolates may not be broadly applicable. Other noteworthy findings were consistent with the previous literature. Colistin-ertapenem was significantly less active than colistin-doripenem, which was expected since ertapenem has preferential affinity for KPC enzymes compared to other carbapenems and is more susceptible to hydrolysis (50). As mentioned earlier, colistin MICs were poorly associated with activity by time-kill assays, as some degree of killing was evident even against resistant isolates. The limitations of colistin MICs are well described and present challenges in defining resistance and accurately identifying resistant isolates (51–53).
In conclusion, our data demonstrate that not all KPC K. pneumoniae isolates are created equal. In the future, effective treatment regimens will likely have to be defined based on a constellation of factors, including the molecular biology of specific isolates, types of infection being treated, underlying diseases, host factors, and pharmacokinetic/pharmacodynamic parameters. It is imperative to accurately identify patients for whom combination antimicrobial regimens are most likely to be effective in order to both optimize the outcomes and minimize the potential for toxicity. Colistin, for example, is among the most toxic antimicrobials currently in use, and combinations of carbapenems have the potential for synergistic neurotoxicity and other untoward events. Therefore, minimizing exposure to such regimens if they are unlikely to be of benefit will be as important as identifying cases in which they may be useful. Along these lines, the development of rapid molecular tests that identify isolates that are susceptible or resistant to antimicrobial agents or combinations of agents in real time is a top priority for the field. The data from this study attest to the potential feasibility of such tests. Follow-up in vitro, animal model, and clinical studies to corroborate our findings for KPC K. pneumoniae and other difficult-to-treat pathogens are urgently needed.
ACKNOWLEDGMENTS
This study was partially supported by the University of Pittsburgh Department of Medicine and a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155). R.K.S. is supported by the National Institutes of Health through grants KL2RR024154 and KL2TR000146.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 4 March 2013
REFERENCES
- 1. Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430–1435 [DOI] [PubMed] [Google Scholar]
- 2. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 3. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 4. Maltezou HC, Giakkoupi P, Maragos A, Bolikas M, Raftopoulos V, Papahatzaki H, Vrouhos G, Liakou V, Vatopoulos AC. 2009. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece). J. Infect. 58:213–219 [DOI] [PubMed] [Google Scholar]
- 5. Rhee JY, Park YK, Shin JY, Choi JY, Lee MY, Peck KR, Song JH, Ko KS. 2010. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob. Agents Chemother. 54:2278–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. 2010. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin. Infect. Dis. 50:364–373 [DOI] [PubMed] [Google Scholar]
- 8. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67 [DOI] [PubMed] [Google Scholar]
- 10. Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO. 2009. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 30:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. 2011. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin. Microbiol. Infect. 17:1798–1803 [DOI] [PubMed] [Google Scholar]
- 13. Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect. Control Hosp. Epidemiol. 30:972–976 [DOI] [PubMed] [Google Scholar]
- 14. Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J. Antimicrob. Chemother. 65:1119–1125 [DOI] [PubMed] [Google Scholar]
- 15. Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob. Agents Chemother. 56:2108–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55:943–950 [DOI] [PubMed] [Google Scholar]
- 17. Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. 2012. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 56:5103–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jernigan MG, Press EG, Nguyen MH, Clancy CJ, Shields RK. 2012. The combination of doripenem and colistin is bactericidal and synergistic against colistin-resistant, carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:3395–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pankey GA, Ashcraft DS. 2011. Detection of synergy using the combination of polymyxin B with either meropenem or rifampin against carbapenemase-producing Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 70:561–564 [DOI] [PubMed] [Google Scholar]
- 20. Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of beta-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob. Agents Chemother. 54:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clancy CJ, Shields RK, Hong JH, Chen L, Zhao Y, Kreiswirth BN, Park S, Perlin DS, Doi Y, Kwak EJ, Silveira FP, Pasculle AW, Nguyen MH. 2012. Clinical and molecular characterization of carbapenemase-resistant K. pneumoniae (KPC) strains from a tertiary center. Abstr. 52nd Intersci Conf. Antimicrob. Agents Chemother., abstr. C2-1216 [Google Scholar]
- 22. Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39:55–60 [DOI] [PubMed] [Google Scholar]
- 23. Mammina C, Bonura C, Di Bernardo F, Aleo A, Fasciana T, Sodano C, Saporito MA, Verde MS, Tetamo R, Palma DM. 2012. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 17:20248. [PubMed] [Google Scholar]
- 24. Kontopoulou K, Protonotariou E, Vasilakos K, Kriti M, Koteli A, Antoniadou E, Sofianou D. 2010. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 beta-lactamase resistant to colistin. J. Hosp. Infect. 76:70–73 [DOI] [PubMed] [Google Scholar]
- 25. Bulik CC, Nicolau DP. 2011. Double-carbapenem therapy for carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55:3002–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, eighth ed. Approved standard M07-A8. CLSI, Wayne, PA [Google Scholar]
- 27. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. Twenty-second informational supplement M100-S22. CLSI, Wayne, PA [Google Scholar]
- 28. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Essack SY, Hall LM, Pillay DG, McFadyen ML, Livermore DM. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum beta-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 32. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 33. National Committee for Clinical Laboratory Standards 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 34. Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob. Agents Chemother. 55:3284–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Wart SA, Andes DR, Ambrose PG, Bhavnani SM. 2009. Pharmacokinetic-pharmacodynamic modeling to support doripenem dose regimen optimization for critically ill patients. Diagn. Microbiol. Infect. Dis. 63:409–414 [DOI] [PubMed] [Google Scholar]
- 36. Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J. Med. Microbiol. 58:1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hernandez-Alles S, Alberti S, Alvarez D, Domenech-Sanchez A, Martinez-Martinez L, Gil J, Tomas JM, Benedi VJ. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673–679 [DOI] [PubMed] [Google Scholar]
- 39. Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333–1341 [DOI] [PubMed] [Google Scholar]
- 40. Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 25:11–25 [DOI] [PubMed] [Google Scholar]
- 41. Zhang L, Dhillon P, Yan H, Farmer S, Hancock RE. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3317–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659–667 [DOI] [PubMed] [Google Scholar]
- 44. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang XD, Cai JC, Zhou HW, Zhang R, Chen GX. 2009. Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated beta-lactamase production and OmpK36 porin deficiency. J. Med. Microbiol. 58:1196–1202 [DOI] [PubMed] [Google Scholar]
- 46. Yang D, Guo Y, Zhang Z. 2009. Combined porin loss and extended spectrum beta-lactamase production is associated with an increasing imipenem minimal inhibitory concentration in clinical Klebsiella pneumoniae strains. Curr. Microbiol. 58:366–370 [DOI] [PubMed] [Google Scholar]
- 47. Bergen PJ, Forrest A, Bulitta JB, Tsuji BT, Sidjabat HE, Paterson DL, Li J, Nation RL. 2011. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob. Agents Chemother. 55:5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giamarellou H, Antoniadou A, Kanellakopoulou K. 2008. Acinetobacter baumannii: a universal threat to public health? Int. J. Antimicrob. Agents 32:106–119 [DOI] [PubMed] [Google Scholar]
- 49. Yoon J, Urban C, Terzian C, Mariano N, Rahal JJ. 2004. In vitro double and triple synergistic activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 48:753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xia Y, Liang Z, Su X, Xiong Y. 2012. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Annals Lab. Med. 32:270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gales AC, Reis AO, Jones RN. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J. Clin. Microbiol. 39:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sader HS, Rhomberg PR, Flamm RK, Jones RN. 2012. Use of a surfactant (polysorbate 80) to improve MIC susceptibility testing results for polymyxin B and colistin. Diagn. Microbiol. Infect. Dis. 74:412–414 [DOI] [PubMed] [Google Scholar]
- 53. Shields RK, Clancy CJ, Press EG, Hong JH, Updike CA, Nguyen MH. 2012. Discrepancies between colistin (COL) MICs and inhibition of Acinetobacter (Ab) and Klebsiella (Kp) during time-kill assays (TKA). Abstr. 52nd Intersci Conf. Antimicrob. Agents Chemother., abstr. E-804 [Google Scholar]