Abstract
The mycobacterial nucleoid-associated protein Lsr2 is a DNA-bridging protein that plays a role in condensation and structural organization of the genome and acts as a global repressor of gene transcription. Here we describe experiments demonstrating that zafirlukast inhibits the complexation between Lsr2 and DNA in vitro. Zafirlukast is shown to inhibit growth in two different species of mycobacteria tested but exhibits no growth inhibition of Escherichia coli. The Lsr2 inhibitory activity is reflected in vivo as determined by monitoring of transcription levels in Mycobacterium tuberculosis. These data suggest that zafirlukast inhibits Lsr2 function in vivo, promoting dysregulation of the expression of an array of genes typically bound by Lsr2 and hindering growth. Since zafirlukast likely operates by a mechanism distinct from current M. tuberculosis drugs and is currently used as a prophylactic treatment for asthma, it offers an intriguing lead for development of new treatments for tuberculosis.
INTRODUCTION
Tuberculosis (TB) is the leading cause of mortality due to a bacterial pathogen, Mycobacterium tuberculosis. There were 8.7 million new cases of TB in 2011, which claimed 1.4 million lives (1). The complexity and length of the current TB treatment often lead to patient noncompliance, and M. tuberculosis has adapted to this suboptimal pharmacological stress by evolving multidrug-resistant strains (MDR-TB) and extensively drug-resistant strains (XDR-TB) (2). Of the 8.7 million cases in 2011, 60,000 were reported to be MDR-TB, and roughly 5,000 of those cases were XDR-TB (1). Several cases of patients infected with strains that are resistant to all of the available first-line drugs and most of the second-line drugs were recently reported (3). Another major concern is that the most recent first-line TB drug discovery occurred almost 50 years ago with rifampin in 1963, and since then the panel of drugs has changed only modestly, while treatments have gained in complexity because of the emergence of resistant strains (4). Therefore, the need for new classes of drugs that use diverse mechanisms to prevent bacterial growth continues to increase in urgency.
Lsr2 is a nucleoid-associated protein encoded by M. tuberculosis and other actinobacteria that binds AT-rich DNA. Lsr2 appears to be functionally analogous to H-NS nucleoid-associated proteins found in Escherichia coli and related bacteria as well as eukaryotic histone proteins (5, 6). This functionality imparts to the genomic DNA resistance to oxidative damage and is essential for M. tuberculosis viability (7). Specifically, Lsr2 binds AT-rich regions in the otherwise GC-rich mycobacterial genome and thereby represses transcription of a large number of genes and is shown to play a major role in suppressing expression of exogenous DNA from transposons and mycobacterial phages (8, 9). Cross-linking studies showed that Lsr2 binds to a portion of more than 20% of the genes identified in the M. tuberculosis genome (9). In addition to identifying the lsr2 gene as a target for Lsr2 binding, the same study showed that many of the genes bound by Lsr2 are important for the mycobacterial response to environmentally induced stress, such as the presence of antibiotics, oxidative stress, and production of virulence factors, as well as genes that affect cell proliferation and persistence (10–14). Therefore, targeting Lsr2 with small molecule inhibitors of DNA binding may severely dysregulate the transcription of a wide array of important M. tuberculosis genes. This would decrease the fitness of the mycobacteria and make them unable to mount their typical defenses against the human immune system during infection or the current treatment regimen. To date, no research groups have described compounds that inhibit DNA binding by Lsr2.
Here we report the development of a high-throughput binding assay that monitors complex formation between Lsr2 and AT-rich DNA in vitro. This assay afforded the identification of zafirlukast as an inhibitor of the complexation between Lsr2 and DNA. Zafirlukast is a cysteinyl leukotriene receptor (CysLT1) antagonist that readily migrates to lung tissue and is currently used as a prophylactic treatment for asthma. Experiments are described showing that zafirlukast inhibits Lsr2 function in a dose-dependent manner and affects both gene transcription in mycobacterial cultures and the growth of mycobacteria.
MATERIALS AND METHODS
Molecular cloning, expression, and purification.
The lsr2 gene from M. tuberculosis H37Rv was placed between the NcoI- and HindIII-cut sites of a modified pET32 plasmid. This plasmid encodes a thioredoxin-Lsr2 fusion containing a polyhistidine tag near the N terminus of the fusion protein. Recombinant Lsr2 was expressed in E. coli BL21(DE3) using Luria-Bertani medium. The culture was grown at 37°C until it reached an optical density at 600 nm (OD600) of between 0.6 and 0.8 followed by induction with IPTG (isopropyl-β-d-thiogalactopyranoside) at 16°C for 36 h. Lsr2 was purified using immobilized metal affinity chromatography with wash buffer (20 mM Tris [pH 7.5], 1 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 25 mM imidazole) and elution buffer (20 mM Tris [pH 7.5], 1 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 250 mM imidazole) on a 5-ml HisTrap FF column (GE Healthcare). Following tag cleavage by application of Prescission protease during dialysis to remove excess imidazole, the thioredoxin-polyhistidine tag and protease were removed using immobilized metal affinity chromatography. Fractions containing Lsr2 were pooled and further purified using cation-exchange chromatography with wash buffer [20 mM MES (morpholineethanesulfonic acid; pH 6.0), 10% glycerol, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), 1 mM EDTA, 150 mM NaCl] and elution buffer [20 mM MES (pH 6.0), 10% glycerol, 0.5 mM TCEP, 1 mM EDTA, 1 M NaCl] on a 5-ml HiTrap SP FF column (GE Healthcare).
Determination of Lsr2/DNA complex Kd.
All polarization experiments were performed using a Biotek synergy H4 plate reader and analyzed with Gen5 and Prism 4.0 software. Zafirlukast was purchased from Sigma-Aldrich. 5′-fluorescein-labeled stem-loop DNA with the sequence 5′-CCTAATTATAACGAAGTTATAATTAGG-3′ was purchased from Integrated DNA Technologies (Coralville, IA) and dissolved in deionized water at a concentration of 100 μM, incubated at 95°C for 15 min, cooled on ice, and stored at −20°C until needed.
The dissociation constant (Kd) for the Lsr2/DNA complex was obtained using a 384-well microplate format fluorescence polarization (FP) assay performed at 37°C. Each well contained fluorescently labeled DNA (10 nM) and concentrations of Lsr2 ranging from 0 to 200 μM. Additional buffer solution (20 mM Tris [pH 7.5], 50 mM NaCl) was added as needed to reach a final volume of 20 μl. An excitation wavelength of 485 nm and an emission wavelength of 528 nm were used to measure fluorescence polarization.
Screening of NCC.
The tested compounds for the library screening were from the National Institutes of Health Clinical Collection (NCC) purchased from BioFocus. The NCC compounds were screened in a black Corning 384-well microplate at 37°C. Each well contained a total volume of 25 μl with the following components in a reaction buffer of 20 mM Tris (pH 7.5) and 50 mM NaCl: Lsr2 (13.65 μM), fluorescently labeled DNA probe (10 nM), dimethyl sulfoxide (DMSO) (4% [vol/vol]), and the tested drug (0.4 mM). Controls were performed by combining all of the above reagents except the inhibitor compounds. Any compound showing a decrease in the FP signal that was greater than 3 times the standard deviation was considered a hit.
Determination of Ki.
The Ki was obtained using the same FP assay format. Each well contained 1 μl of fluorescently labeled DNA (200 nM) and 20 μl of Lsr2 (200 μM), and the concentration of zafirlukast was varied by addition of a constant volume (1 μl) with increasing concentrations from 0 to 500 μM. Ki was calculated with Prism (GraphPad software) using the equation Ki = [[Lb](IC50)(Kd)]/[[Lo][Ro] + [Lb](−[Ro] − [Lo] + [Lb] − Kd)], where [Lb] is bound DNA concentration, [Lo] is the total DNA concentration, [Ro] is the total Lsr2 concentration, and IC50 is the 50% inhibitory concentration.
Kirby-Bauer disc diffusion assays.
Carbenicillin-containing agar plates were inoculated with M. smegmatis ATCC 14468 from a liquid culture. Following the initial inoculation, wafers impregnated with various amounts of zafirlukast dissolved in DMSO (49, 98, 196, and 392 μg) were positioned on the agar plate. The plate was incubated for 48 h at 37°C followed by examination of the zones of inhibition. Analogous experiments using E. coli in LB medium were performed using the same protocol but with incubation for only 16 h. For M. tuberculosis strain mc26230, medium from a Middlebrook 7H9 liquid culture containing pantothenic acid and incubated at 37°C to mid-log phase was used to inoculate a Middlebrook 7H9 agar plate augmented with pantothenic acid. Filter paper wafers impregnated with either 10 μl of DMSO or 10 μl of DMSO containing 500 μg of zafirlukast were added to these plates, which were incubated at 37°C until growth was sufficient to clearly observe the zones of inhibition.
Test of the bactericidal activity of zafirlukast.
A liquid culture of M. smegmatis ATCC 14468 in Luria-Bertani medium containing ampicillin (50 μg/ml) was incubated at 37°C until it reached an OD600 near 0.3. Zafirlukast was added to the liquid culture at a final concentration of 20 μM. Aliquots from this culture were removed at different time points and diluted 500-fold, and 250 μl of this diluted sample was applied to carbenicillin-containing agar plates. Each plate was incubated for 2 days at 37°C followed by a colony count to determine the number of CFU.
Semiquantitative RT-PCR.
For semiquantitative reverse transcriptase (RT)-PCR, mid-log-phase cells of the attenuated M. tuberculosis strain mc26230 were induced with zafirlukast at a final concentration of 12.5 μM and incubated for 16 h. Five or 10 ng of total RNA, purified with the RNeasy kit (Qiagen), was used for reverse transcription with SuperScript III by following the manufacturer's protocol (Invitrogen). The PCR step was performed using Taq DNA polymerase. As an internal control, we determined the transcript level of sigA in M. tuberculosis. Control reactions without SuperScript III or RNA were also performed. PCR products were run on a 1% agarose gel and stained with SYBR Safe DNA gel stain (Invitrogen), followed by band intensity quantification with the Quantity One software (Bio-Rad). The primers used for these experiments are shown in Table S1 in the supplemental material.
Intensity-fading MALDI-TOF mass spectrometry assay.
All mass spectrometry experiments were performed using a Bruker ultrafleXtreme matrix-assisted laser desorption ionization–tandem time of flight (MALDI TOF/TOF) mass spectrometer. Zafirlukast and ritonavir were dissolved in DMSO and combined at a ratio to produce ion peaks of similar intensity. This zafirlukast-ritonavir solution was diluted 10-fold with a 20 mM Tris (pH 8.5) buffer and then combined with solutions of Lsr2 ranging in concentration from 0 to 60 μM and incubated for 12 h at 4°C. From this solution, 2 μl was added to each of the solutions containing increasing concentrations of Lsr2 in 20 mM Tris (pH 8.5). The sample was mixed using the dried-droplet method with a matrix solution (1:2 [vol/vol]) of sinapinic acid (10 mg/ml) containing 30% acetonitrile (vol/vol) in deionized water.
Determination of the MIC.
A bacterial inoculum of M. smegmatis grown in LB medium containing ampicillin (50 μg/ml) was adjusted to an OD600 of 0.5 and diluted to yield a final bacterial density of 5 × 105CFU/ml. Zafirlukast was dissolved in DMSO and added to 2-ml aliquots of the bacterial suspension to give a final concentration range from 3 to 12 μM. Each concentration of zafirlukast was tested on three aliquots. Following a 48-h incubation period at 37°C, 0.2 ml from each culture was transferred into a 96-well plate and 10 μl of thiazolyl blue tetrazolium bromide (10 mg/ml in methanol) was added to each well. The plate was incubated at 37°C for 30 min. The MIC was recorded as the lowest concentration that did not result in any color change.
Test of the bactericidal activity of zafirlukast.
A liquid culture of M. smegmatis in LB medium containing ampicillin (50 μg/ml) was incubated at 37°C until it yielded a bacterial density near 5 × 105 CFU/ml. Zafirlukast was added to 2-ml aliquots of the bacterial suspension to reach a final concentration of 10 μM. The cultures were incubated at 37°C, removed at different time points, and centrifuged at 3,500 × g for 20 min. Each pellet was resuspended in 200 μl of LB medium and diluted 2,000-fold. A total of 250 μl of this diluted sample was applied to carbenicillin-containing agar plates. Each plate was incubated for 2 days at 37°C followed by a colony count to determine the number of CFU.
RESULTS
Lsr2-DNA complexation is rapidly quantified using fluorescence polarization.
To better understand Lsr2-DNA complexation and identify compounds that modulate Lsr2 function, we employed a fluorescence polarization (FP) assay using a fluorescein-labeled AT-rich DNA probe previously shown to bind Lsr2 (9). In a series of binding experiments where the concentration of probe was held constant while the concentration of Lsr2 increased, the FP signal showed a concomitant increase that reflected complexation of Lsr2 with the DNA probe. Fitting data from these binding experiments gave a Kd of 0.60 ± 0.06 μM (R2 = 0.97) for the Lsr2/DNA complex, which corresponds well with previously determined values (Fig. 1A) (11). This information was then used to develop a high-throughput FP assay amenable to library screening for identification of compounds that inhibit Lsr2-DNA complexation. In this assay, Lsr2 was preincubated with the DNA probe, and each compound from the NIH Clinical Collection was added to each well containing the Lsr2/DNA complex. The FP signal was measured for every mixture of Lsr2, DNA, and drug. The mean polarization from triplicate experiments was then calculated for each concentration of Lsr2.
Fig 1.
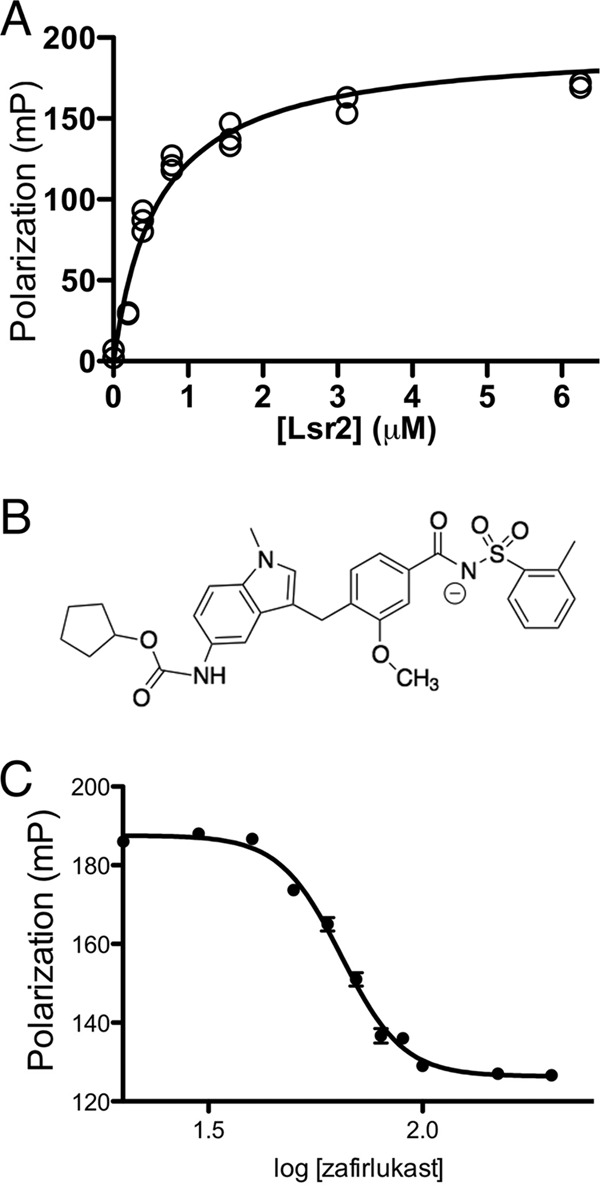
Lsr2-DNA complexation and inhibition by zafirlukast. (A) Shown are the normalized data from the FP experiments in millipolarization units (mP). Each Lsr2 concentration was performed in triplicate. Because the standard deviation at each was very small, all of the data points are shown instead of using error bars. (B) Shown is the anticipated structure of zafirlukast at physiological pH based on previous studies measuring the pKa of the secondary amine within the N-acyl sulfonamide moiety (34). (C) Data from FP experiments where the DNA probe and Lsr2 concentrations were fixed while the zafirlukast concentration was varied. The IC50 was found to be equal to 64.55 μM.
Zafirlukast inhibits Lsr2-DNA complexation through interactions with Lsr2.
Screening of the NIH Clinical Collection identified seven compounds as potential leads based on a criterion of an FP signal decrease greater than 3 times the standard deviation from the mean polarization measurement. Of these, six are known DNA intercalators or groove binders, so they were not studied further, since the purpose of this study is to identify a compound that specifically inhibits Lsr2 function. The single remaining lead from this screening study, zafirlukast, is a leukotriene receptor antagonist commonly prescribed as a prophylactic treatment for asthma (Fig. 1B). To determine the inhibitory constant (Ki) for zafirlukast, a dose dependence study using the same basic FP assay as described above was used (Fig. 1C). However, the concentrations of both the DNA probe and Lsr2 were changed slightly from those concentrations used for library screening to better assess inhibitory activity. The probe and Lsr2 concentrations were held constant, while the concentration of zafirlukast was varied. These experiments exhibited a consistent dose-dependent inhibition of Lsr2-DNA complexation that yielded an IC50 of 64.55 μM (R2 = 0.989) and a calculated Ki value of 13.9 μM.
Zafirlukast directly binds Lsr2.
To support the hypothesis that zafirlukast interacts specifically with Lsr2 and not DNA, an intensity-fading MALDI-TOF mass spectrometry assay was performed (15, 16). Samples containing fixed concentrations of ritonavir as an internal standard and zafirlukast were exposed to various Lsr2 concentrations. Subsequent MALDI-TOF experiments monitored the intensity of the zafirlukast and ritonavir ion peaks (Fig. 2A). Comparing the relative ion abundance of the two drugs from each of the mixtures shows that the ratio of free zafirlukast to ritonavir ions decreases as the concentration of Lsr2 increases (Fig. 2B). The decrease in the relative ion intensity of zafirlukast can be directly attributed to its binding with Lsr2 and its removal from the pool of unbound zafirlukast.
Fig 2.
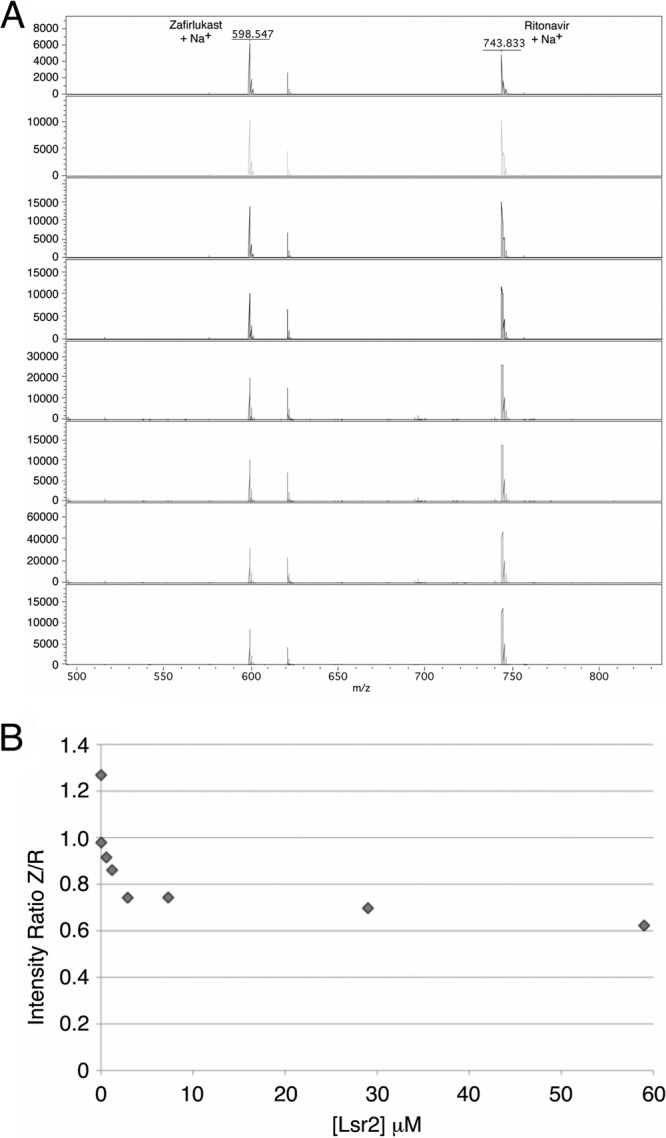
Direct binding of zafirlukast to Lsr2 using intensity-fading MALDI mass spectrometry. (A) Shown are the MALDI-TOF spectra of different mixtures of zafirlukast, ritonavir, and Lsr2. The concentration of Lsr2 in each drop increases from top to bottom, promoting a decrease in the relative intensity of the zafirlukast-plus-Na+ ion peak (m/z 598.5) relative to the ritonavir-plus-Na+ ion peak (m/z 743.9). (B) Plot of the ion ratio of zafirlukast (Z) over ritonavir (R) versus the concentration of Lsr2. As the concentration of Lsr2 is increased, a larger portion of the zafirlukast is bound to the protein, thereby suppressing the amount of the free ion and decreasing the intensity of the zafirlukast signal.
Zafirlukast inhibits the growth of mycobacteria.
To assess the ability of zafirlukast to enter the mycobacterial cell, inhibit Lsr2 function, and affect cell growth, a Kirby-Bauer disk diffusion assay was performed using either Mycobacterium smegmatis or the attenuated M. tuberculosis strain mc26230 (Fig. 3A and B). Various amounts of zafirlukast (49 to 392 μg) were used since it was unknown to what extent the drug would inhibit growth. Following incubation, the plate exhibited clear zones of inhibition whose sizes are concomitant with increasing amounts of drug.
Fig 3.
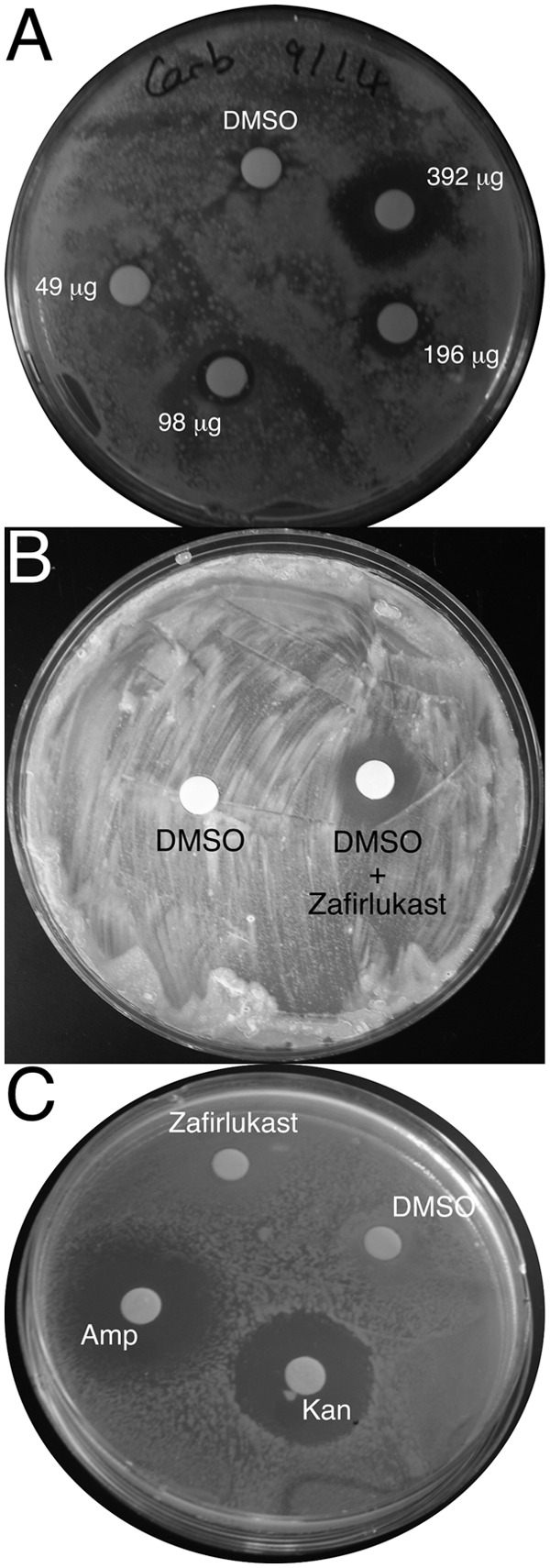
Zafirlukast inhibits the growth of various mycobacterial species. M. smegmatis was cultured in the presence of carbenicillin (50 μg/ml) using wafers impregnated with various amounts of zafirlukast to show the concentration dependence of growth inhibition. (B) M. tuberculosis strain mc26230 grown in the presence of zafirlukast (500 μg). (C) E. coli BL21(DE3) cells were grown on agar plates in the presence of wafers impregnated with kanamycin, ampicillin, or zafirlukast (150 μg). Both kanamycin and ampicillin exhibited the anticipated zones of inhibition, while zafirlukast produces no zone of inhibition.
It is typical for antimycobacterial compounds to have better efficacy against M. smegmatis than M. tuberculosis, so it was expected that M. tuberculosis would not respond as strongly to zafirlukast. To account for this, a zafirlukast dose of 500 μg was used for the M. tuberculosis mc26230 Kirby-Bauer growth assay. As expected, the zone of inhibition when testing M. smegmatis growth in the presence of 392 μg drug was larger than that observed when using 500 μg for M. tuberculosis, whereas exposure of a laboratory strain of E. coli to the same amount of compound exhibits no growth inhibition (Fig. 3C).
These data clearly show that E. coli, which does not encode an Lsr2 homolog but instead uses the H-NS protein to modulate genome structure, is unaffected by zafirlukast. In contrast, the effect on growth of two different species of mycobacteria suggests that the activity of zafirlukast may be specific to organisms that encode Lsr2, which includes Mycobacterium leprae, Mycobacterium ulcerans, and other disease-causing actinobacteria. Experiments have been initiated to assess the effects of zafirlukast on the growth of both Streptomyces coelicolor and Corynebacterium glutamicum.
To further characterize the antimycobacterial activity of zafirlukast, attempts were made to determine the MIC. Zafirlukast at concentrations between 0.5 and 100 μM was added to mid-log-phase cultures of M. smegmatis, and bacterial growth was assessed by measuring the OD600. At zafirlukast concentrations of 10 μM or higher, the liquid culture medium clarified after 3 days and a viscous material accumulated at the bottom of the culture flask. Agar plates inoculated with the culture fluid exhibited no growth, suggesting a lack of viable planktonic M. smegmatis bacteria. However, when assessing the viability of the whole M. smegmatis culture by addition of MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide], a modest MIC of 6 μM was exhibited for zafirlukast.
Zafirlukast exhibits bactericidal activity.
To determine whether zafirlukast is bactericidal or bacteriostatic, a plating assay was performed using material from an M. smegmatis culture incubated with 10 μM zafirlukast. A colony count was performed at different time points after the addition of zafirlukast. The number of CFU from suspended cells decreased after only 24 h of incubation, with clarification of the liquid medium after 3 days of incubation with the drug. As previously mentioned concomitant with the clarification of the culture, cellular debris accumulated at the bottom of the flask. Since this is common for mycobacteria, it was necessary to determine if this material contained any living M. smegmatis cells.
To determine if any of the precipitated cell debris contained viable bacteria, the pelleted detritus was resuspended in culture medium lacking zafirlukast but containing 0.2% (vol/vol) Tween 80 to minimize bacterial agglutination. This resuspended sample from each time point was diluted and plated on agar plates. The experiment was performed in triplicate, so each time point was analyzed based on three different cultures incubated at the same time under the same conditions. Following addition of zafirlukast, a time course over a 7-day period shows a significant decrease in CFU (Fig. 4). After 7 days, zero CFU were observed from each of the three different cultures. Although the insoluble material contains viable M. smegmatis in the days immediately following application of the drug, these numbers are lower than expected if zafirlukast were simply bacteriostatic. Ultimately, the lack of viable bacteria after 7 days suggests that the activity of zafirlukast is bactericidal for M. smegmatis.
Fig 4.

Killing of M. smegmatis by zafirlukast. Shown is a plot illustrating the number of M. smegmatis CFU following application of zafirlukast to a liquid culture. Error bars are from experiments performed in triplicate. The day 7 and day 10 plates had no colonies after 2 days of incubation at 37°C.
Zafirlukast increases gene expression in genes bound by Lsr2.
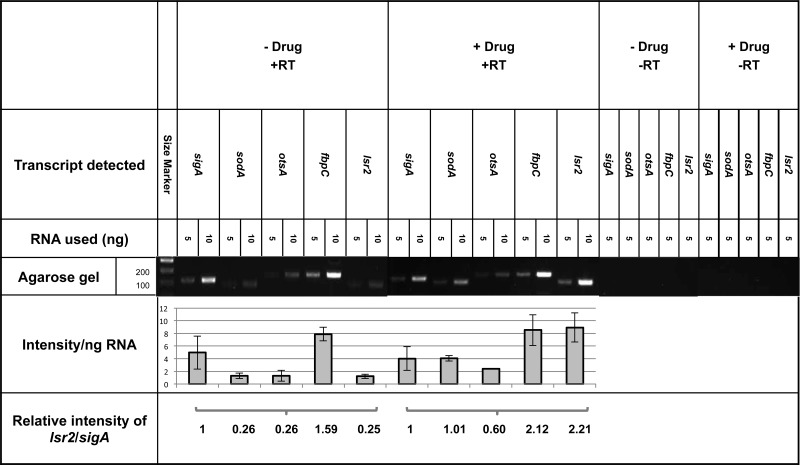
To gain information concerning the mechanism of growth inhibition by zafirlukast, semiquantitative reverse transcriptase (RT)-PCR was used to examine the effects of administering the drug on transcription of five M. tuberculosis genes previously shown to be bound by Lsr2 (9) (Fig. 5). Two of the inspected genes, otsA and fbpC, were chosen because they play important roles in synthesizing trehalose and the mycobacterial outer membrane, respectively (14, 17, 18). The third gene, sodA, encodes the superoxide dismutase virulence factor (19). As it is known that the Lsr2 protein binds its own gene, lsr2, the mRNA levels of that gene were also quantified (9). A constitutively expressed housekeeping gene, sigA, was examined as a standard. Two different cultures were exposed to zafirlukast, and RNA was extracted from each sample. Two different concentrations of RNA, 5 and 10 ng, were used for amplification of the selected genes, and the intensities of the resulting bands from two replicates from each of the two cultures were quantified. These experiments consistently show that addition of zafirlukast promotes the increase of gene expression in the four mycobacterial genes known to be bound by Lsr2.
Fig 5.
Zafirlukast stimulates transcription in vivo. Electrophoretic gels and quantitation of the reverse transcriptase-PCR products are shown. Each major column indicates an experiment either with or without application of zafirlukast. Specific genes are labeled, and results obtained with either 5 or 10 ng of RNA are shown. The intensities relative to the sigA band intensities are shown in the bottom row. For each RNA concentration, two replicates were performed in each of two different experiments.
The largest increase observed was in the mRNA levels of lsr2, which exhibits an 8.8-fold increase in lsr2 expression. Transcription of the otsA and fbpC genes exhibited modest effects with 2.3- and 1.3-fold increases, respectively, while sodA exhibited a 4-fold increase in mRNA levels. The sigA mRNA levels decreased slightly, which is consistent with previous data (20). All of these changes are consistent with the hypothesis that zafirlukast inhibits Lsr2 activity.
DISCUSSION
Lsr2 plays an important role in regulating the transcription of a large portion of the M. tuberculosis genome, and this essential function makes it an attractive drug target. This study aimed to identify small molecules that inhibit the DNA-binding activity of the mycobacterial Lsr2 protein. The employment of a sensitive FP assay that monitors Lsr2 binding of AT-rich DNA provided evidence that zafirlukast is an inhibitor of the complexation between Lsr2 and DNA. This inhibition occurs through direct interactions between the drug and the protein, which is supported by the intensity-fading MALDI-TOF mass spectrometry data. We have shown using Kirby-Bauer disk diffusion assays that zafirlukast was able to enter the mycobacterial cell and effectively inhibit the growth of M. smegmatis and M. tuberculosis in a concentration-dependent manner.
Since it is known that mycobacteria can form drug-insensitive clusters or biofilms even in the presence of bactericidal compounds such as isoniazid, it was necessary to determine the number of CFU in the bacterial detritus (21). Bacterial death of M. smegmatis was observed after 3 days of incubation in the presence of zafirlukast and extends to longer time points, which implies that this inhibitor possesses bactericidal activity.
The semiquantitative reverse transcriptase-PCR assay also revealed that the addition of zafirlukast into the culture medium of M. tuberculosis had a measureable effect on transcription of the tested mycobacterial genes. As expected, an increase in gene expression was observed after incubation with the drug for all of the tested genes, with lsr2 responding most prominently. Although Lsr2 expression levels are known to increase slightly in response to heat shock and application of iron, that change is modest with respect to the almost 9-fold change observed when zafirlukast is applied. This suggests that the response triggered by the administration of zafirlukast results from a mechanism different from the antibiotic-induced stress caused by known antitubercular drugs. Therefore, it is reasonable to suggest that zafirlukast is acting through a novel molecular mechanism (22–25).
The first of two possible mechanisms by which zafirlukast inhibition of Lsr2 could lead to growth inhibition of mycobacteria is by strongly stimulating transcription of the lsr2 gene. This overexpression would saturate the mycobacterial genomic DNA with Lsr2 protein, which would strongly inhibit expression of genes important for growth. However, that hypothesis is not consistent with the presented RT-PCR data that show a general increase in transcription of the studied genes.
A second mechanism could be from the overexpression of one or more “toxic” genes due to the inhibition of Lsr2 activity. Such proteins may benefit the bacteria at low to moderate levels in the cell but are detrimental to growth or viability at higher concentrations due to their enzymatic activity or the overproduction of toxic metabolites. Each of these phenomena has been observed in M. tuberculosis. The production of ChiZ (Rv2719c), a cell wall hydrolase involved in cell division, increases dramatically upon exposure of M. tuberculosis to DNA-damaging agents and by an unknown mechanism promotes the survival of the bacterium (26). However, the overexpression of ChiZ under nominal, nonstressing conditions compromises cell division and decreases overall viability of the bacteria, possibly by promoting the degradation of cell wall components (26, 27).
A recent example of mycobacterial toxicity due to metabolite accumulation in mycobacteria was shown by Kalscheuer et al. (28). This paper showed that in a GlgE knockout, the accumulation of maltose 1-phosphate promotes the increased expression of enzymes necessary for production of the toxic metabolite, thereby producing a positive-feedback loop that promotes rapid killing of M. tuberculosis. However, the mechanism of killing is not yet known.
In E. coli, it is known that NADH dehydrogenase I expression is upregulated in response to bactericidal drugs. This promotes hydroxyl radical production that causes DNA damage and a subsequent SOS response (29). It has been shown that M. smegmatis NADH dehydrogenase mutants with decreased enzymatic activity afford resistance to isoniazid by altering the ratio of reduced and oxidized NAD (30). Lsr2 binds to 3 of the M. tuberculosis genes encoding NADH dehydrogenase I (Rv3145, Rv3152, and Rv3158) (9). If Lsr2 promotes an increase in NADH dehydrogenase expression resulting in a concomitant increase in the production of hydroxyl radicals, this would suggest a possible mode of killing. Furthermore, as Lsr2 is known to protect the mycobacterial genome from oxidative damage, inhibition of DNA binding activity by zafirlukast could enhance the effects of endogenous hydroxyl radical formation. If mycobacterial death is indeed a consequence of dysregulating transcription of a large number of genes in the mycobacterial genome affected by Lsr2 DNA binding activity, other detrimental metabolic effects are possible.
Dysregulation of transcription may help explain why growth inhibition is observed at drug concentrations slightly below the determined Ki value for the Lsr2/zafirlukast complex. The strong induction of lsr2 transcription exhibited in the RT-PCR data suggests that the expression of lsr2 is highly regulated by Lsr2 activity and therefore highly sensitive to inhibitors of Lsr2-DNA binding activity. In such a case, inhibition of only half of the Lsr2 proteins in a mycobacterial cell may still have extreme effects on transcription of lsr2 or other genes whose overexpression may lead to cell death. Ultimately, a global analysis of mycobacterial transcription levels in response to zafirlukast application is required to more clearly assess the mechanism of growth inhibition.
This study aimed to identify inhibitors of Lsr2-DNA binding activity for the development of novel antitubercular compounds. Even though the exact mechanism of action by which growth inhibition occurs is still under study, the presented data suggest inhibition of Lsr2 activity by zafirlukast and a concomitant physiological response by mycobacteria to this inhibition. Not only is zafirlukast now identified as a specific inhibitor of Lsr2 function, but also the application of this drug is clearly bactericidal to M. smegmatis.
Multiple obstacles can hinder M. tuberculosis drug development. The first, penetration of the mycobacterial cell, does not appear to be a barrier for zafirlukast as suggested by the growth study (31). The second major obstacle is ensuring that a bioavailable drug can get to the site of M. tuberculosis infection. Some antibiotics such as imipenem have a poor oral bioavailability and are administered as an intravenous or intramuscular injection that requires stringent treatment and access to a clinic (32). Zafirlukast is orally bioavailable and is currently used as a treatment for asthma, which suggests that the molecule is able to access the primary site of tuberculosis infections: the lungs (33). The fact that zafirlukast is a commercially available generic drug and safe for daily use could potentially have an immediate impact on TB treatment if used alongside other available therapies and possibly paves the way for the development of new classes of compounds affecting the same target but with improved efficacy.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a National Institutes of Health grant to D.R.R. (AI089653) and by grants from the KORDI (Korea Ocean Research and Development Institute) in-house program (PE98402) and KRDA (Korea Rural Development Agency) to C.M.K.
We thank Bill Jacobs for the generous gift of M. tuberculosis strain mc26230.
Footnotes
Published ahead of print 25 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02407-12.
REFERENCES
- 1. WHO 2012. Global tuberculosis report 2012. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Moonan PK, Quitugua TN, Pogoda JM, Woo G, Drewyer G, Sahbazian B, Dunbar D, Jost KC, Jr, Wallace C, Weis SE. 2011. Does directly observed therapy (DOT) reduce drug resistant tuberculosis? BMC Public Health 11:19 doi:10.1186/1471-2458-11-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. 2012. Totally drug-resistant tuberculosis in India. Clin. Infect. Dis. 54:579–581 [DOI] [PubMed] [Google Scholar]
- 4. Chang KC, Yew WW. 2013. Management of difficult multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis: update 2012. Respirology 18:8–21 [DOI] [PubMed] [Google Scholar]
- 5. Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, Tran V, Berbenetz NM, Kocincova D, Yip CM, Reyrat JM, Liu J. 2008. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 36:2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. 2008. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 190:7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR, Jr, Salgame P, Alland D. 2009. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc. Natl. Acad. Sci. U. S. A. 106:4414–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ali SS, Xia B, Liu J, Navarre WW. 2012. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 15:175–181 [DOI] [PubMed] [Google Scholar]
- 9. Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. 2010. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 107:5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen JM, German GJ, Alexander DC, Ren H, Tan T, Liu J. 2006. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 188:633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colangeli R, Helb D, Vilcheze C, Hazbon MH, Lee CG, Safi H, Sayers B, Sardone I, Jones MB, Fleischmann RD, Peterson SN, Jacobs WR, Jr, Alland D. 2007. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3:e87 doi:10.1371/journal.ppat.0030087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamichhane G, Tyagi S, Bishai WR. 2005. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun. 73:2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 98:12712–12717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 15. Villanueva J, Yanes O, Querol E, Serrano L, Aviles FX. 2003. Identification of protein ligands in complex biological samples using intensity-fading MALDI-TOF mass spectrometry. Anal. Chem. 75:3385–3395 [DOI] [PubMed] [Google Scholar]
- 16. Yanes O, Villanueva J, Querol E, Aviles FX. 2007. Detection of non-covalent protein interactions by ‘intensity fading’ MALDI-TOF mass spectrometry: applications to proteases and protease inhibitors. Nat. Protoc. 2:119–130 [DOI] [PubMed] [Google Scholar]
- 17. Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420–1422 [DOI] [PubMed] [Google Scholar]
- 18. De Smet KA, Weston A, Brown IN, Young DB, Robertson BD. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199–208 [DOI] [PubMed] [Google Scholar]
- 19. Jackett PS, Aber VR, Lowrie DB. 1978. Virulence and resistance to superoxide, low pH and hydrogen peroxide among strains of Mycobacterium tuberculosis. J. Gen. Microbiol. 104:37–45 [DOI] [PubMed] [Google Scholar]
- 20. Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715–724 [DOI] [PubMed] [Google Scholar]
- 21. Kulka K, Hatfull G, Ojha AK. 2012. Growth of Mycobacterium tuberculosis biofilms. J. Vis. Exp. 60:e3820 doi:10.3791/3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731 [DOI] [PubMed] [Google Scholar]
- 23. Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., III 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174–40184 [DOI] [PubMed] [Google Scholar]
- 24. Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129–3138 [DOI] [PubMed] [Google Scholar]
- 25. Wong DK, Lee BY, Horwitz MA, Gibson BW. 1999. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67:327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. 2006. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62:132–147 [DOI] [PubMed] [Google Scholar]
- 27. Vadrevu IS, Lofton H, Sarva K, Blasczyk E, Plocinska R, Chinnaswamy J, Madiraju M, Rajagopalan M. 2011. ChiZ levels modulate cell division process in mycobacteria. Tuberculosis (Edinb.) 91(Suppl 1):S128–S135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalscheuer R, Syson K, Veeraraghavan U, Weinrick B, Biermann KE, Liu Z, Sacchettini JC, Besra G, Bornemann S, Jacobs WR., Jr 2010. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an alpha-glucan pathway. Nat. Chem. Biol. 6:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 30. Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR., Jr 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J. Bacteriol. 180:2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jarlier V, Nikaido H. 1990. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J. Bacteriol. 172:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mouton JW, Touzw DJ, Horrevorts AM, Vinks AA. 2000. Comparative pharmacokinetics of the carbapenems: clinical implications. Clin. Pharmacokinet. 39:185–201 [DOI] [PubMed] [Google Scholar]
- 33. Dekhuijzen PN, Koopmans PP. 2002. Pharmacokinetic profile of zafirlukast. Clin. Pharmacokinet. 41:105–114 [DOI] [PubMed] [Google Scholar]
- 34. Su SC, Hartkopf AV, Karger BL. 1976. High-performance ion-pair partition chromatography of sulfa drugs. Study and optimization of chemical parameters. J. Chromatogr. 119:523–537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.