Abstract
Bacterial virulence factors have been increasingly regarded as attractive targets for development of novel antibacterial agents. Virulence inhibitors are less likely to generate bacterial resistance, which makes them superior to traditional antibiotics that target bacterial viability. Salmonella enterica serovar Typhimurium, an important food-borne human pathogen, has type III secretion system (T3SS) as its major virulence factor. T3SS secretes effector proteins to facilitate invasion into host cells. In this study, we identified several analogs of cytosporone B (Csn-B) that strongly block the secretion of Salmonella pathogenicity island 1 (SPI-1)-associated effector proteins, without affecting the secretion of flagellar protein FliC in vitro. Csn-B and two other derivatives exhibited a strong inhibitory effect on SPI-1-mediated invasion to HeLa cells, while no significant toxicity to bacteria was observed. Nucleoid proteins Hha and H-NS bind to the promoters of SPI-1 regulator genes hilD, hilC, and rtsA to repress their expression and consequently regulate the expression of SPI-1 apparatus and effector genes. We found that Csn-B upregulated the transcription of hha and hns, implying that Csn-B probably affected the secretion of effectors through the Hha–H-NS regulatory pathway. In summary, this study presented an effective SPI-1 inhibitor, Csn-B, which may have potential in drug development against antibiotic-resistant Salmonella.
INTRODUCTION
The type III secretion system (T3SS) is a highly specialized virulent protein nano-injector, which traverses the cell wall of Gram-negative bacteria and transports effector proteins from bacterial cytoplasm into eukaryotic host cells to facilitate bacterial invasion and dissemination (1, 2). As an important virulence factor, T3SS is highly conserved in structure and function among different Gram-negative bacteria (3–5), such as Salmonella spp., Yersinia spp., Shigella spp., Pseudomonas spp., enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), Chlamydia spp., and Burkholderia spp., and some specific plant-pathogenic bacteria, Erwinia spp., Xanthomonas campestris, Ralstonia solanacearum, and Rhizobium spp. (3, 6, 7). Similar to flagella genetically and structurally (8–10), T3SS consists of approximately 20 proteins (3–5). T3SS is often absent in nonpathogenic bacteria (2, 11–13) and not essential for bacterial growth (2, 14–16). Traditional antibiotics, which directly kill pathogenic microorganisms, have a high likelihood of bacterial resistance (17). In contrast, T3SS-blocking agents can reduce the pathogenicity of Gram-negative pathogens while having no inhibitory effect on bacterial growth. This property of T3SS-blocking agents makes them ideal for new antibiotic development (17). T3SS is increasingly being proposed and explored as an attractive drug target for developing novel antibacterial agents (17, 18).
Several series of T3SS inhibitors have been identified by whole-cell-based screening methods, such as the reporter gene screening system (19), enzyme-linked immunosorbent assay (ELISA) (20), contact hemolysis screening system (21, 22), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (23, 24). Salicylidene acylhydrazides have a broad spectrum to inhibit T3SS in several important Gram-negative pathogens, i.e., Yersinia spp. (14, 25–27), Chlamydia spp. (28–34), Salmonella spp. (35–37), Shigella spp. (38), and EHEC (39). A variety of other synthesized compounds, N-phenylbenzamides (19, 25, 40), thiazolidinones (41, 42), and salicylideneanilines (20) and several natural products, caminosides (23, 24), and phenolic acids (43, 44) were also identified as T3SS inhibitors.
Salmonella enterica is an important pathogen of humans and animals. It can cause diseases ranging from mild gastroenteritis to severe systemic infection and is a serious problem for public health, as antibiotic-resistant strains are constantly emerging worldwide. The development of new control and treatment regimens, therefore, is an urgent affair (45). The pathogenicity of S. enterica mainly depends on two T3SSs, encoded by Salmonella pathogenicity island 1 (SPI-1) and SPI-2. SPI-1 mediates invasion of the intestinal epithelium and induction of proinflammatory responses through injecting effector proteins into host cells, while SPI-2 regulates the replication of bacteria in host phagocytic cells (46).
To exploit novel antibacterial agents targeting SPI-1, we have used SDS-PAGE and Western blotting in this study to assess the effect of a series of synthetic compounds on secretion of SPI-1 effector proteins in vitro. We identified several strong and selective inhibitors that blocked the secretion of SPI-1 effector proteins and invasion of Salmonella into HeLa cells while exhibiting minimal cytotoxicity. The preliminary structure-activity relationship (SAR) and mechanism of action were also discussed in the study.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study were listed in Table 1. S. enterica serovar Typhimurium UK-1 χ8956 (47) was grown in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) or on LB agar plates supplemented with 0.2% l-arabinose at 37°C or 25°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium UK-1 χ8956 | ΔPrpoS183::TT araC pBAD rpoS | 47 |
| E. coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| Plasmids | ||
| pKD46 | Vector containing the promoter of araC pBAD activator used in λ Red recombination, Apr | 50 |
| pWSK29 | Low-copy-no. vector, Apr | 51 |
| pBAD-hilA-pWSK29 | pWSK29 carrying araC pBAD and hilA, Apr | This work |
Apr, resistant to ampicillin.
Sources of the compounds tested as T3SS inhibitors.
INP0403 was chosen as the positive-control compound used in this study (31, 36–39). The activities of cytosporone B (Csn-B) and its derivatives (curvulin, C1, C2, and C3; secocurvularin, C5, C8, E0, E1, E2, E3, E4, E5, E6, A2, A3, A4 and A5) as T3SS inhibitors were assessed in this study. All compounds were synthesized through the routes described in the supplemental information (see Schemes S1 and S2 in the supplemental material) and fully characterized by 1H nuclear magnetic resonance (NMR), 13C NMR, and mass spectrometry. All compounds were dissolved in dimethyl sulfoxide (DMSO) (Sigma) to concentrations of 100 mM as stock solutions. The compounds were added to bacterial or cell culture medium at a final concentration of 100, 50, 25, 12.5, or 6.25 μM as indicated. A negative control containing 0.1% (vol/vol) DMSO was added in all experiments.
Isolation and detection of secreted proteins.
SPI-1-associated effector proteins were isolated and detected as described by Negrea et al. (37) and Hudson et al. (36). In short, S. enterica serovar Typhimurium cells were grown overnight in LB broth (0.2% l-arabinose) at 25°C. The overnight culture was diluted 10 times in fresh LB (0.2% l-arabinose) and grown for 4 h at 37°C with agitation in the presence or absence of different compound at a final concentration of 100 μM. The cultures were diluted 10-fold to measure the optical density at 600 nm (OD600) using an Eppendorf Biophotometer Plus. The bacteria were pelleted by repeated centrifugation (14,000 × g, 10 min), and the secreted proteins in the supernatant were precipitated using trichloroacetic acid (TCA) at a final concentration of 10%. The precipitated proteins were collected by centrifugation at 10,000 × g for 10 min at 4°C. Pellets were washed by ice-cold acetone twice and dissolved in an appropriate volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, based on OD600 values of the cultures. This was to confirm that the protein samples were derived from cultures containing equivalent numbers of bacteria. The samples were immediately heated for 10 min at 95°C to denature the proteins and subsequently analyzed by SDS-PAGE. Proteins were visualized either by staining with Coomassie blue or by Western blots as detailed below.
Western blots.
S. enterica serovar Typhimurium cells were cultured and treated using the same procedure described under “Isolation and detection of secreted proteins.” Proteins were separated by 9% SDS-PAGE and then electrotransferred to a polyvinylidene fluoride (PVDF) (Millipore Immobilon-P) membrane using a wet transfer apparatus (Bio-Rad). The blotted membrane was incubated in blocking solution (5% [wt/vol] bovine serum albumin [BSA] in Tris-buffered saline mixed with Tween 20 [TBST]) for 1 h at room temperature. The membrane was incubated in 5% BSA supplemented with anti-SipC monoclonal antibody or anti-FliC monoclonal antibody for 2 h at room temperature or overnight at 4°C and then washed three times with TBST for 15 min. The membrane was incubated in TBST supplemented with an anti-mouse IgG conjugated with horseradish peroxidase (HRP) (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature and then washed three times with TBST. The membrane was then incubated in ECLA reaction buffer (0.1 M Tris-HCl, pH 8.5, 25 μM luminol, 4 μM p-coumaric acid) with an equal volume of ECLB reaction buffer (0.064% [vol/vol] H2O2 in 0.1 M Tris-HCl, pH 8.5) for 2 min and then detected by the enhanced chemiluminescence (ECL) method (Molecular Imager ChemiDoc XRS+; Bio-Rad, Hercules, CA). The results were analyzed by the relative intensity of the secreted protein level obtained from Image Lab Software.
MTT assay.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) method was used to determine the effect of compounds on the growth rate of host cells (48). HeLa cells (4,000 to 5,000) were seeded in 96-well plates (flat-bottom, tissue-culture-treated plates; Costar) in triplicate, incubated in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) for 12 h, and then treated with compounds at final concentrations ranging from 12.5 to 100 μM. After 4 h of treatment, MTT solution (10 μl, 5 mg/ml in PBS; Sigma) was added to each well. Cells were incubated for 3 h at 37°C, and then the supernatant was discarded. DMSO (200 μl; Sigma) was added to each well, and the wells were incubated for 10 min at 37°C. The absorbance at 570 nm was measured using a microplate reader (Bio-Rad 680). The data represented the percent viability compared to that of the control. All the experiments were done in triplicate.
Gentamicin protection assay.
To determine the effect of compounds on S. enterica serovar Typhimurium invasion of host cells, a gentamicin protection assay was performed according to the method described before (37) with minor modifications. S. enterica serovar Typhimurium was cultured from 25°C to 37°C to induce SPI-1 as described in reference 36. HeLa cells (2 × 104; in DMEM, 10% FBS) were seeded in 24-well plates (flat-bottom, tissue-culture-treated plates; Costar), and the plates were incubated for 12 h at 37°C and 5% CO2. Meanwhile, S. enterica serovar Typhimurium cells were cultured overnight at 25°C with agitation. The overnight culture of S. enterica serovar Typhimurium was diluted 10-fold in LB in the presence or absence of compounds at a final concentration of 100 μM and cultured for 1 h at 37°C. HeLa cells were precultured with DMEM without FBS for 30 min and then infected with S. enterica serovar Typhimurium at a multiplicity of infection (MOI) of 10. One hour after infection at 37°C and 5% CO2, the culture medium was discarded and the cells were washed with PBS. DMEM supplemented with 100 μg/ml gentamicin was added to each well to kill noninvasive S. enterica serovar Typhimurium cells, and incubation was continued for another 1 h. After incubation, the HeLa cells were washed three times with PBS and lysed with 1% Triton X-100 solution. The CFU of bacteria were counted by plating the appropriate dilution in LB with 0.2% l-arabinose. All experiments were done in triplicate.
Measurement of bacterial growth.
S. enterica serovar Typhimurium χ8956 was cultured in LB supplemented with 0.2% l-arabinose at 37°C and then diluted at 1:100 in new LB broth and incubated for 5 h at 37°C in the presence or absence of compounds. At the time points indicated, the OD570 of the culture was measured using a microplate reader (Bio-Rad 680). Two independent experiments were performed, and three replicate samples were used in each experiment.
Bacterial cell fractionation.
S. enterica serovar Typhimurium cells were cultured and treated by compounds or DMSO under the same procedure described under “Isolation and detection of secreted proteins.” The cells were pelleted by centrifugation at 12,000 × g for 5 min, and the culture supernatant was collected to detect extracellular proteins. The pellet was resuspended in PBS with the same volume of LB medium and ultrasonicated until the opaque suspension became clear. The lysis mixture was centrifuged at 12,000 × g for 5 min and divided into two fractions: intracellular soluble proteins and cell debris. A bacterial culture with the same volume as that fractionated previously was directly ultrasonicated until the opaque suspension was clear. The lysis mixture was centrifuged at 12,000 × g for 5 min, and the supernatant was collected for detection of total soluble proteins. To study whether the effect of Csn-B on SPI-1 was linked to protein degradation, different protease inhibitors were used in this assay. The protease inhibitor PMSF at a final concentration of 1 mM or leupeptin at a final concentration of 20 μg/ml was added to the PBS-resuspended solution of pellets. Proteins from different fractions were gathered under the same procedure described earlier. Proteins SipC and FliC in different fractions were analyzed by Western blots.
Real-time quantitative PCR (qPCR).
To study the induced or repressed genes by Csn-B, the relative transcriptional levels were determined by real-time PCR. Primers (see Table S1 in the supplemental material) used in this study were designed by Primer Premier 5.0 software. An overnight culture of S. enterica serovar Typhimurium at 25°C with agitation was diluted 100-fold in LB (0.2% l-arabinose) and grown in the presence of 100 μM Csn-B or DMSO at the same volume for 4 h at 37°C. The overnight culture was diluted 100-fold and continued to grow for 4 h at 25°C with agitation as the noninduced group. S. enterica serovar Typhimurium cells were collected, and total bacterial RNA was isolated using a TRIzol Plus RNA purification kit (Invitrogen). The cDNA was synthesized using the RevertAid first-strand cDNA synthesis kit (Fermentas) with random hexamer as a primer following the manufacturer's instructions. The cDNA (25 ng) was amplified by the use of SYBR Premix Ex Taq (TaKaRa, Dalian, China) and 10-pmol primers for target sequences on a real plex2 Mastercycler (Eppendorf, Hamburg, Germany). After completion of 35 PCR cycles, melt curve data were generated to ascertain template-independent amplification. The comparative CT (cycle threshold) method was used for analysis of relative changes in transcriptional levels (49). The change in gene transcription was calculated using the formula sample/control = 2−ΔΔCT, where ΔΔCT is the ΔCT of the sample minus the ΔCT of the control and ΔCT is the CT of the target gene minus the CT of the 16S rRNA (internal control) (49). All PCR experiments were performed with three replicates to obtain standard deviations.
Gene cloning and overexpression of HilA.
To construct the HilA-overexpressing strain, the gene sequence ranging from 15 bp upstream of the hilA initiation codon to its stop codon was amplified from genomic DNA of S. enterica serovar Typhimurium UK-1 χ8956 by PCR. The primers used were as follows: hilA forward, 5′-CGGAATTCGAATACACTATTATCATGC-3′ (EcoRI); hilA reverse, 5′-GCTCTAGATTACCGTAATTTAATCAAGC-3′ (XbaI). Plasmid pKD46 was digested by ClaI and EcoRI to obtain the araC pBAD activator-promoter sequence (50); at the same time, plasmid pWSK29 (51) was digested by ClaI and XbaI. Then these three DNA fragments were purified by a gel extraction kit (OMEGA) and ligated by a DNA ligation kit (TaKaRa, Dalian, China) to construct plasmid pBAD-hilA-pWSK29. The constructed plasmid was introduced into electrocompetent S. enterica serovar Typhimurium UK-1 χ8956 to obtain a HilA-overexpressing strain. The strain was cultured in LB broth supplemented with l-arabinose (0.02%) and ampicillin (100 μg/ml) to induce the overexpression of HilA.
Statistical analysis.
Means and standard deviations were calculated using GraphPad Prism 5 (GraphPad Software, La Jolla, CA), and the statistical significance was determined using the two-way analysis of variance (ANOVA) method in this software.
RESULTS AND DISCUSSION
Csn-B and its derivatives inhibited the secretion of SPI-1 effector proteins and invasion of S. enterica serovar Typhimurium into HeLa cells.
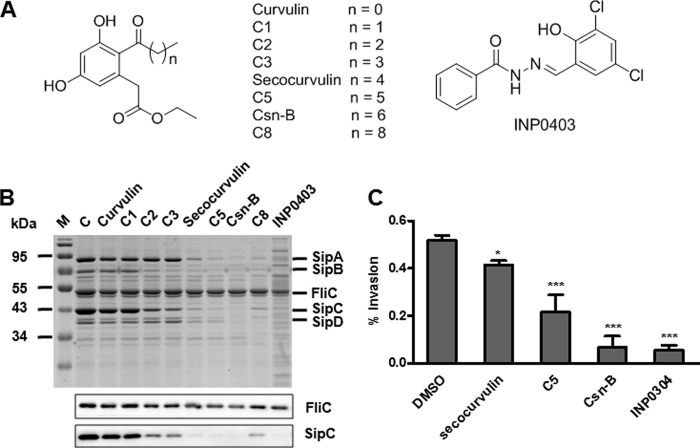
Our screening work was based on the fact that S. enterica serovar Typhimurium secreted SPI-1 effector proteins into the culture supernatant when they were grown between 25°C and 37°C, the SPI-1-induced condition in vitro (36). INP0403 (Fig. 1A; see Scheme S1 in the supplemental material), which strongly affected T3SS of several Gram-negative bacteria (31, 36–39), was chosen as the positive-control compound. Through screening the chemical library of our laboratory by SDS-PAGE assay, we found that several compounds showed inhibitory effects on the secretion of SPI-1 effectors (data not shown). Most interestingly, cytosporone B (Csn-B), an octaketide, exhibited a strong inhibitory effect on the secretion of SPI-1 effectors (SipA, SipB, SipC, SipD) without an evident effect on the flagellar protein FliC (Fig. 1B and Table 2; see Fig. S1 in the supplemental material). Csn-B, isolated from the endophytic fungus Dothiorella sp. strain HTF3, is a known agonist for nuclear orphan receptor Nur77 (52, 53). Several Csn-B derivatives synthesized in our lab (see Scheme S2 in the supplemental material) also had this kind of inhibitory activity (see Fig. S1). The identities of the protein bands in Fig. 1B, Fig. 2A, Fig. 3C, and Fig. S1 were determined by protein molecular weights, Western blots, and review of the literature (54). The results (Fig. 1A and B and Table 2) indicated that the alkyl chain linked to the ketone carbonyl group in Csn-B derivatives could be responsible for their activities on SPI-1. However, compound C8, with nine carbon atoms linked to the ketone group, exhibited weaker activity than Csn-B (Fig. 1A and B). Moreover, the modification of the phenyl acetic acid ester group of Csn-B derivatives with methyl ester or carboxylic acid (E0∼E6 and A2∼A5) (see Fig. S1) significantly decreased their effects, suggesting that the ester group may also be vital to the inhibitory effect. Overall, Csn-B showed the best inhibitory activity against the secretion of SPI-1 effectors.
Fig 1.
Csn-B and its derivatives inhibited the secretion of SPI-1 effectors and invasion of S. enterica serovar Typhimurium into HeLa cells. (A) Chemical structures of Csn-B derivatives and positive control INP0403. (B) Compounds inhibited secretion of SPI-1 effector proteins. SipA/B/C/D, SPI-1 effector proteins; FliC, flagellar filament protein; M, marker; C, DMSO control. (C) Compounds secocurvularin, C5, Csn-B, and INP0403 inhibited the invasion of S. enterica serovar Typhimurium into HeLa cells. The values represent percentages of invading bacteria as related to the initial input, and error bars indicate standard deviations from the means. The levels of statistical significance is indicated as follows: *, P ≤ 0.05; ***, P ≤ 0.001. The P value was calculated by comparing the value for the DMSO negative control.
Table 2.
Effects of Csn-B derivatives on the secretion of SipC in S. enterica serovar Typhimurium
| Compound no. | Name | % secretiona (mean ± SD) |
|---|---|---|
| 1 | Curvulin | 67.2 ± 3.6 |
| 2 | C1 | 58.3 ± 14.5 |
| 3 | C2 | 50.6 ± 8.2 |
| 4 | C3 | 35.2 ± 12.1 |
| 5 | Secocurvularin | 13.3 ± 4.1 |
| 6 | C5 | 10.5 ± 2.4 |
| 7 | Csn-B | 2.1 ± 1.4 |
| 8 | C8 | 16.2 ± 6.7 |
| 9 | E0 | 104.2 ± 12.6 |
| 10 | E1 | 99.5 ± 21.4 |
| 11 | E2 | 101.5 ± 3.4 |
| 12 | E3 | 80.2 ± 5.5 |
| 13 | E4 | 53.5 ± 16.7 |
| 14 | E5 | 64.5 ± 6.5 |
| 15 | E6 | 28.3 ± 10.8 |
| 16 | A2 | 60.3 ± 6.4 |
| 17 | A3 | 96.3 ± 3.7 |
| 18 | A4 | 92.5 ± 9.6 |
| 19 | A5 | 70.3 ± 19.0 |
| 20 | INP0403 | 7.1 ± 1.3 |
% secretion: the percentage of SipC protein from culture supernatants treated by different compounds was calculated by Western blots compared with DMSO control for three independent replicates, and standard deviations are shown. Compounds were tested at a final concentration of 100 μM.
Fig 2.

Csn-B inhibited the secretion of SPI-1 effectors in a dose-dependent manner and did not affect growth of S. enterica serovar Typhimurium. (A) Csn-B inhibited secretion of SPI-1 effector proteins at a range of concentrations from 6.25 to 100 μM under an SPI-1-induced condition in vitro. Culture supernatant proteins were subjected to 12% SDS-PAGE followed by Coomassie blue staining or Western blotting. M, marker; C, DMSO control. Inhibition percentage was calculated by comparing the SipC blot with that of the DMSO control. (B) Csn-B at a concentration of 100 μM did not significantly inhibit the bacterial growth.
Fig 3.
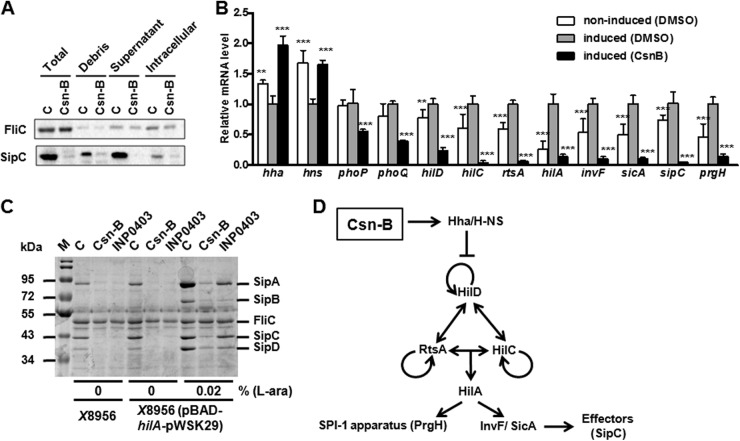
Csn-B inhibited SPI-1 effectors through the Hha–H-NS regulatory pathway. (A) Effects of Csn-B at a final concentration of 100 μM on SipC in different culture fractions were analyzed by Western blotting. Total, supernatant of bacterial culture after ultrasonic treatment, i.e., supernatant fraction and cytoplasmic fraction; Debris, cell debris of bacteria after ultrasonic treatment; Supernatant, culture supernatant; Intracellular, intracellular fraction of bacterial culture; C, DMSO control. (B) Effect of Csn-B on relative mRNA levels of the SPI-1 regulatory factors, effector gene, and apparatus protein gene. RNA was extracted from bacteria treated with Csn-B at a final concentration of 100 μM or by the same volume of DMSO under an SPI-1-induced condition in vitro or from bacteria treated with the same volume of DMSO under a noninduced condition. The y axis represents the relative transcriptional level of each gene (real-time qPCR) by the relative quantification method. Significant differences: **, P ≤ 0.01; ***, P ≤ 0.001. The P value was calculated by comparing the value for the DMSO control under an induced condition. (C) Effect of Csn-B on the secretion of SPI-1 effectors in the HilA-overexpressing strain. S. enterica serovar Typhimurium χ8956 or strain χ8956 carrying plasmid pBAD-hilA-pWSK29 was cultured in LB broth at 25°C overnight; then the cultures were diluted 10 times in LB broth or LB broth supplemented with l-arabinose (0.02%) and ampicillin (100 μg/ml). The diluted cultures were incubated with Csn-B or INP0403 at a final concentration of 100 μM at 37°C for 4 h. The proteins isolated from culture supernatants were analyzed by SDS-PAGE. M, Marker; C, DMSO control. (D) Regulatory pathway of Csn-B on SPI-1. Arrows indicate transcriptional activation. Blunt lines indicate transcriptional inhibition.
The compounds with strong inhibitory effects on the secretion of SPI-1 effectors were further evaluated in terms of their effects on SPI-1-mediated invasion, as the invasion of Salmonella into host cells was predominately related to SPI-1 (55, 56). Because the invasion assay was cell based, we first determined the cytotoxicity of the compounds secocurvularin, C5, and Csn-B on HeLa cells by MTT assay (see Fig. S2 in the supplemental material). The MTT results showed that these three compounds had no obvious toxic effects on cell growth at various concentrations during a 4-h incubation. Then their inhibitory effects on the invasion of Salmonella into HeLa cells were assessed in a gentamicin protection assay (Fig. 1C). The results indicated that all three compounds inhibited bacterial invasion compared to the DMSO control. As expected, their inhibitory activities in the invasion assay shared the same preliminary SAR as their effects on the secretion of SPI-1 effectors revealed by SDS-PAGE and Western blotting (Fig. 1B). Since all the results (Fig. 1B and C) showed that Csn-B exhibited the strongest inhibitory effect on SPI-1, it was chosen for further investigation.
Csn-B inhibited the secretion of SPI-1 effectors in a dose-dependent manner and did not affect bacterial growth.
The inhibitory rates of Csn-B at concentrations of 100, 50, and 25 μM were all higher than 90%, while those of 12.5 μM and 6.25 μM were substantially lower, only 78% and 59%, respectively (Fig. 2A). The results showed that Csn-B inhibited the secretion of SipC in a dose-dependent manner. Moreover, the 50% inhibitory concentration (IC50) of Csn-B was estimated to be less than 6.25 μM, implying that Csn-B was a strong inhibitor of SPI-1. On the other hand, Csn-B did not show an inhibitory effect on flagellar protein FliC, suggesting that Csn-B has no obvious influence on the flagella of the pathogen.
To determine whether the inhibitory activity of Csn-B on SPI-1 resulted from directly killing the bacteria, the growth curve of S. enterica serovar Typhimurium in LB broth was developed in the presence or absence of Csn-B at 100 μM (Fig. 2B). No inhibitory effect on bacterial growth was observed at all time points examined; instead, a modest promoting effect was observed. Therefore, Csn-B at a final concentration of 100 μM was adopted for further investigation to study the mechanism of Csn-B's action on SPI-1.
Csn-B inhibited the secretion of SPI-1 effectors through the Hha–H-NS regulatory pathway.
The inhibitory level of Csn-B on SPI-1 effectors in different fractions of bacterial culture was deeply investigated here. A bacterial culture was fractionated into cell debris, culture supernatant, and intracellular fraction. The results (Fig. 3A) showed that Csn-B significantly decreased SipC levels in different fractions of bacterial culture, while it did not obviously influence the level of flagellar protein FliC in all fractions. It was speculated that the decrease of SipC level may be due to protein degradation or downregulation by Csn-B. Different protease inhibitors were added to bacterial culture fractions to identify whether or not the low SipC level resulted from degradation. There was no significant difference observed between DMSO control and protease inhibitor-treated samples (see Fig. S3 in the supplemental material). SipC was significantly inhibited in both the Csn-B group and the Csn-B-plus-protease-inhibitor group (see Fig. S3). The results suggested that the inhibitory effect of Csn-B on SPI-1 effectors was unlikely to be due to protein degradation. Instead, Csn-B is likely to affect the expression of SPI-1 effectors, based on the results of Fig. 3A (see Fig. S3).
To further determine the mechanism of Csn-B's action, the relative mRNA levels of several genes regulating the transcription of SPI-1 were assessed by real-time qPCR (Fig. 3B). HilA, the key regulator of SPI-1, combining with the promoters of AraC-like regulator genes invF and sicA, directly activates the transcription of its downstream genes, such as SPI-1 apparatus genes prg and org, inv and spa operon genes, and effector genes (57). In addition, AraC-like proteins HilC, HilD, and RtsA form a feed-forward regulatory loop to control the transcription of hilA. These three proteins can not only autoinduce the transcription of their genes, but also activate hilA independently (58). The mRNA levels of hilD, hilC, rtsA, and the downstream SPI-1 genes were obviously higher in the DMSO group that was induced by temperature change than in the noninduced DMSO group (Fig. 3B), which suggested that temperature change could activate the transcription of these genes. The results (Fig. 3B) were consistent with the results described before (37); i.e., temperature change can induce expression of SPI-1 effectors. On the other hand, Csn-B exhibited a remarkable inhibitory effect on transcription of these genes compared with the DMSO control under induced conditions, which showed that Csn-B inhibited the transcription of these genes to reduce the expression of SPI-1 effector proteins.
Furthermore, the effect of Csn-B on SPI-1 was analyzed in a Salmonella strain recombinantly overexpressing HilA, the key regulatory factor of SPI-1. No difference was observed (Fig. 3C) between the wild-type strain (Salmonella χ8956) and the HilA-overexpressing strain without l-arabinose induction, whereas the effector proteins SipA, SipB, SipC, and SipD were all upregulated in the HilA-overexpressing strain under the induction of 0.02% l-arabinose (Fig. 3C). On the other hand, the FilC protein hardly exhibited any difference between groups (Fig. 3C), which documented that HilA could not influence the expression of FliC. Csn-B strongly decreased the level of SPI-1 effectors in the HilA-overexpressing strain induced by l-arabinose, which further demonstrated that Csn-B inhibited the secretion of SPI-1 effectors via affecting the expression of SPI-1 regulator HilA (Fig. 3C). Interestingly, the positive-control compound INP0403 had an inhibitory effect on SPI-1 effectors as strong as that of Csn-B in both the wild-type strain and the HilA-overexpressing strain without induction of l-arabinose, whereas it exerted a significantly weaker inhibition on SPI-1 effectors in the HilA-overexpressing strain than Csn-B when induced by l-arabinose, which may suggest an INP0403 mechanism of inhibition other than the HilA pathway. Because compound INP0010 (D9), the analog of INP0403, could significantly affect transport of SPI-1 effectors as well as transcription of SPI-1 regulators (37), with further analysis the result of this research (Fig. 3C) may lead to the discovery that INP0403 has the same mechanism of action as INP0010.
Nucleoid proteins Hha and H-NS bond with the promoter regions of hilD, hilC, hilA, and rtsA to inhibit the transcription of SPI-1 genes and expression of SPI-1 effectors (59). In addition, proteins PhoP/and PhoQ directly or indirectly inhibit the transcription of hilA (60, 61). Comparing the induced group with the noninduced group, the results (Fig. 3B) indicated that temperature change led to downregulation of transcription of hns and hha genes, but no significant influence on transcription of phoP and phoQ was found, suggesting that nucleoid protein complex Hha–H-NS but not PhoP and PhoQ may be the predominant regulator of SPI-1 effector genes under temperature change. Nucleoid protein Hha undergoes a conformational change when binding to H-NS to form heterocomplexes (59, 62), while a conformational change of the H-NS dimer from 25°C to 37°C promotes the transcription of a large number of SPI-1-regulated genes (63). The results (Fig. 3B) were consistent with what was described in the literature (59, 62, 63). In this research, Csn-B evidently promoted the transcription of hns and hha, while it inhibited the transcription of phoP and phoQ (Fig. 3B). All in all, it could be concluded from the results (Fig. 3A, B, and C) that the Hha–H-NS–HilD–HilC–RtsA–HilA pathway, not the PhoP-PhoQ pathway, may be the main regulatory pathway for Csn-B to exert its activity on SPI-1 effector proteins (Fig. 3D).
Conclusions.
In this research, Csn-B and its derivatives exhibited inhibitory effects on the secretion of SPI-1 effectors. The preliminary SAR analysis suggested that the compounds with a long carbon chain and phenyl acetic acetate ester group could most likely result in strong activity. Csn-B affected the transcription of SPI-1-related genes through the Hha–H-NS–HilD–HilC–RtsA–HilA regulatory pathway. The results are important for understanding the molecular mechanism of these inhibitors' action on T3SS. Because T3SS is conserved in many Gram-negative pathogens, Csn-B and its derivatives may also have the same effect on other T3SSs. Thus, Csn-B could be a useful lead compound for development of new antibiotics targeting T3SS.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roy Curtiss III (School of Life Sciences, Arizona State University) for the S. enterica serovar Typhimurium strain and S. R. Kushner (Department of Genetics, University of Georgia) for plasmid pWSK29. We also express our gratitude to Liangcheng Du (Department of Chemistry, University of Nebraska—Lincoln) for helpful advice and critical reading of the manuscript.
This work was supported by 973 programs (2010CB833802 and 2012CB721005), the NSFC (31028019), and a Distinguished Young Scholars grant 30325044 to Yuemao Shen from the NSFC.
Footnotes
Published ahead of print 4 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02421-12.
REFERENCES
- 1. Aiello D, Williams JD, Majgier-Baranowska H, Patel I, Peet NP, Huang J, Lory S, Bowlin TL, Moir DT. 2010. Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54:1988–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kimura K, Iwatsuki M, Nagai T, Matsumoto A, Takahashi Y, Shiomi K, Omura S, Abe A. 2011. A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot. 64:197–203 [DOI] [PubMed] [Google Scholar]
- 3. Stamm LM, Goldberg MB. 2011. Microbiology. Establishing the secretion hierarchy. Science 331:1147–1148 [DOI] [PubMed] [Google Scholar]
- 4. Marlovits TC, Stebbins CE. 2010. Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Worrall LJ, Lameignere E, Strynadka NC. 2011. Structural overview of the bacterial injectisome. Curr. Opin. Microbiol. 14:3–8 [DOI] [PubMed] [Google Scholar]
- 6. Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun GW, Gan YH. 2010. Unraveling type III secretion systems in the highly versatile Burkholderia pseudomallei. Trends Microbiol. 18:561–568 [DOI] [PubMed] [Google Scholar]
- 8. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 9. Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M. 2004. Conserved features of type III secretion. Cell.Microbiol. 6:805–816 [DOI] [PubMed] [Google Scholar]
- 10. Blocker A, Komoriya K, Aizawa S. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U. S. A. 100:3027–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Layton AN, Hudson DL, Thompson A, Hinton JC, Stevens JM, Galyov EE, Stevens MP. 2010. Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302:114–122 [DOI] [PubMed] [Google Scholar]
- 12. Patel JC, Rossanese OW, Galan JE. 2005. The functional interface between Salmonella and its host cell: opportunities for therapeutic intervention. Trends Pharmacol. Sci. 26:564–570 [DOI] [PubMed] [Google Scholar]
- 13. Keyser P, Elofsson M, Rosell S, Wolf-Watz H. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17–29 [DOI] [PubMed] [Google Scholar]
- 14. Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swietnicki W, Carmany D, Retford M, Guelta M, Dorsey R, Bozue J, Lee MS, Olson MA. 2011. Identification of small-molecule inhibitors of Yersinia pestis type III secretion system YscN ATPase. PLoS One 6:e19716 doi:10.1371/journal.pone.0019716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crabill E, Joe A, Block A, van Rooyen JM, Alfano JR. 2010. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 154:233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gauthier A, Finlay BB. 2002. Type III secretion system inhibitors are potential antimicrobials. ASM News 68:383–387 [Google Scholar]
- 18. Marra A. 2006. Targeting virulence for antibacterial chemotherapy: identifying and characterising virulence factors for lead discovery. Drugs R. D. 7:1–16 [DOI] [PubMed] [Google Scholar]
- 19. Kauppi AM, Andersson CD, Norberg HA, Sundin C, Linusson A, Elofsson M. 2007. Inhibitors of type III secretion in Yersinia: design, synthesis and multivariate QSAR of 2-arylsulfonylamino-benzanilides. Bioorg. Med. Chem. 15:6994–7011 [DOI] [PubMed] [Google Scholar]
- 20. Gauthier A, Robertson ML, Lowden M, Ibarra JA, Puente JL, Finlay BB. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49:4101–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan NJ, Brady MJ, Leong JM, Goguen JD. 2009. Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miki T, Okada N, Shimada Y, Danbara H. 2004. Characterization of Salmonella pathogenicity island 1 type III secretion-dependent hemolytic activity in Salmonella enterica serovar Typhimurium. Microb. Pathog. 37:65–72 [DOI] [PubMed] [Google Scholar]
- 23. Linington RG, Robertson M, Gauthier A, Finlay BB, MacMillan JB, Molinski TF, van Soest R, Andersen RJ. 2006. Caminosides B-D, antimicrobial glycolipids isolated from the marine sponge Caminus sphaeroconia. J. Nat. Prod. 69:173–177 [DOI] [PubMed] [Google Scholar]
- 24. Linington RG, Robertson M, Gauthier A, Finlay BB, van Soest R, Andersen RJ. 2002. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4:4089–4092 [DOI] [PubMed] [Google Scholar]
- 25. Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241–249 [DOI] [PubMed] [Google Scholar]
- 26. Kauppi AM, Nordfelth R, Hagglund U, Wolf-Watz H, Elofsson M. 2003. Salicylanilides are potent inhibitors of type III secretion in Yersinia. Adv. Exp. Med. Biol. 529:97–100 [DOI] [PubMed] [Google Scholar]
- 27. Dahlgren MK, Zetterstrom CE, Gylfe S, Linusson A, Elofsson M. 2010. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg. Med. Chem. 18:2686–2703 [DOI] [PubMed] [Google Scholar]
- 28. Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 103:14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bailey L, Gylfe A, Sundin C, Muschiol S, Elofsson M, Nordstrom P, Henriques-Normark B, Lugert R, Waldenstrom A, Wolf-Watz H, Bergstrom S. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581:587–595 [DOI] [PubMed] [Google Scholar]
- 31. Slepenkin A, Enquist PA, Hagglund U, de la Maza LM, Elofsson M, Peterson EM. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75:3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu H, Slepenkin A, Elofsson M, Keyser P, de la Maza LM, Peterson EM. 2010. Candidate vaginal microbicides with activity against Chlamydia trachomatis and Neisseria gonorrhoeae. Int. J. Antimicrob. Agents 36:145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muschiol S, Normark S, Henriques-Normark B, Subtil A. 2009. Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol. 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slepenkin A, Chu H, Elofsson M, Keyser P, Peterson EM. 2011. Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J. Infect. Dis. 204:1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yount JS, Tsou LK, Dossa PD, Kullas AL, van der Velden AW, Hang HC. 2010. Visible fluorescence detection of type III protein secretion from bacterial pathogens. J. Am. Chem. Soc. 132:8244–8245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hudson DL, Layton AN, Field TR, Bowen AJ, Wolf-Watz H, Elofsson M, Stevens MP, Galyov EE. 2007. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51:2631–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Negrea A, Bjur E, Ygberg SE, Elofsson M, Wolf-Watz H, Rhen M. 2007. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 51:2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Veenendaal AK, Sundin C, Blocker AJ. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tree JJ, Wang D, McInally C, Mahajan A, Layton A, Houghton I, Elofsson M, Stevens MP, Gally DL, Roe AJ. 2009. Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77:4209–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dahlgren MK, Kauppi AM, Olsson I-M, Linusson A, Elofsson M. 2007. Design, synthesis, and multivariate quantitative structure-activity relationship of salicylanilides—potent inhibitors of type III secretion in Yersinia. J. Med. Chem. 50:6177–6188 [DOI] [PubMed] [Google Scholar]
- 41. Baron C. 2010. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13:100–105 [DOI] [PubMed] [Google Scholar]
- 42. Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, Bronstein PA, Kline T, Miller SI. 2008. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y, Peng Q, Selimi D, Wang Q, Charkowski AO, Chen X, Yang CH. 2009. The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microbiol. 75:1223–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamazaki A, Li J, Zeng Q, Khokhani D, Hutchins WC, Yost AC, Biddle E, Toone EJ, Chen X, Yang CH. 2012. Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob. Agents Chemother. 56:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhan MK, Bahl R, Bhatnagar S. 2005. Typhoid and paratyphoid fever. Lancet 366:749–762 [DOI] [PubMed] [Google Scholar]
- 46. Hensel M. 2004. Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 294:95–102 [DOI] [PubMed] [Google Scholar]
- 47. Curtiss R, III, Wanda S-Y, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, Mo H, Wang S, Kong W. 2009. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77:1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Preet R, Mohapatra P, Mohanty S, Sahu SK, Choudhuri T, Wyatt MD, Kundu CN. 2012. Quinacrine has anticancer activity in breast cancer cells through inhibition of topoisomerase activity. Int. J. Cancer 130:1660–1670 [DOI] [PubMed] [Google Scholar]
- 49. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 50. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 52. Brady SF, Wagenaar MM, Singh MP, Janso JE, Clardy J. 2000. The cytosporones, new octaketide antibiotics isolated from an endophytic fungus. Org. Lett. 2:4043–4046 [DOI] [PubMed] [Google Scholar]
- 53. Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, Li G, Xu Q, Zhang M, Weimer BC, Chen D, Cheng Z, Zhang L, Li Q, Li S, Zheng Z, Song S, Huang Y, Ye Z, Su W, Lin SC, Shen Y, Wu Q. 2008. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 4:548–556 [DOI] [PubMed] [Google Scholar]
- 54. Mizusaki H, Takaya A, Yamamoto T, Aizawa S. 2008. Signal pathway in salt-activated expression of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bueno SM, Wozniak A, Leiva ED, Riquelme SA, Carreno LJ, Hardt WD, Riedel CA, Kalergis AM. 2010. Salmonella pathogenicity island 1 differentially modulates bacterial entry to dendritic and non-phagocytic cells. Immunology 130:273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Collazo CM, Galan JE. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691–705 [DOI] [PubMed] [Google Scholar]
- 59. Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189:6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aguirre A, Cabeza ML, Spinelli SV, McClelland M, Garcia Vescovi E, Soncini FC. 2006. PhoP-induced genes within Salmonella pathogenicity island 1. J. Bacteriol. 188:6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Queiroz MH, Madrid C, Paytubi S, Balsalobre C, Juarez A. 2011. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology 157:2504–2514 [DOI] [PubMed] [Google Scholar]
- 62. De Alba CF, Solorzano C, Paytubi S, Madrid C, Juarez A, Garcia J, Pons M. 2011. Essential residues in the H-NS binding site of Hha, a co-regulator of horizontally acquired genes in Enterobacteria. FEBS Lett. 585:1765–1770 [DOI] [PubMed] [Google Scholar]
- 63. Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.