Abstract
High-throughput phenotypic screening against the yeast Saccharomyces cerevisiae revealed a series of triazolopyrimidine-sulfonamide compounds with broad-spectrum antifungal activity, no significant cytotoxicity, and low protein binding. To elucidate the target of this series, we have applied a chemogenomic profiling approach using the S. cerevisiae deletion collection. All compounds of the series yielded highly similar profiles that suggested acetolactate synthase (Ilv2p, which catalyzes the first common step in branched-chain amino acid biosynthesis) as a possible target. The high correlation with profiles of known Ilv2p inhibitors like chlorimuron-ethyl provided further evidence for a similar mechanism of action. Genome-wide mutagenesis in S. cerevisiae identified 13 resistant clones with 3 different mutations in the catalytic subunit of acetolactate synthase that also conferred cross-resistance to established Ilv2p inhibitors. Mapping of the mutations into the published Ilv2p crystal structure outlined the chlorimuron-ethyl binding cavity, and it was possible to dock the triazolopyrimidine-sulfonamide compound into this pocket in silico. However, fungal growth inhibition could be bypassed through supplementation with exogenous branched-chain amino acids or by the addition of serum to the medium in all of the fungal organisms tested except for Aspergillus fumigatus. Thus, these data support the identification of the triazolopyrimidine-sulfonamide compounds as inhibitors of acetolactate synthase but suggest that targeting may be compromised due to the possibility of nutrient bypass in vivo.
INTRODUCTION
The increasing incidence of life-threatening systemic fungal infections, associated with high mortality in immunocompromised patients, poses a significant unmet medical need. As part of a program to identify novel antifungal agents, we have run a phenotypic screen using Saccharomyces cerevisiae and tested hits for their activity spectrum in a panel of fungal pathogens. Further investigation of one such hit led to identification of a series of triazolopyrimidine-sulfonamide compounds with broad-spectrum antifungal activity. Yeast chemogenomic profiling indicated that the probable target of these compounds was ILV2, encoding acetolactate synthase (ALS), a key enzyme in the biosynthesis of branched-chain amino acids.
Amino acid biosynthesis pathways are potential antifungal targets as they are highly conserved in fungi, while humans lack biosynthetic pathways for de novo synthesis of “essential” amino acids. The branched-chained amino acid biosynthetic pathway (isoleucine, valine, and leucine) is of particular interest as auxotrophic mutants of both Candida albicans and Cryptococcus neoformans have been reported to show reduced virulence in mouse models (1, 2). Acetolactate synthase (ALS, also termed acetohydroxyacid synthase) catalyzes the first common step in branched-chained amino acid synthesis in fungi as well as in plants and bacteria (3, 4). Interestingly, several classes of commercially used herbicides, including sulfonylureas, imidazolinones, sulfonanilides, and some others, target this pathway in plants through the inhibition of ALS (4, 5). The triazolopyrimidine and sulfonanilide herbicides were shown to be submicromolar inhibitors of ALS from both plants and yeast (6, 7). We report here the identification and characterization of a structurally similar series of putative ALS inhibitors and their evaluation as potential leads for antifungal drug discovery.
MATERIALS AND METHODS
Fungal strains and medium conditions.
The following fungal strains were used in this study: Saccharomyces cerevisiae BY4741 (Open Biosystems catalog no. YSC1056), Saccharomyces cerevisiae ATCC 9763, Candida albicans ATCC 24433, Aspergillus fumigatus ATCC MYA-3627, Rhizopus oryzae ATCC 4621, and Cryptococcus neoformans ATCC 36556. The strains used in the expanded MIC panel are described in Table S1 in the supplemental material. For analysis of the amino acid rescue using defibrinated sheep blood (DSB), 5,000 conidia were spotted onto AMM (Aspergillus minimal medium minus supplement solution) agar (8) or water-agarose (1% agarose in distilled water) supplemented with 5% DSB (Remel, R54012) containing the indicated concentrations of compound 1. The plates were incubated at 35°C for 3 days before being photographed. Compounds were dissolved in 90% dimethyl sulfoxide (DMSO) and stored at 4°C for up to 8 weeks. Further scientific studies with the chemical and biological material published in this study are encouraged and can be obtained under material transfer agreement.
Primary screen and antifungal susceptibility testing.
The primary compound screen against S. cerevisiae was performed as described previously (9). Antifungal susceptibility testing was performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution M27-A3 and M38-A2 (10, 11). Testing was performed in RPMI 1640 (SH30011.03; HyClone) with 2.05 mM glutamine and phenol red, without bicarbonate, and buffered with 0.165 mol/liter 3-(N-morpholino)propanesulfonic acid (MOPS) (A1076,0250; AppliChem). The medium was adjusted to pH 7.0 and filter sterilized. Where indicated, assay medium was supplemented with the following: 10% human serum (S7023, 115K89091; Sigma), 10% fetal calf serum (R92157; Remel), 10% fetal bovine serum (FBS) (10082-139; Gibco), 10% bovine serum albumin (BSA) (BP671-10; Fisher), leucine (328-39-2; Acros Organics), isoleucine (73-32-5; Acros Organics), and/or valine (72-18-4; Acros Organics). Two endpoints were recorded: MIC-0 (complete suppression of visible fungal growth) and MIC-2 (prominent, >50% growth inhibition). All experiments were repeated at least 3 times with similar results.
Assay for mammalian cell toxicity.
The WST-1 cytotoxicity and proliferation assay was performed according to manufacturer's instructions (Roche catalog no. 11 644 807 001) to test compounds for growth inhibition over 72 h in the mammalian cell lines K562 (CRL-1573) and HepG2 (HB-8065).
Chemogenomic profiling.
The growth-inhibitory potency of test substances was determined using wild-type S. cerevisiae strain BY4743. Optical density at 600 nm (OD600) values of exponentially growing S. cerevisiae cultures in rich medium were recorded with a robotic system. Twelve-point serial dilutions were assayed in 96-well plates with a reaction volume of 150 μl. Solutions containing dimethyl sulfoxide (DMSO) were normalized to 2%. Thirty percent inhibitory concentration (IC30) values were calculated using logistic regression curve fits generated by TIBCO Spotfire v3.2.1 (TIBCO Software, Inc.).
The Saccharomyces haploinsufficiency profiling (HIP) and homozygous profiling (HOP) and microarray analysis were performed as published previously (12). The basic concept behind this assay is that HIP identifies genes where one functional copy, compared to two, confers hypersensitivity to inhibition by the compound. This indicates pathways directly affected by the compound. HOP (with both gene copies deleted) indicates synthetic lethality and identifies compensating pathways to those directly affected by the compound. Thus, genome-wide hetero- and homozygous deletion libraries of S. cerevisiae strains were purchased (OpenBiosystems, catalog no. YSC1056 and YSC1055) and pools were constructed as published previously (12). Each HIP strain is heterozygous and each HOP strain completely null for one gene (with each strain being identified by a unique DNA sequence, called a “bar-code” or “tag” inserted into the deleted gene). For each HIP and HOP experiment, each test substance was assayed in duplicate (2 wells) at its IC30, in 24-well plates (Greiner 662102), with 1,600 μl/well yeast extract-peptone-dextrose (YPD). DMSO was normalized to 2%. At the onset of the HIP experiment ∼250 yeast cells/strain (100 μl of a 1.5-OD600/ml culture) from an overnight log-phase preculture were added to the compound- and DMSO-containing wells. Plates were incubated for 16 h in a robotic shaking incubator at 30°C at 550 rpm allowing for ∼5 doublings. Approximately 250 yeast cells/strain (120 μl of a 1.2-OD600/ml culture) were subsequently transferred into a new 24-well plate. Once inoculated, the new plate was incubated at 30°C at 550 rpm to allow the next 5 yeast generations (generations 6 to 10). This procedure was repeated twice more until the final plates containing the yeast with ∼20 generations were stored at 4°C. The HOP assay was performed similar to the HIP experiment, but the duration was reduced to 16 h/∼5 doublings, and thus no back-dilutions were necessary. Before the experiment, aliquots of the HOP pool were thawed and recovered for 3 h in YPD, and then ∼320 yeast cells/strain (110 μl of a 1.5-OD600/ml culture) were transferred into each well. The cell material from the final HIP and HOP plates was harvested, the genomic DNA (gDNA) was extracted, and the tags were amplified as published previously (12). Next, the relative abundance of each strain in the compound-treated wells was compared to that of eight no-drug control samples that were produced in the same experiment.
For the experimental analysis, we used the same computation of normalized tag intensities, outlier masking, and saturation correction published previously (12). Sensitivity was computed as the median absolute deviation logarithmic (MADL) score for each compound-concentration combination, and then gene-wise z-scores (across all experiments), which are based on a robust parametric estimation of gene variability allowing for up to 15% outliers, were computed as described in detail by D. Hoepfner et al. (Dominic Hoepfner, Stephen B. Helliwell, Heather Sadlish, Sven Schuierer, Ireos Filipuzzi, et al., submitted for publication). The similarity of the obtained HOP profiles was compared by calculating the Pearson correlation coefficients of a compound HOP profile with all other compound profiles. In order to assess the significance of the correlation coefficients, we applied the Fisher z′ transformation and fitted a normal distribution to the transformed values. This empirical normal distribution allowed us to assign a P value to each correlation coefficient. The (negative) logarithm of the false discovery rate (FDR)-corrected P value is then displayed. The complete data set for all compounds presented in this study is provided in the supplemental material (File S1). The raw microarray data will be provided upon request.
Selection of drug-resistant S. cerevisiae.
Strain BY4743Δ8, derived from BY4741 but with eight genes involved in drug resistance deleted (efflux pump genes SNQ2, PDR5, and YOR1 and transcription factor genes PDR1, PDR2, PDR3, YAP1, YRM1) was incubated in 2.5% ethyl methanesulfonate until only 50% of the cells formed colonies. A total of 2 × 107 mutagenized cells were plated on two 14-cm dishes with synthetic complete medium (0.7 g/liter Difco yeast nitrogen base without amino acids, 0.79 g/liter MPbio CSM amino acid mixture, 2% glucose) containing 100 nM compound 1. After 4 days, 13 resistant colonies with mutations in ILV2 (as identified by direct sequencing) could be isolated. Resistance by mutated ILV2 was confirmed by cloning the corresponding mutations into fresh BY4743Δ8 cells and recording dose-response curves in YPD medium with serial dilutions of compound 1, chlorimuron-ethyl, and sulfometuron-methyl (PS1081, 34224; Sigma-Aldrich) at a 200 μM maximum concentration and 11 serial dilutions. DMSO was normalized to 2%. Curves were calculated by taking the 22-h OD600 measurements and applying a logistic regression curve fit.
In silico docking.
The published S. cerevisiae Ilv2-chlorimuron-ethyl cocrystal structure (13) was used to perform the molecular docking of compound 1. The docking was performed using ICM (Molsoft, version 3.6-1) using default parameters. The obtained docking mode has been further minimized in the binding pocket using Macromodel (implemented in Maestro, version 9.3) using OPLS_2005 force field and implicit water. The digital file is available on the Dryad website (http://dx.doi.org/10.5061/dryad.qb753).
Amino acid rescue experiments.
OD600 values of exponentially growing S. cerevisiae cultures in YPD medium were recorded with a robotic system. Twelve-point serial dilutions were assayed in 96-well plates with a reaction volume of 150 μl. Solutions containing DMSO were normalized to 2%. Logistic regression curve fits were used to generate dose-response curves using TIBCO Spotfire v3.2.1 (TIBCO Software, Inc.). Cells were grown in YPD medium in the absence or presence of additional branched-chain amino acids (50 mM valine, 50 mM leucine, 50 mM isoleucine), and the Erg11 inhibitor voriconazole (PZ0005; Sigma-Aldrich) was used as a control.
RESULTS
Triazolopyrimidine-sulfonamides have broad-spectrum in vitro activity and minimal cytotoxicity.
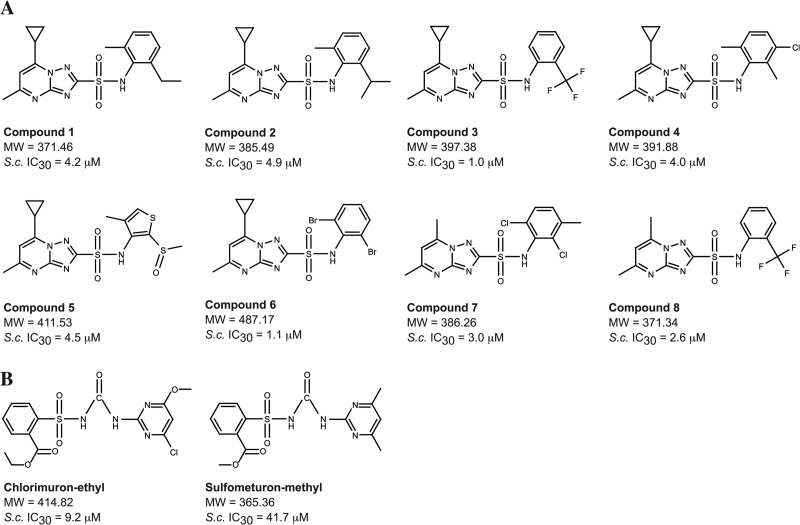
To identify novel compounds with antifungal activity, high-throughput screening was performed using the Novartis compound archive against S. cerevisiae cells in a miniaturized 1,536-well plate assay (9). Compound 1 showed complete growth inhibition at the screening concentration of 20 μM. Using compound 1 as a lead tool molecule, similarity searches revealed 7 structurally similar substances of the triazolopyrimidine-sulfonamide class (Fig. 1), which had not been part of the primary screening collection. All compounds displayed low μM IC30s for S. cerevisiae (Fig. 1) and significant broad-spectrum antifungal activity in vitro, as shown in Table 1. In most cases, the compounds achieved a complete suppression of fungal growth (MIC-0). Robustness of the obtained MIC values was confirmed when we repeated testing on a larger panel of strains, mostly clinical isolates (for details, see Tables S1 and S2 in the supplemental material). Additionally, the WST-1 cytotoxicity and proliferation assay demonstrated that all 8 compounds had no significant cytotoxicity activity against two common cell lines used for measuring the cytotoxicity and antiproliferative properties of xenobiotics (Table 2). These characteristics, coupled with the observation that the substances fall nicely within the drug-like chemical space, as predicted by Lipinski (not more than 5 hydrogen bond donors, not more than 10 hydrogen bond acceptors, a molecular mass below 500 Da, and an octanol-water partition coefficient log P [measure of lipophilicity] not greater than 5) (14), suggested an attractive starting point for an antifungal program.
Fig 1.
Compounds used in this study. (A) Structures and S. cerevisiae (S.c.) IC30s of substances tested. (B) Reference substances with a known target.
Table 1.
MICs of identified Ilv2p inhibitors
| Compound | MIC (μg/ml) of Ilv2p inhibitor fora: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
S. cerevisiae NF3201 |
C. albicans NF2103 |
A. fumigatus NF4905 |
R. oryzae NF4801 |
C. neoformans NF3102 |
||||||
| MIC-2 | MIC-0 | MIC-2 | MIC-0 | MIC-2 | MIC-0 | MIC-2 | MIC-0 | MIC-2 | MIC-0 | |
| 1 | 4 | 4 | 1 | 4 | 4 | 4 | 1 | 1 | 2 | 8 |
| 2 | 4 | 8 | >128 | >128 | 4 | 16 | 2 | ≤0.25 | 0.5 | 16 |
| 3 | 8 | 16 | 32 | >128 | 4 | 8 | 8 | 16 | 8 | >64 |
| 4 | 8 | 16 | 4 | 16 | 16 | 16 | 2 | 2 | 8 | 64 |
| 5 | 4 | 8 | 2 | 16 | 8 | 8 | 4 | 4 | 8 | >64 |
| 6 | 4 | 8 | 8 | 16 | 4 | 8 | 16 | 16 | >64 | >64 |
| 7 | 8 | 16 | 32 | 64 | 64 | 64 | >128 | >128 | 32 | >128 |
| 8 | 32 | 64 | 64 | >128 | 64 | 64 | >128 | >128 | 32 | 128 |
MIC-0, complete suppression of visible fungal growth; MIC-2, >50% growth inhibition.
Table 2.
Comparison of the cytotoxicity values of compound 1, compounds 2 to 7, and puromycin
| Compound or puromycin | CC50a in μg/ml (μM) of compound 1 for cell line: |
|||||||
|---|---|---|---|---|---|---|---|---|
| K562 |
HepG2 |
|||||||
| 1% FBS | 2% FBS | 5% FBS | 10% FBS | 1% FBS | 2% FBS | 5% FBS | 10% FBS | |
| Compounds | ||||||||
| 1 | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) |
| 2 | >128 (>332) | >128 (>332) | >128 (>332) | >128 (>332) | >128 (>332) | >128 (>332) | >128 (>332) | >128 (>332) |
| 3 | >128 (>322) | >128 (>322) | >128 (>322) | >128 (>322) | >128 (>322) | >128 (>322) | >128 (>322) | >128 (>322) |
| 4 | 77 (198) | >128 (>327) | 90 (230) | 95 (243) | >128 (>327) | >128 (>327) | >128 (>327) | >128 (>327) |
| 5 | >128 (>311) | >128 (>311) | >128 (>311) | >128 (>311) | >128 (>311) | >128 (>311) | >128 (>311) | >128 (>311) |
| 6 | >128 (>263) | >128 (>263) | >128 (>263) | >128 (219) | >128 (>263) | >128 (>263) | >128 (>263) | >128 (>263) |
| 7 | >128 (>331) | >128 (>331) | >128 (>331) | >128 (>331) | >128 (>331) | >128 (>331) | >128 (>331) | >128 (>331) |
| 8 | >128 (>345) | >128 (>345) | >128 (>345) | >128 (>345) | NDb | ND | ND | ND |
| Puromycin | 0.39 (0.83) | 0.39 (0.83) | 0.28 (0.60) | 0.16 (0.34) | 0.36 (0.77) | 0.34 (0.73) | 0.32 (0.69) | 0.24 (0.51) |
CC50, 50% cytotoxic concentration.
ND, not determined.
Chemogenomic profiling identifies acetolactate synthase Ilv2p as potential target.
While target identification is not absolutely essential for drug development, it facilitates the optimization of a compound's inhibitory activity. To investigate the mechanism of action, we subjected all 8 compounds of the series to chemogenomic profiling using the S. cerevisiae heterozygous and homozygous deletion collections. Together, haploinsufficiency profiling (HIP) and homozygous profiling (HOP) are a gene-dosage-dependent method that assesses the effect of compounds against potential targets encoded by the S. cerevisiae genome (15). HIP indicates pathways directly affected by the compound. HOP (with both gene copies deleted) indicates synthetic lethality and identifies compensating pathways to those directly affected by the compound.
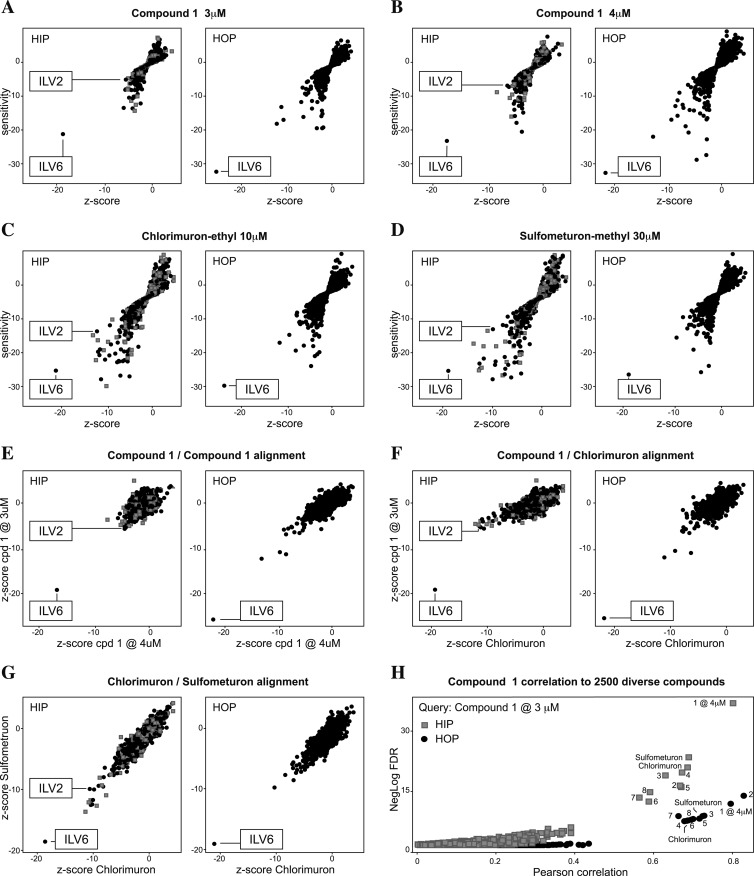
Compound 1 was tested in two independent experiments at slightly different concentrations (3 and 4 μM), and both experiments yielded very similar results (Fig. 2A and B). The heterozygous ILV6/ilv6 strain showed the most pronounced hypersensitivity against compound 1 at both concentrations tested. ILV6 codes for the regulatory subunit of ALS (16). Interestingly, ILV6 was also the most pronounced hit in the HOP experiments. As in the HOP strains, the genes are completely deleted; this indicates that Ilv6p cannot be the target of compound 1 as the protein is not present in the ilv6/ilv6 HOP strain. However, it indicates that ILV6 scores synthetic genetic interactions with the compound target. Such interactions are published for the ILV6/ILV2 pair: Ilv6p regulates activity of the catalytic subunit of ALS, encoded by ILV2 (16, 17). The ILV2 HIP strain did not score as hypersensitive as ILV6, but its sensitivity was scored as relevant by the applied z-score (<−5). Profiling of the established Ilv2p sulfonylurea inhibitors chlorimuron-ethyl and sulfometuron-methyl (Fig. 2C and D) provided further evidence for Ilv2p being the primary target of compound 1 as both substances produced HIP-HOP profiles that were very similar to those of compound 1, with ILV6 scoring in HIP and HOP and ILV2 scoring by the applied z-score in HIP (Fig. 2E and F). Extension of testing to the other 7 compounds resulted in profiles that all shared these characteristics (http://dx.doi.org/10.5061/dryad.qb753). These profiles were specific to this class of compounds: comparison of the profile of compound 1 tested at 3 μM to our database of 2,500 diverse chemical substances identified the compounds from this study, including the established ALS inhibitors, as a distinct group with considerably better clustering than any other profile in our database (Fig. 2H).
Fig 2.
HIP-HOP profiling results suggest inhibition of acetolactate synthase Ilv2p. (A and B) HIP-HOP profiles of compound 1 at the indicated concentrations. The strains with deletions in the genes coding for the catalytic (ILV2) and regulatory (ILV6) subunits of acetolactate synthase are labeled. (C and D) HIP-HOP profiles of reference compounds known to inhibit acetolactate synthase by binding to Ilv2p. (E to G) Pairwise alignment of compound 1 to its replicate experiment and the reference compounds. (H) Correlation of the HIP-HOP profile of compound 1 tested at 3 μM to HIP-HOP profiles of 2,500 diverse chemical structures as plotted by correlation and relevance. Note that the substances discussed in this article are highlighted as a distinct group with significantly better clustering than any of the other tested substances. Gray boxes represent strains with deletions in essential genes, and black dots represent strains with deletions in nonessential genes. Profile data with all strains annotated are provided in File S1 in the supplemental material.
Mutagenesis identifies resistance-conferring mutations in Ilv2p at the chlorimuron-ethyl binding pocket.
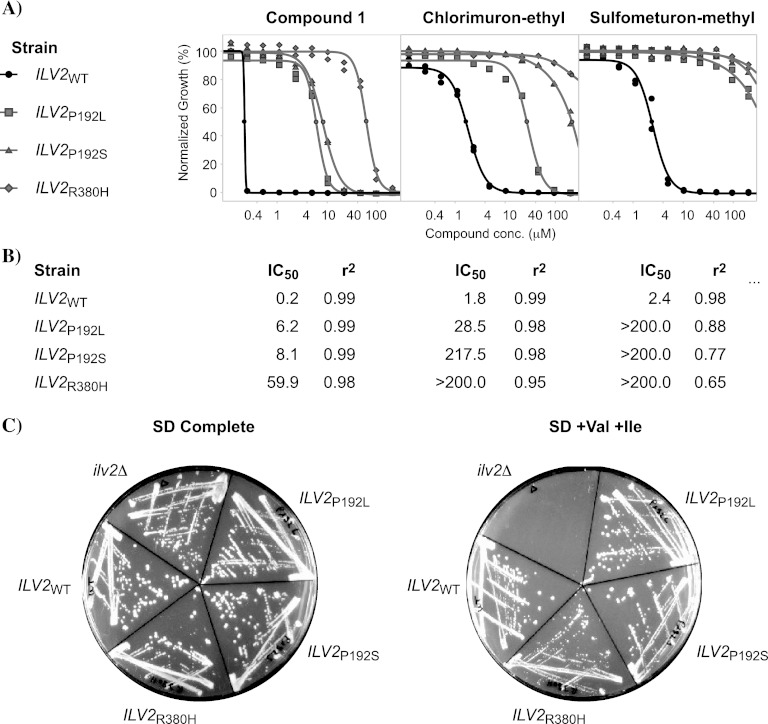
To further strengthen the observed genetic link between the S. cerevisiae ALS and compound 1, we examined whether mutations that confer compound resistance would map into the ILV2 gene. We used BY4741Δ8 cells, with 8 genes involved in drug resistance deleted (see Materials and Methods), as they were exquisitely sensitive to compound 1. The cells were mutagenized with ethyl methanesulfonate and then selected against 100 nM compound 1, a concentration that completely inhibited growth of wild-type cells on plates. After 72 h, colonies appeared on plates and direct sequencing revealed nonsynonymous mutations in the ILV2 gene, whereas the ILV6 gene was wild type. We identified 6 clones with the P192L mutation, 3 clones with the P192S mutation, and 4 clones with the R380H mutation. To validate these mutations and to exclude the contribution of second site mutations, we engineered all three mutations into new BY4741Δ8 wild-type cells and tested for resistance by measuring dose-response curves (Fig. 3A). All mutations confirmed and shifted the IC50 between 30- and 300-fold (Fig. 3B). When the validation was extended to chlorimuron-ethyl and sulfometuron-methyl, the mutations also mediated significant cross-resistance to these established Ilv2p sulfonylurea inhibitors (Fig. 3A and B). The identified mutations not only yield resistance but also retain functionality of the Ilv2 protein, as shown by the ability to support growth on media lacking valine and isoleucine (Fig. 3C).
Fig 3.
Validation of identified resistance-conferring mutations and testing for cross-resistance to known acetolactate synthase inhibitors. (A) IC50 curves based on duplicates of BYΔ8 strains with the introduced ILV2 point mutations indicated. (B) IC50s and r2 curve parameters for the tested substances. (C) The identified ILV2 point mutations result in a functional Ilv2 protein, as shown by the ability to grow on medium lacking valine and isoleucine.
In silico docking allows the placement of compound 1 into the Ilv2p chlorimuron-ethyl binding pocket.
The genetic data indicated that Ilv2p is the primary binding target of the triazolopyrimidine-sulfonamide compounds presented in this study. The observed cross-resistance of identified mutations with sulfonylurea inhibitors could suggest a common binding site. As cocrystal structures of S. cerevisiae Ilv2p with chlorimuron-ethyl and sulfometuron-methyl have been solved (13, 16, 18), we first mapped the position of the resistance-conferring sites relative to the bound inhibitors. The Ilv2p crystal structure is published as a homodimer with two active sites formed at the dimer interface. The identified positions from the mutagenesis study (P192 and R380) outline the published inhibitor-binding pocket (Fig. 4A and B). Analyzing the published data for chlorimuron-ethyl showed that the sulfonyl and keto group of the cocrystallized inhibitor formed H bonds with R380. P192 forms hydrophobic interactions with the phenyl group of the crystallized ligand. In addition to these interactions, the pyrimidine moiety occupies a hydrophobic region formed by W586, F201, M582, and V583. As a next step, we performed molecular docking of compound 1 into the pocket (Fig. 4C). In Fig. 4D, we show the superimposition of the docked compound 1 with the X-ray ligand. Compound 1 could fit in the binding pocket in a similar way to the cocrystallized ligand. The interactions of compound 1 with the neighboring amino acids are very similar to those of the sulfonylurea inhibitors: the compound 1 sulfonamide group forms an H bond with arginine 380, the triazolopyrimidine group occupies the same hydrophobic region as the X-ray pyrimidine group, and the phenyl group forms hydrophobic contacts with P192. (The Protein Data Bank [PDB] file of the docking model is provided at http://dx.doi.org/10.5061/dryad.qb753.)
Fig 4.
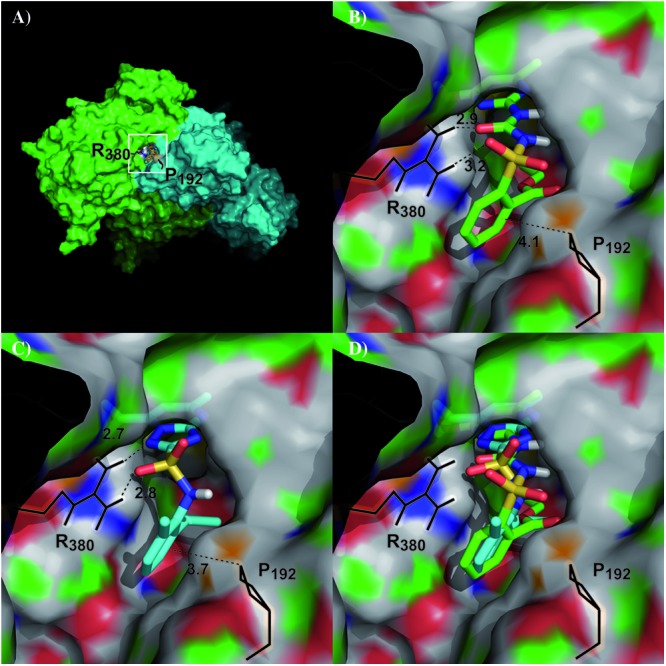
Mapping of identified resistance-conferring residues into the Ilv2 protein structure and in silico docking of compound 1. (A) The structure of the yeast Ilv2 protein was solved and published as a dimeric complex forming two inhibitor binding sites at the interface of the two chains at opposite poles (PDB no. 1N0H, with chain A represented in green, chain B in cyan, and the visible binding site labeled). Residues 192 and 380 identified in the resistance screen point toward the cocrystallized inhibitor chlorimuron-ethyl and outline the opening of the binding pocket. (B) Close-up (white square in panel A) of the Ilv2 dimer with the cocrystallized chlorimuron-ethyl inhibitor and the identified amino acid positions that can yield resistance when mutated. (C) Identified docking solution of compound 1 in the chlor1muron ethyl-binding pocket. Compound 1 (cyan) was aligned with chlorimuron-ethyl (green) and docked into the binding pocket. (D) Comparison of identified docking solution of compound 1 (cyan) with the cocrystallized ligand chlorimuron-ethyl (green). All distances are in Å.
Addition of exogenous amino acids or serum abrogates antifungal activity of compound 1.
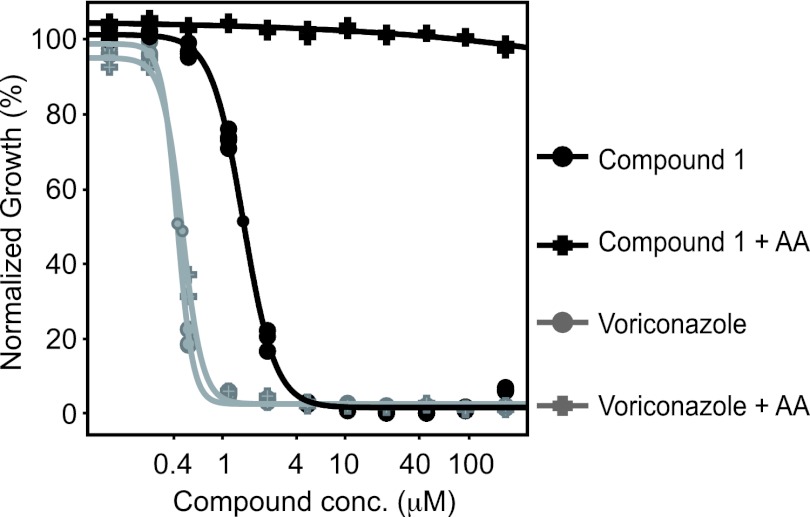
The gene coding for acetolactate synthase is not an essential gene, but deletion renders cells auxotrophic for branched-chain amino acids. To determine whether Ilv2p is the sole target of the triazolopyrimidine-sulfonamide compound series, we tried to rescue compound 1 growth inhibition by supplementing the medium with leucine, isoleucine, and valine. As shown in Fig. 5, addition of 50 mM the indicated amino acids abolished growth inhibition up to the highest tested concentration of compound 1 (200 μM), whereas no rescue effect was observed with the antifungal agent voriconazole, which targets Erg11p. Together with the genetic data presented above, this strongly suggests that Ilv2p is the only target in S. cerevisiae. Supplementation assays were also performed in a range of pathogenic fungi. Growth was rescued in all of the fungal organisms challenged with compound 1 (Table 3; see Table S2 in the supplemental material). In contrast, the addition of lysine, an unrelated amino acid not synthesized via the branched-chained amino acid pathway, was unable to rescue growth. Additionally, supplementation of the medium with FBS was able to partially abrogate growth inhibition in all of the fungal organisms tested except for A. fumigatus. To confirm growth rescue was not due to serum binding of the compound, BSA (4 mg/ml), equivalent to 10% of the serum concentration, was used as a protein binding control (Table 4). The RPMI-only and BSA media displayed similar MIC profiles, suggesting that the growth rescue was primarily due to the amino acids present in FBS and not protein binding of the compound.
Fig 5.
S. cerevisiae amino acid rescue experiment. Addition of extra leucine, isoleucine, and valine amino acids (AA) shifts the IC50 of compound 1 but not of the Erg11 inhibitor voriconazole. Curves were fitted on experiments run in triplicates.
Table 3.
Amino acid supplementation can rescue growth inhibition by compound 1
| Strain | MIC typea | MIC (μg/ml) with additive(s)b |
||
|---|---|---|---|---|
| None | Val + Ile | Lys | ||
| S. cerevisiae NF3201 | MIC-2 | 4 | >128 | 4 |
| MIC-0 | 4 | >128 | 4 | |
| C. albicans NF2103 | MIC-2 | 2 | >128 | 2 |
| MIC-0 | 2 | >128 | 2 | |
| A. fumigatus NF4905 | MIC-2 | 2 | >128 | 2 |
| MIC-0 | 2 | >128 | 2 | |
| R. oryzae NF4801 | MIC-2 | 0.5 | >128 | 0.5 |
| MIC-0 | 0.5 | >128 | 0.5 | |
MIC-0, complete suppression of visible fungal growth; MIC-2, >50% growth inhibition.
Valine, 150 μg/ml; isoleucine, 30 μg/ml; lysine, 30 μg/ml.
Table 4.
Effect of serum on the antifungal activity of compound 1
| Strain | MIC typea | MIC (μg/ml) with additive(s) |
||
|---|---|---|---|---|
| None | 10% FBS | BSA (4 mg/ml) | ||
| S. cerevisiae NF3201 | MIC-2 | 16 | 16 | 32 |
| MIC-0 | 32 | >128 | 64 | |
| C. albicans NF2103 | MIC-2 | 2 | 8 | 2 |
| MIC-0 | 4 | >128 | 8 | |
| A. fumigatus NF4905 | MIC-2 | 4 | 8 | 8 |
| MIC-0 | 8 | 16 | 16 | |
| R. oryzae NF4801 | MIC-2 | 1 | >128 | 2 |
| MIC-0 | 1 | >128 | 2 | |
MIC-0, complete suppression of visible fungal growth; MIC-2, >50% growth inhibition.
Amino acid rescue of acetolactate synthase inhibition in A. fumigatus is medium dependent.
From the above data, amino acid supplementation could rescue ALS inhibition in A. fumigatus, while the addition of serum (more relevant to the in vivo situation) did not, suggesting that medium conditions may play an important role in amino acid uptake in A. fumigatus. In C. neoformans, ilv2 mutants are auxotrophic for branched-chained amino acids and could be rescued with the addition of valine and isoleucine only when proline, but not ammonium, was the nitrogen source, indicating nitrogen regulation of amino acid transport (1). In addition, C. neoformans ilv2 mutants were temperature sensitive and unable to grow at 37°C, even in the presence of sufficient amino acids (1). To determine if a similar phenomenon occurs in A. fumigatus, we challenged the organism to overcome ALS inhibition in different media supplemented with amino acids, fetal calf serum, or human serum. The RPMI medium contains 20 μg/ml valine and 50 μg/ml leucine and isoleucine, which are comparable to recorded levels in plasma of 27.2, 8, and 16 μg/ml, respectively (10, 19). The addition of valine and isoleucine was able to rescue cells from ALS inhibition in RMPI medium, but not in Aspergillus minimal medium (AMM), which contains no additional amino acids, or under the poor-nitrogen-source conditions of AMM plus sodium nitrate (Table 5), suggesting A. fumigatus may possess different amino acid uptake requirements than C. neoformans. Finally, to rule out temperature effects on amino acid uptake, the assays were also performed at 30°C with similar results (data not shown).
Table 5.
Supplementation of A. fumigatus NF4905 growth inhibition by compound 1 in various growth mediaa
| MIC-2 (μg/ml) | MIC-0 (μg/ml) | Medium |
|---|---|---|
| 2 | 4 | RPMI only |
| >128 | >128 | RPMI + valine (150 μg/ml) + isoleucine (30 μg/ml) |
| 4 | 8 | RPMI + 10% FCS |
| 4 | 8 | RPMI + 10% human serum |
| 2 | 2 | AMM (ammonium) only |
| 2 | 4 | AMM (ammonium) + valine (150 μg/ml) + isoleucine (30 μg/ml) |
| 8 | 8 | AMM (ammonium) + 10% FCS |
| 4 | 4 | AMM (ammonium) + 10% human serum |
| 2 | 2 | AMM (sodium nitrate) only |
| 4 | 8 | AMM (sodium nitrate) + valine (150 μg/ml) + isoleucine (30 μg/ml) |
| 8 | 8 | AMM (sodium nitrate) + 10% FCS |
| 4 | 8 | AMM (sodium nitrate) + 10% human serum |
The experiment was performed at 35°C. MIC-0, complete suppression of visible fungal growth; MIC-2, >50% growth inhibition.
It has previously been shown that an A. fumigatus mutant defective in lysine biosynthesis can scavenge amino acids from blood, and in subsequent in vivo studies, the mutant retained full virulence when injected intravenously through the tail vein (20). To determine if branched-chain amino acids can similarly be scavenged from blood to overcome ALS inhibition, equal numbers of conidia were spotted on AMM or 1% agarose supplemented with 5% defibrinated sheep blood. After 72 h of growth, the wells containing sheep blood displayed an increased MIC compared to the normally more favorable AMM growth medium, suggesting that A. fumigatus can scavenge branched-chain amino acids from blood (Fig. 6).
Fig 6.
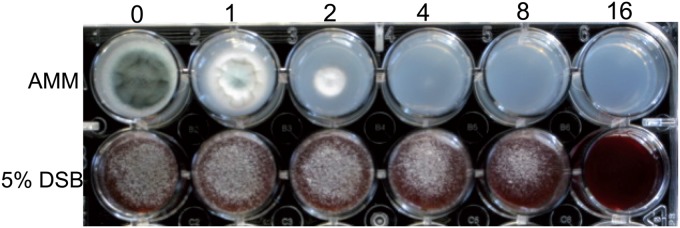
Defibrinated sheep blood rescues compound 1 inhibition in A. fumigatus. Equal numbers of conidia were spotted onto AMM or 1% agarose supplemented with 5% DSB medium containing the indicated concentrations of compound 1 (μg/ml). The plate was incubated for 72 h at 35°C.
DISCUSSION
The triazolopyrimidine series of compounds described here appear initially to represent an attractive starting point for antifungal drug discovery, with broad-spectrum antifungal activity, potential for cidal action, no significant in vitro cytotoxicity, and low protein binding. Using chemogenomic profiling as a first assay to investigate the target, we observed strains compromised for both the regulatory (Ilv6p) and catalytic (Ilv2p) subunits of acetolactate synthase in the profiles. Ilv6p has been published as stimulating Ilv2p activity severalfold (16), and in agreement with this, Ilv6p scored in both HIP and HOP profiles, showing that reduction (HIP) or loss (HOP) of Ilv6p has a profound effect on strain viability in the presence of the compounds. The raising of resistant mutants revealed point mutations in the active site of Ilv2p, further confirming that the catalytic subunit is the primary binding target for the compounds of this series.
However, identification of the target as ALS immediately raises the concern of nutrient bypass in vivo, a potential issue for any target in amino acid or other biosynthetic pathways common to fungi and mammals. In vitro, the compounds were clearly active in RPMI, a medium containing levels of branched-chain amino acids comparable to those of plasma. However, supplementation of RPMI with exogenous amino acids or serum could rescue growth, indicating the risk of nutrient bypass in vivo. The situation is more complex in Aspergillus, with the effects of exogenous amino acids dependent on the medium conditions, and nitrogen sensing may play a major role in influencing amino acid uptake (1). However, our data showed that A. fumigatus is capable of scavenging branched-chain amino acids from sheep blood. In vivo, there are mixed reports of the outcome of amino acid inhibition. Disruption of the ALS complex through deletion of ILV2 resulted in decreased virulence in C. albicans and C. neoformans, suggesting that free amino acid pools are limited in vivo and de novo branched-chain amino synthesis is required for full pathogenesis (1, 2). On the other hand, Becker et al. recently reported that ILV5 (downstream of ILV2/ILV6) was nonessential for C. albicans in a murine infection model (21). Effects can also vary between infection models; for example, an A. fumigatus mutant auxotrophic for lysine through disruption of hcsA, a gene encoding homocitrate synthase, resulted in attenuated virulence in a murine model of bronchopulmonary aspergillosis, but retained full virulence when injected intravenously (20).
Valine, leucine, and isoleucine are essential amino acids in mammalian cells and have to be supplemented in the medium. Thus, nutrient bypass could account for the observed low cytotoxicity of the compounds, even though no good homolog of ALS can be identified in mammalian cells using available in silico methods. However, lowering the levels of FBS and thus available branched-chain amino acids as presented in Table 2 had no obvious effect on the cytotoxicity, supporting the notion that the compounds do not act on mammalian cells due to differences in amino acid biosynthesis pathways and the absence of ALS.
Taken together, the evidence is still inconclusive as to whether targeting branched-chained amino acid synthesis in vivo could be efficacious in an antifungal context, but there is significant potential for abrogation by nutrient bypass. This concern is presumably the reason that no ALS inhibitors have been developed as antimicrobials despite the existence of suitable lead molecules, although they have been suggested as a novel approach to antituberculosis agents (22). The observation that significant resistance against compounds from this study and 2 known Ilv2 inhibitors can be acquired by a single-amino-acid change while maintaining functionality of the Ilv2 protein adds an additional concern. It suggests that the probability for resistance in clinical settings will be high. It is to be hoped that further study of the mechanisms by which amino acids are scavenged in vivo may provide insights into how to effectively target amino acid biosynthesis pathways for the treatment of systemic fungal infections.
Supplementary Material
ACKNOWLEDGMENTS
We give special thanks to Roger Fujimoto for chemistry support, including similarity search by inventory, David Cocozziello for cytotoxicity profiling, and Uwe Plikat and Luc Dobler for handling and storage of the microarray data.
Footnotes
Published ahead of print 11 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01809-12.
REFERENCES
- 1. Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. 2004. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology 150:1547–1558 [DOI] [PubMed] [Google Scholar]
- 2. Kingsbury JM, McCusker JH. 2010. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2) mutants is influenced by the carbon source and rapamycin. Microbiology 156:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chipman D, Barak Z, Schloss JV. 1998. Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta 1385:401–419 [DOI] [PubMed] [Google Scholar]
- 4. McCourt JA, Duggleby RG. 2006. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids 31:173–210 [DOI] [PubMed] [Google Scholar]
- 5. Singh BA, Shaner DL. 1995. Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babczinski P, Zelinski T. 1991. Mode of action of herbicidal ALS inhibitors on acetolactate synthase from green plant cell cultures, yeast and Escherichia coli. Pestic. Sci. 31:305–323 [Google Scholar]
- 7. Gerwick BC, Subramanian MV, Loney-Gallant VI, Chandler DP. 1990. Mechanism of action of the 1,2,4-triazolo (1,5-a) pyrimidines. Pestic. Sci. 29:357–364 [Google Scholar]
- 8. Cove DJ. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51–56 [DOI] [PubMed] [Google Scholar]
- 9. Hoepfner D, Karkare S, Helliwell SB, Pfeifer M, Trunzer M, Bonnechose SD, Zimmerlin A, Tao J, Richie D, Hofmann A, Reinker S, Frederiksen M, Movva NR, Porter JA, Ryder NS, Parker CN. 2012. An integrated approach for identification and target validation of antifungal compounds active against Erg11p. Antimicrob. Agents Chemother. 56:4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed MA38-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard, 3rd ed M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Pierce SE, Fung EL, Jaramillo DF, Chu AM, Davis RW, Nislow C. 2006. A unique and universal molecular barcode array. Nat. Methods 3:601–603 [DOI] [PubMed] [Google Scholar]
- 13. Pang SS, Guddat LW, Duggleby RG. 2003. Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase. J. Biol. Chem. 278:7639–7644 [DOI] [PubMed] [Google Scholar]
- 14. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26 [DOI] [PubMed] [Google Scholar]
- 15. Hoon S, Onge RP, Giaever G, Nislow C. 2008. Yeast chemical genomics and drug discovery: an update. Trends Pharmacol. Sci. 29:499–504 [DOI] [PubMed] [Google Scholar]
- 16. Pang SS, Duggleby RG, Schowen RL, Guddat LW. 2004. The crystal structures of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. J. Biol. Chem. 279:2242–2253 [DOI] [PubMed] [Google Scholar]
- 17. Falco SC, Dumas KS, Livak KJ. 1985. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 13:4011–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCourt JA, Pang SS, Guddat LW, Duggleby RG. 2005. Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase. Biochemistry 44:2330–2338 [DOI] [PubMed] [Google Scholar]
- 19. Cynober LA. 2002. Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition 18:761–766 [DOI] [PubMed] [Google Scholar]
- 20. Schobel F, Jacobsen ID, Brock M. 2010. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot. Cell 9:878–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, Jiang B, Roemer T. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. U. S. A. 107:22044–22049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grandoni JA, Marta PT, Schloss JV. 1998. Inhibitors of branched-chain amino acid biosynthesis as potential antituberculosis agents. J. Antimicrob. Chemother. 42:475–482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.