Abstract
Within recent years, it has been established that extracellular DNA is a key constituent of the matrix of microbial biofilms. In addition, it has recently been demonstrated that DNA binds positively charged antimicrobials such as aminoglycosides and antimicrobial peptides. In the present study, we provide evidence that extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. We show that exogenously supplemented DNA integrates into P. aeruginosa biofilms and increases their tolerance toward aminoglycosides. We provide evidence that biofilms formed by a DNA release-deficient P. aeruginosa quorum-sensing mutant are more susceptible to aminoglycoside treatment than wild-type biofilms but become rescued from the detrimental action of aminoglycosides upon supplementation with exogenous DNA. Furthermore, we demonstrate that exposure to lysed polymorphonuclear leukocytes, which are thought to be a source of extracellular DNA at sites of infections, increases the tolerance of P. aeruginosa biofilms toward aminoglycosides. Although biofilm-associated aminoglycoside tolerance recently has been linked to extracellular DNA-mediated activation of the pmr genes, we demonstrate that the aminoglycoside tolerance mediated by the presence of extracellular DNA is not caused by activation of the pmr genes in our P. aeruginosa biofilms but rather by a protective shield effect of the extracellular DNA.
INTRODUCTION
Work done in the last decade has shown that bacteria in natural, industrial, and clinical settings most often live in biofilms, i.e., sessile-structured microbial communities encased in extracellular matrix materials. One of the most important characteristics of microbial biofilms is that the resident bacteria display a remarkable increased resistance to antimicrobial attack (1, 2). Accordingly, biofilms formed by opportunistic pathogenic bacteria are involved in highly problematic chronic infections and in devastating medical device-associated infections. Because the present-day armory of antimicrobial compounds in many cases cannot fully eradicate biofilm infections, there is an urgent need to develop alternative measures which may function to either boost the activity of conventional antimicrobials or restore proper action of the immune system against biofilms. Knowledge about the molecular mechanisms involved in biofilm formation and biofilm-associated antimicrobial tolerance will form the basis for the development of drugs which can cure otherwise recalcitrant infections.
The extracellular matrix, which is essential for interconnecting the bacteria in biofilms, can be composed of polysaccharides, proteins, and extracellular DNA (eDNA) (3–10). We have shown that eDNA functions as a cell-to-cell interconnecting matrix compound in P. aeruginosa biofilms (3, 7, 11–13). Subsequently, evidence was provided that eDNA functions as a matrix component in biofilms formed by many other bacterial species, e.g., Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus intermedius, and Streptococcus mutans (8, 14–16). Evidence has been provided that the quorum-sensing system plays a role in the formation of eDNA in P. aeruginosa biofilms (7, 11, 13) and that DNA release from P. aeruginosa populations involves lysis of a small subpopulation of the cells (7). However, in the case of infectious biofilms that develop inside the human body, the eDNA that stabilizes the biofilms may also be provided by lysed human cells (17). The P. aeruginosa biofilms present in medical settings, such as in the lungs of cystic fibrosis patients or in the wounds of chronic wound sufferers, evidently produce virulence factors, in particular rhamnolipids, that lyse attacking polymorphonuclear leukocytes (PMNs) (18, 19), and the eDNA liberated from the lysed PMNs can subsequently be incorporated into the biofilms (17).
Biofilm bacteria's robustness to antimicrobials is caused by a number of different mechanisms: (i) certain components of the extracellular biofilm matrix can bind the antimicrobial and limit its penetration, (ii) differential physiological activities in the biofilm population can provide insurance effects to specific subpopulations, (iii) expression of specific genes can increase antibiotic tolerance, (iv) a subpopulation of differentiated persister cells in the biofilm is particularly tolerant to antibiotic treatments (2).
It has previously been shown that eDNA plays a role in the tolerance of P. aeruginosa biofilms toward antimicrobial peptides and aminoglycosides (20). In that study, it was demonstrated that eDNA binds cations and creates a cation-limited environment that results in induction of the pmr genes in P. aeruginosa and thereby increased resistance toward antimicrobial peptides and aminoglycosides. It was noted that eDNA also caused aminoglycoside tolerance in biofilms formed by a P. aeruginosa pmr mutant, and it was concluded that DNA-induced resistance to aminoglycosides is not limited to pmr gene induction, but a mechanism accounting for this was not suggested. It is well established that DNA can bind positively charged antibiotics such as aminoglycosides and antimicrobial peptides (21–23). Therefore, it is highly likely that eDNA may contribute to biofilm-associated antimicrobial resistance by acting as a shield that binds aminoglycosides and antimicrobial peptides.
In the present study, we provide evidence that eDNA contributes to aminoglycoside tolerance in P. aeruginosa biofilms by acting as an antimicrobial shield. DNA supplemented to the perfusion medium of biofilm flow chambers was shown to be incorporated into the resident P. aeruginosa biofilms and increase their tolerance to aminoglycoside treatment. Biofilms formed in flow chambers by a DNA release-deficient P. aeruginosa quorum-sensing mutant were shown to be susceptible to aminoglycoside treatments, but they displayed increased aminoglycoside tolerance if they were supplied with exogenous DNA prior to antibiotic treatments. Moreover, we found that supplementation of the perfusion medium of biofilm flow chambers with lysed PMNs increased the tolerance of the resident P. aeruginosa biofilms toward subsequent aminoglycoside treatment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For routine strain manipulations, P. aeruginosa and Escherichia coli strains were grown in LB medium at 37°C. Where appropriate, antibiotics were used for bacterial cultures at the following concentrations: for P. aeruginosa strains, gentamicin (Biochrome AG, Germany) at 30 μg/ml, streptomycin (Sigma, Germany) at 300 μg/ml, carbenicillin (Sigma, Germany) at 200 μg/ml, and tetracycline (Sigma, Germany) at 15 μg/ml; for E. coli strains, ampicillin (Vepidan ApS, Denmark) at 100 μg/ml, chloramphenicol (Sigma, Germany) at 25 μg/ml, kanamycin (Sigma, Germany) at 50 μg/ml, tetracycline (Sigma, Germany) at 10 μg/ml, and gentamicin at 15 μg/ml. AB medium (33) supplemented with 30 mM glucose was used for cultivation of biofilms in microtiter trays. FAB medium (34) supplemented with 0.3 mM glucose was used for cultivation of biofilms in flow chambers.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) or sequence | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild type | 24 (J. S. Mattick laboratory) |
| PAO1-GFP | P. aeruginosa PAO1 tagged with enhanced GFP (EGFP) in a mini-Tn7 construct; Gmr | 25 |
| PAO1 pmrF-GFP | P. aeruginosa PAO1 transposon mutant ID35399-PA3553 (Washington Genome Center); tagged with EGFP in a mini-Tn7 construct; Gmr | 26 |
| PAO1 lasR rhlR-GFP | P. aeruginosa lasR rhlR tagged with EGFP in a mini-Tn7 construct; Smr | This study |
| E. coli | ||
| HB101 | recA thi pro leu hsdRM; Smr; strain used for maintenance and proliferation of plasmids | 27 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| S17-1 | pro thi recA hsdR (r− m+) Tpr Smr Kmr ΩRP4-2-Tc::Mu-Km::Tn7 | 28 |
| Plasmids | ||
| pUX-BF13 | mob+ ori-R6K; helper plasmid providing the Tn7 transposition functions in trans; Ampr | 29 |
| pBK-miniTn7(Smr)-gfp | Delivery plasmid for miniTn7-PA1/04/03-GFP; Ampr Smr | 30 |
| pEX18ApGW | Gateway compatible gene replacement vector; Sucs Ampr | 31 |
| pPS856 | 0.83-kb blunt-ended SacI fragment from pUCGM ligated into the EcoRV site of pPS854; Ampr Gmr | 32 |
| pDONR221 | Gateway donor vector; Kmr | Invitrogen |
| pDONR221-lasR | lasR entry clone; Kmr Gmr | This study |
| pDONR221-rhlR | rhlR entry clone; Kmr Gmr | This study |
| pEX18Ap-lasR | lasR knockout vector; Sucs Ampr Gmr | This study |
| pEX18Ap-rhlR | rhlR knockout vector; Sucs Ampr Gmr | This study |
| pRK600 | ori-ColE1 RK2-mob+ RK2-tra+; helper plasmid for conjugation; Cmr | 27 |
Construction of a P. aeruginosa lasR rhlR mutant.
lasR and rhlR knockout fragments containing a gentamicin resistance cassette were generated by PCR overlap extension essentially as described by Choi and Schweizer (31). Primers (whose sequence will be supplied upon request) were used to amplify chromosomal regions upstream and downstream of lasR and rhlR and to amplify a gentamicin resistance cassette from plasmid pPS856 (32). The PCR fragments were fused together and amplified with primers GW-attB1 and GW-attB2 incorporating the attB1 and attB2 recombination sites at either end of the knockout cassettes. Through use of the Gateway cloning system (Invitrogen), the resulting knockout fragments were first transferred by the BP reaction into pDONR221, generating entry plasmids pDONR221-lasR and pDONR221-rhlR, and subsequently transferred by the LR reaction into pEX18ApGW, generating the knockout plasmids pEX18Ap-lasR and pEX18Ap-rhlR.
A P. aeruginosa lasR rhlR mutant was constructed as follows: The knockout plasmids were transferred into the PAO1 wild type by two-parental mating using the donor strain E. coli S17-1 with selection done on AB-citrate plates supplemented with gentamicin. Resolution of single-crossover events was achieved by streaking on 5% sucrose plates via the counterselectable sacB marker on the knockout plasmid. In order to construct the lasR rhlR double mutant, Flp-mediated excision of the gentamicin resistance cassette in the rhlR mutant was performed using the pFLP2 plasmid as described by Choi and Schweizer (31). The resulting double crossovers in the mutants were confirmed by PCR. The P. aeruginosa lasR rhlR mutant was green fluorescent protein (GFP) tagged by inserting a mini-Tn7-gfp-Smr cassette into a neutral site of the genome, using four-parental mating, essentially as described previously by Klausen et al. (25).
Assessment of the minimal bactericidal antibiotic concentration for biofilm-grown cells in microtiter trays.
We used a quantitative assay of biofilm antimicrobial tolerance essentially as described by Mah et al. (35). P. aeruginosa AB-glucose overnight cultures were diluted 100-fold in fresh AB-glucose medium or AB-glucose medium supplemented with 4 mg dialyzed salmon sperm/ml, and inocula with or without exogenous DNA were transferred to microtiter trays (100 μl/well). The microtiter trays were incubated at 37°C for 24 h to allow biofilm formation in the wells. Subsequently, the liquid medium with planktonic cells was removed, and fresh AB-glucose medium (125 μl/well) with various concentrations of tobramycin or gentamicin was added to the wells. After 24 h of incubation with antibiotic exposure, the liquid medium was substituted with fresh medium lacking antibiotic (150 μl/well), and the biofilms were allowed to recover in this medium for an additional 24 h, upon which their viability was assessed through Wallac microplate reader (PerkinElmer, USA) quantification of the optical density (OD) of the liquid medium in the wells. The values of minimal bactericidal antibiotic concentration for biofilm-grown cells (MBC-B) correspond to the minimal antibiotic concentration required to kill the biofilm bacteria so that the OD590 of the liquid medium of the wells did not exceed 0.1 after 24 h of incubation at 37°C.
Cultivation of flow chamber biofilms.
Biofilms were cultivated at 37°C in flow chambers which were assembled and prepared as described by Crusz et al. (36). Flow chambers were inoculated with P. aeruginosa overnight cultures diluted to an OD600 of 0.01 in FAB-glucose medium as described by Pamp et al. (34). Where indicated, salmon sperm DNA (Life Technologies), at a final concentration of 40 μg/ml, was added to the medium irrigated to the flow chambers after 2 days of cultivation. Where indicated, lysed human PMNs, corresponding to a final concentration of 104 PMNs/ml, were added to the medium irrigated to the flow chambers after 2 days of cultivation. Human PMNs (purified as described by Bjarnsholt et al. [37]) that had been lysed by subjecting them to freezing at −20°C followed by thawing at room temperature were a kind gift from Lars Christophersen (Department of Clinical Microbiology, University Hospital, Rigshospitalet, Copenhagen, Denmark). After 4 days of cultivation, biofilms were exposed to 25 μg/ml tobramycin (Sigma, Germany) by addition of the antibiotic to the medium irrigated to the flow chambers. In order to monitor bacterial killing in the antimicrobial-treated biofilms, the fluorescent dead-cell indicator propidium iodide (Sigma, Germany) was added at a final concentration of 0.3 μM. An inverse correlation between staining of P. aeruginosa cells with 0.3 μM propidium iodide and CFU on agar plates has previously been demonstrated (26). The experiments were done with the velocity of the laminar flow in the flow chamber channels at 0.2 mm/s, using a Watson Marlow 205S peristaltic pump (Watson Marlow, United Kingdom). In each case, three replicate experiments were done with essentially the same results.
Microscopy and image processing.
Microscopic observation and image acquisition of biofilms were performed with a Zeiss LSM 710 confocal laser scanning microscope (Carl Zeiss, Germany) equipped with an argon and an NeHe laser and detectors and filter sets for simultaneous monitoring of GFP (excitation, 488 nm; emission, 517 nm) and propidium iodide (excitation, 543 nm; emission, 565 nm). Images were obtained using a 63×/1.4 objective. Simulated fluorescence projections were generated using the IMARIS software package (Bitplane AG, Switzerland).
RESULTS
Exogenously added DNA is incorporated into P. aeruginosa biofilms.
Initially, we investigated if externally added non-P. aeruginosa DNA becomes incorporated into flow-chamber-grown P. aeruginosa biofilms. We grew P. aeruginosa wild-type biofilms for 2 days in flow chambers, supplemented the medium irrigated to half of the flow chambers with salmon sperm DNA, continued biofilm growth with or without DNA supplementation for two more days, stained the biofilms with propidium iodide, and acquired CLSM micrographs of the biofilms. As shown in Fig. 1, the biofilms that were supplemented with salmon sperm DNA contained a high concentration of eDNA throughout the microcolonies, whereas the nonsupplemented biofilms showed areas of high concentration of eDNA, especially in the inner part of the microcolonies, in agreement with previous investigations (7, 11, 13). From these experiments, we conclude that exogenously supplemented DNA is capable of incorporating into P. aeruginosa biofilms and physically contributes to the composition of the biofilm matrix.
Fig 1.
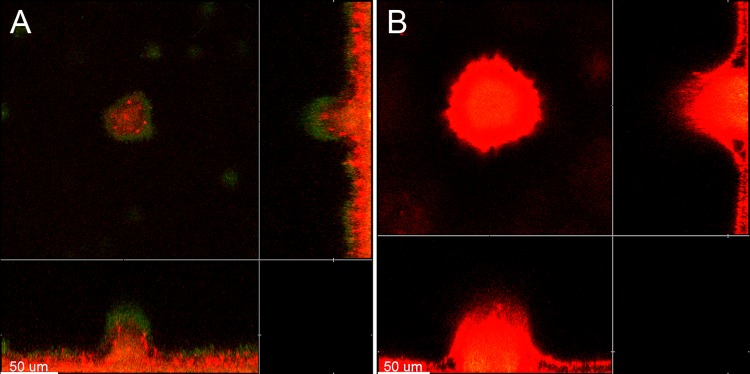
Visualization of extracellular DNA in 4-day-old P. aeruginosa PAO1-GFP biofilms grown in flow chambers without addition of exogenous DNA (A) or with addition of salmon sperm DNA (B) to the medium irrigated to the flow chambers after 2 days of cultivation. The bacteria appear green due to expression of GFP, whereas the extracellular DNA surrounding the bacteria appears red due to staining with propidium iodide and visualization with ultrasensitive confocal laser scanning microscopy. Scale bars, 50 μm.
We have previously shown that addition of genomic P. aeruginosa DNA to P. aeruginosa wild-type biofilms promotes the formation of huge microcolonies, much bigger than those found in biofilms that were not supplemented with DNA (13). Similar to our previous work, we found in the present study that addition of exogenous DNA led to the formation of larger microcolonies in the biofilms (data not shown), presumably because the number of detaching bacteria was reduced in the DNA-supplemented biofilms. However, the purpose of the present investigation was to compare the effect of eDNA on antimicrobial tolerance in P. aeruginosa biofilms, and it was therefore essential to compare equally sized microcolonies in the DNA-supplemented and nonsupplemented flow chamber biofilms. Accordingly, throughout the present study, we have deliberately selected microcolonies of roughly equal size in the DNA-supplemented and nonsupplemented biofilms during microscopy.
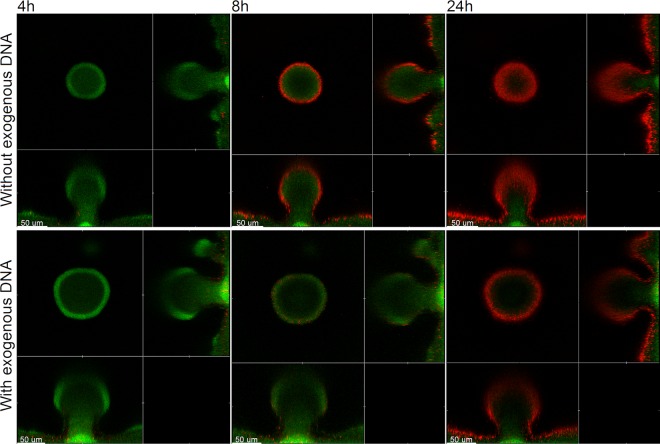
Exogenously added DNA increases the survival of flow-chamber-grown P. aeruginosa wild-type biofilms toward tobramycin treatment.
After having shown that externally added DNA gets incorporated into our P. aeruginosa flow chamber biofilms, we next investigated if this extra eDNA can increase the tolerance of the biofilms to tobramycin exposure. For visualization of eDNA in biofilms as shown in Fig. 1, we combined propidium iodide staining with ultrahigh sensitivity settings of the CLSM as published previously (7). However, with normal CLSM sensitivity settings, propidium iodide staining is routinely used to visualize dead bacteria in biofilms (11). To investigate if the presence of extra eDNA can increase the tolerance of flow-chamber-grown biofilms to tobramycin treatment, we grew P. aeruginosa biofilms in flow chambers with or without addition of salmon sperm DNA (as described above) and then subsequently irrigated the flow chambers with DNA-free medium supplemented with tobramycin and propidium iodide. Figure 2 shows CLSM images acquired in such biofilms 4, 8, and 24 h after the shift to the tobramycin-containing, DNA-free perfusion medium. The 4-hour image in Fig. 2 (as well as in Fig. 3, 4, and 5) shows that the biofilms contained only few dead bacteria initially, and it also shows that eDNA is not visualized by the propidium iodide staining with normal CLSM settings, confirming that propidium iodide staining can visualize eDNA when the sensitivity of the CLSM is ultrahigh (as in Fig. 1) but only dead bacteria when the sensitivity of the CLSM is normal (as in Fig. 2, 3, 4, and 5), in agreement with our previous studies (7, 11). After 8 h of tobramycin treatment, the outer layer of the biofilm microcolonies that had not received extra eDNA were killed, whereas virtually all the bacteria in the biofilms that were supplemented with DNA appeared alive. After 24 h of continuing tobramycin treatment, a larger fraction of the bacteria in the outer part of the biofilm microcolonies both with and without extra eDNA were killed, presumably due to saturation of the eDNA by the constant delivery of fresh tobramycin in the perfusion medium. From the experiments described here, we conclude that exogenously added DNA can increase the tolerance of flow-chamber-grown P. aeruginosa wild-type biofilms to tobramycin treatments.
Fig 2.
Spatiotemporal distribution of live and dead bacteria in tobramycin-treated P. aeruginosa PAO1-GFP biofilms that were grown with or without exogenous DNA. Biofilms were grown for 4 days and were then continuously exposed to tobramycin (25 μg/ml) and propidium iodide. In the case of the biofilm shown in the lower panel, the medium irrigated to the flow chambers was supplied with salmon sperm DNA after 2 days of cultivation. Confocal laser scanning micrographs were acquired 4, 8, and 24 h after the beginning of tobramycin treatment. The images show a horizontal xy section, with two flanking images representing sections in the xz and yz planes. Live cells appear green due to expression of GFP, and dead cells appear red due to staining with the dead-cell indicator propidium iodide. Scale bars, 50 μm.
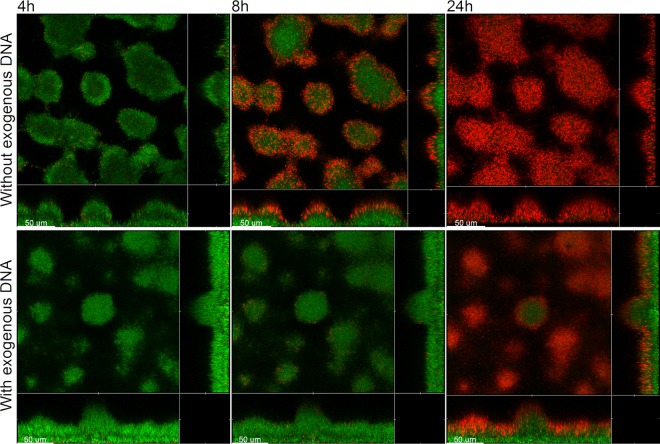
Fig 3.
Spatiotemporal distribution of live and dead bacteria in tobramycin-treated P. aeruginosa lasR rhlR-GFP biofilms that were grown with or without exogenous DNA. Biofilms were grown for 4 days and were then continuously exposed to tobramycin (25 μg/ml) and propidium iodide. In the case of the biofilm shown in the lower panel, the medium irrigated to the flow chambers was supplied with salmon sperm DNA after 2 days of cultivation. Confocal laser scanning micrographs were acquired 4, 8, and 24 h after the beginning of tobramycin treatment. The images show a horizontal xy section, with two flanking images representing sections in the xz and yz planes. Live cells appear green due to expression of GFP, and dead cells appear red due to staining with the dead-cell indicator propidium iodide. Scale bars, 50 μm.
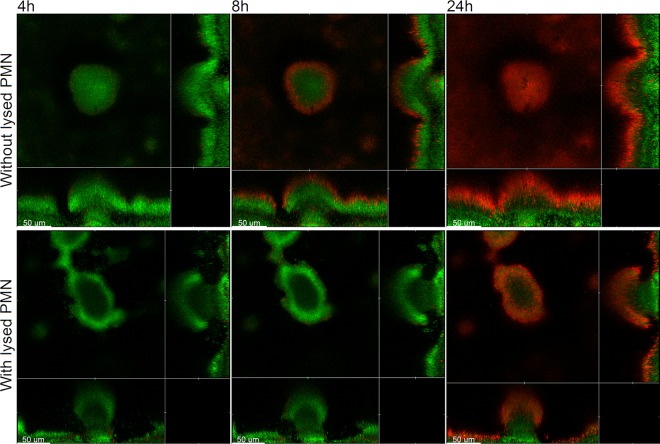
Fig 4.
Spatiotemporal distribution of live and dead bacteria in tobramycin-treated P. aeruginosa PAO1-GFP biofilms that were grown with or without addition of lysed PMNs. Biofilms were grown for 4 days and were then continuously exposed to tobramycin (25 μg/ml) and propidium iodide. In the case of the biofilm shown in the lower panel, the medium irrigated to the flow chambers was supplied with lysed PMNs after 2 days of cultivation. Confocal laser scanning micrographs were acquired 4, 8, and 24 h after the beginning of tobramycin treatment. The images show a horizontal xy section, with two flanking images representing sections in the xz and yz planes. Live cells appear green due to expression of GFP, and dead cells appear red due to staining with the dead-cell indicator propidium iodide. Scale bars, 50 μm.
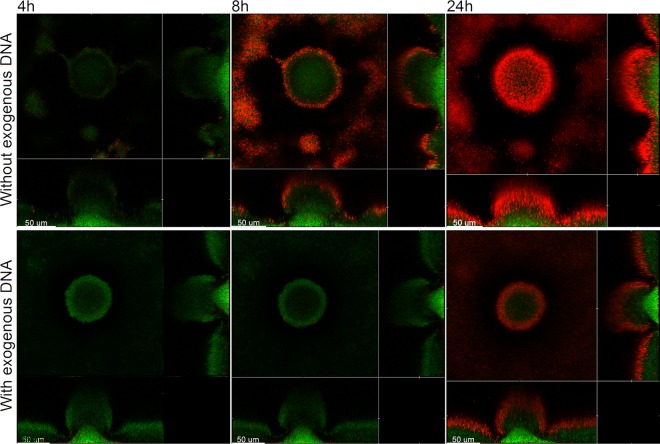
Fig 5.
Spatiotemporal distribution of live and dead bacteria in tobramycin-treated P. aeruginosa pmrF-GFP biofilms that were grown with or without exogenous DNA. Biofilms were grown for 4 days and were then continuously exposed to tobramycin (25 μg/ml) and propidium iodide. In the case of the biofilm shown in the lower panel, the medium irrigated to the flow chambers was supplied with salmon sperm DNA after 2 days of cultivation. Confocal laser scanning micrographs were acquired 4, 8, and 24 h after the beginning of tobramycin treatment. The images show a horizontal xy section, with two flanking images representing sections in the xz and yz planes. Live cells appear green due to expression of GFP, and dead cells appear red due to staining with the dead-cell indicator propidium iodide. Scale bars, 50 μm.
The effect of DNA supplementation on biofilm-associated tobramycin tolerance shown in Fig. 2 is consistent but relatively modest. However, in the experiments reported here, the DNA-supplemented medium irrigated to the flow cells contained approximately 40 μg salmon sperm DNA/ml, which is roughly two orders of magnitude lower than the concentration of eDNA expected at sites of biofilm infections (e.g., see references 38 and 39 for eDNA concentrations in infected cystic fibrosis [CF] lungs). The cost of salmon sperm DNA in combination with the large medium volumes used for flow chamber experiments prohibited the use of higher eDNA concentrations.
Flow chamber biofilms formed by a DNA release-deficient P. aeruginosa mutant are highly susceptible to tobramycin treatment but resist tobramycin treatment if they are supplied with exogenous DNA.
We have previously shown that quorum-sensing mutants, such as P. aeruginosa lasR rhlR and P. aeruginosa pqsA mutants, are deficient in DNA release and form biofilms which are more fragile than wild-type biofilms (7). In addition, evidence has been provided that biofilms formed by P. aeruginosa quorum-sensing mutants display reduced tolerance to tobramycin treatment (37). In the present study, we investigated how addition of exogenous DNA affects the tobramycin tolerance of biofilms formed in flow chambers by a P. aeruginosa lasR rhlR quorum-sensing mutant. After 4 h of tobramycin treatment, a few dead cells appeared in the 4-day-old biofilms that did not receive extra eDNA, whereas virtually all the bacteria in the DNA-supplemented biofilms were alive (Fig. 3). After 8 h of tobramycin treatment, the outer layer of the microcolonies in the P. aeruginosa lasR rhlR biofilms that did not receive extra eDNA were killed, whereas almost all the bacteria in the DNA-supplemented P. aeruginosa lasR rhlR biofilms were alive (Fig. 3). After 24 h of tobramycin treatment, virtually all the bacteria in the P. aeruginosa lasR rhlR biofilms that were not supplemented with DNA were killed, whereas a substantial subpopulation in the inner part of the DNA-supplemented P. aeruginosa lasR rhlR biofilms survived the tobramycin treatment (Fig. 3). From these experiments, we conclude that biofilms formed in flow chambers by a DNA release-deficient P. aeruginosa mutant are susceptible to tobramycin treatment but display increased tobramycin tolerance if they are supplied with exogenous DNA.
Addition of lysed PMNs increases the tolerance of P. aeruginosa biofilms toward tobramycin treatment.
Walker et al. (17) provided evidence that DNA and actin polymers released from lysed PMNs can be used as extracellular matrix components in P. aeruginosa biofilms. To investigate if components from lysed PMNs can increase the tolerance of biofilms toward tobramycin, we grew P. aeruginosa biofilms in flow chambers in which the perfusion medium was either supplemented with lysed PMNs or not and then subsequently irrigated the flow chambers with PMN-free medium supplemented with tobramycin and propidium iodide. Figure 4 shows CLSM images acquired in the biofilms 4, 8, and 24 h after the shift to the tobramycin-containing medium. After 4 h of tobramycin treatment, virtually all the cells in the PMN-supplemented and nonsupplemented biofilms were alive. After 8 h of tobramycin treatment, the outer layers of the microcolonies of those biofilms that did not receive lysed PMNs prior to the antibiotic treatment were killed, whereas almost all the bacteria in the biofilms that had been supplemented with lysed PMNs prior to the antibiotic treatment were alive. After 24 h of tobramycin treatment, a larger fraction of the bacteria in the outer part of the microcolonies in the biofilms both with and without lysed PMNs were killed, presumably due to saturation of the biofilm matrix by the continuous addition of tobramycin. The PMN-supplemented medium contained approximately 104 lysed PMNs/ml, which is considerably less than the concentration of PMNs expected at sites of biofilm infections. For example, the concentration of PMNs in sputum from CF lungs has been reported to be in the range of 106 to 108 PMNs/ml (40, 41). From the experiments described above, we conclude that components from lysed PMNs, most likely eDNA, can increase the tolerance of P. aeruginosa biofilms toward tobramycin.
Exogenously added DNA increases the tobramycin tolerance of P. aeruginosa pmrF mutant biofilms.
Mulcahy et al. (20) provided evidence that eDNA through cation binding can create a cation-limited environment that results in induction of the cationic antimicrobial peptide resistance operon pmrHFIJKLME in P. aeruginosa. It was shown that the DNA-induced expression of the pmrHFIJKLME operon resulted in up to a 2,560-fold increased resistance to cationic antimicrobial peptides and a 640-fold increased resistance to aminoglycosides. Thus, the aminoglycoside tolerance mediated by the presence of eDNA in the biofilms that we report here might, as an alternative to our interpretation, be caused by activation of the pmr genes mediated via binding and withdrawal of cations by the eDNA. In order to investigate if the pmr genes have a role in the eDNA-mediated aminoglycoside tolerance observed in our study, we investigated the response of a P. aeruginosa pmrF mutant to tobramycin treatment. We have previously shown that biofilms formed by this P. aeruginosa pmrF mutant, unlike wild-type biofilms, are highly susceptible to colistin treatment (34), which is in accordance with a role of the pmr genes in cationic antimicrobial peptide resistance (42–44). In the present study, we found that exogenously added DNA also increased the aminoglycoside tolerance of biofilms formed by the P. aeruginosa pmrF mutant (Fig. 5). From this experiment, we conclude that the biofilm-associated aminoglycoside tolerance mediated by the presence of eDNA is not caused by activation of the pmr genes under our conditions.
Exogenously added DNA increases the minimal bactericidal concentration of tobramycin and gentamicin toward P. aeruginosa biofilms grown in microtiter trays.
The experiments with live/dead-stained flow cell biofilms described above were performed three times with essentially the same results, but they nevertheless suffer from being qualitative. We included microtiter tray-based assays of the minimal bactericidal antibiotic concentrations for biofilm-grown cells (MBC-B) in our study in order to obtain quantitative data in addition to the flow cell experiments. The use of a microtiter tray-based assay also allowed us to use eDNA concentrations in the range found for biofilm infections. The concentration of eDNA in sputum from CF lungs was reported to be within 2 to 20 mg/ml (38, 39), and accordingly we used a salmon sperm DNA concentration of 4 mg/ml in our MBC-B assays. We did not use higher eDNA concentrations than 4 mg/ml, since it has been reported that eDNA at concentrations of 5 mg/ml or higher can kill P. aeruginosa in LB cultures (20). We first grew the biofilms for 24 h in microtiter trays containing AB-glucose medium with or without salmon sperm DNA. The planktonic bacteria in the microtiter tray wells reached the same optical density with or without salmon sperm DNA, confirming that the eDNA did not have antimicrobial activity at the concentration we used. We then removed the liquid medium with the planktonic cells and added fresh DNA-free medium with various concentrations of either tobramycin or gentamicin, and after 24 h of exposure to one or the other antibiotic, the biofilms were allowed to recover in fresh medium without antibiotics for an additional 24 h, upon which their viability was assessed. The microtiter tray-based MBC-B assays were carried out with tobramycin for the wild-type, lasR rhlR, and pmrF strains, whereas the experiment with gentamicin was done only with the P. aeruginosa wild type, since the pmrF and lasR rhlR mutants harbor gentamicin resistance genes. As shown in Table 2, the MBC-B values were substantially higher for the biofilms grown with salmon sperm DNA added to the medium in comparison to when no exogenous DNA was added. The experiments with the pmrF mutant confirmed that the effect of eDNA on the aminoglycoside tolerance of P. aeruginosa biofilms is independent of the Pmr system also in the microtiter tray-based assay. Contrary to our expectation, the MBC-B value obtained for the lasR rhlR mutant without DNA supplementation was not significantly lower than the MBC-B value obtained for the wild type without DNA supplementation. Currently, we cannot explain this observation, but it is possible that a subpopulation of dead DNA-releasing cells arise during biofilm formation in microtiter trays for both the quorum-sensing mutant and the wild type. The MBC-B values obtained in the present study are lower than those reported by others previously (35), which may indicate that our biofilms were less developed than those investigated by others.
Table 2.
Minimal bactericidal antibiotic concentration for P. aeruginosa biofilms grown for 24 h in microtiter trays with or without dialyzed salmon sperm DNA in the medium
| Strain | DNA | MBC-Ba |
|
|---|---|---|---|
| Tobramycin | Gentamicin | ||
| Wild type | − | 19 (2.0) | 33 (1.4) |
| Wild type | + | 62 (3.8) | 72 (5.8) |
| pmr mutant | − | 15 (1.3) | NA |
| pmr mutant | + | 52 (5.8) | NA |
| lasR rhlR mutant | − | 18 (0.7) | NA |
| lasR rhlR mutant | + | 53 (1.4) | NA |
Averages (μg/ml) and standard deviations (in brackets) from three measurements are shown. NA, not applicable.
DISCUSSION
In the present paper, we demonstrate that exogenously added DNA can be incorporated into P. aeruginosa biofilms and increase their tolerance toward aminoglycosides. We provide evidence that biofilms formed in flow chambers by a DNA release-deficient P. aeruginosa quorum-sensing mutant are susceptible to aminoglycoside treatment but become aminoglycoside tolerant if they are supplied with exogenous DNA. We demonstrate that addition of lysed PMNs increases the tolerance of P. aeruginosa biofilms toward aminoglycosides. Moreover, we show that the biofilm-associated aminoglycoside tolerance mediated by the presence of eDNA is not caused by activation of the pmr genes under our conditions.
In agreement with our results, Mulcahy et al. (20) also found that the presence of eDNA increased the MBC-B values of P. aeruginosa wild-type biofilms toward tobramycin and gentamicin. They also included a pmr mutant (PA3553) in their studies, noted that this mutant also displayed DNA-induced resistance to tobramycin and gentamicin, and suggested that DNA-induced resistance to aminoglycosides is not limited to PA3553 induction (20). The results of Mulcahy et al. therefore support our conclusion that eDNA can mediate aminoglycoside tolerance in P. aeruginosa biofilms both because of activation of the Pmr system and because of a shield effect. The effect of eDNA on the aminoglycoside tolerance of our microtiter tray-grown biofilms was not as dramatic as that found by Mulcahy et al. For example, we found that addition of eDNA increased the tobramycin tolerance of wild-type biofilms 3.3-fold and the tobramycin tolerance of pmr mutant biofilms 3.5-fold, whereas Mulcahy et al. reported that DNA supplementation increased the tobramycin tolerance of wild-type biofilms 128-fold and the tobramycin tolerance of pmr mutant biofilms 16-fold. However, in our experiments, a lot of eDNA most likely was removed from the biofilms when the planktonic bacteria were removed prior to addition of DNA-free medium with antibiotic, whereas Mulcahy et al. challenged the biofilms with antibiotic in the same media in which they were cultivated (i.e., containing eDNA).
DNA binds aminoglycosides and positively charged antimicrobial peptides via electrostatic interactions (23). We anticipate that eDNA in P. aeruginosa biofilms can act like a sink that sequesters the antibiotics. The eDNA-mediated antimicrobial tolerance observed in our flow chamber biofilms lasted only for a limited time period. This finding is consistent with saturation of the eDNA by the constant delivery of fresh antibiotic in the perfusion medium irrigated to the flow cells. However, eDNA-mediated delayed penetration of aminoglycosides in biofilms might play an important role in the antimicrobial tolerance displayed by biofilm infections. An infected CF lung, for example, may have zones with poor access to aminoglycoside aerosols, so that the antibiotic concentration is too low to saturate the biofilm matrix.
Although PMNs normally can clear acute infections caused by planktonic bacteria, the PMNs fail to eradicate bacteria present in biofilms, possibly due to protection offered by the extracellular biofilm matrix and production of certain virulence factors that can abolish proper PMN function (1). Accordingly, P. aeruginosa was found to produce cytolytic rhamnolipids in biofilms (18, 45). The production of rhamnolipid in P. aeruginosa biofilms was demonstrated to be induced by the presence of PMNs, and the rhamnolipid was found to subsequently lyse the PMNs from which DNA was shown to be released (18, 45). Thus, the rhamnolipid may contribute to the ability of P. aeruginosa to persist in biofilm infections. In line with this, microscopic inspection of lung tissues from chronically infected CF patients and from chronic wound tissues revealed that P. aeruginosa biofilms in these tissues were surrounded but not penetrated by PMNs (46, 47), similar to what has been observed in studies with in vitro P. aeruginosa biofilms overlaid with freshly isolated PMNs (37). The persistent PMN accumulation and necrosis associated with biofilm infections result in sites highly enriched with DNA, actin, and granule proteins. Walker et al. (17) provided evidence that the presence of PMNs enhances P. aeruginosa biofilm formation through the lytic release of DNA and actin polymers that reinforces the biofilm matrix. These findings suggest a potential maladaptation of the primary innate immune response against bacterial infection. When the host fails to eradicate the infection, DNA and other cellular components from necrotic PMNs serve as an extracellular matrix to facilitate biofilm formation. As shown here, the DNA provided by the PMNs can increase the tolerance of the biofilms toward aminoglycosides. However, because DNA also binds cationic antimicrobial peptides (48, 49), it is possible that the DNA in biofilms can protect the bacteria against many of the antimicrobials produced by the host, e.g., lysozyme, lactoferrin, β-defensin, and LL37. The activity of the bacteria and PMNs during a biofilm infection may thus result in a vicious cycle: PMNs are attracted by the bacteria, and when they get in contact with the biofilm, they are lysed. The DNA liberated from the lysed PMNs is then used by the bacteria to reinforce the biofilm and shield against antimicrobial peptides produced by the host and aminoglycosides administered by the physician.
In addition to eDNA, other components of the extracellular matrix may confer tolerance to aminoglycosides in biofilms. Evidence has been provided that alginate in the biofilm matrix and cyclic glucans in the periplasm of the bacteria may protect P. aeruginosa biofilms from aminoglycosides by binding the antibiotics (35, 50). Moreover, Colvin et al. (51) recently provided evidence that the Pel exopolysaccharide plays a role in aminoglycoside tolerance in P. aeruginosa biofilms. However, contrary to the study by Colvin et al. (51), Khan et al. (52) presented evidence that the presence of Pel polysaccharide is not important for aminoglycoside tolerance in P. aeruginosa biofilms grown in microtiter dishes.
The work described here has provided knowledge about the molecular mechanisms which are involved in antimicrobial tolerance in P. aeruginosa biofilms. A detailed understanding of the mechanisms underlying the recalcitrance of biofilms toward antimicrobial therapy will ultimately enable us to develop efficient treatments against a wide range of persistent infections.
ACKNOWLEDGMENTS
We thank Morten Theil Rybtke for designing the primers used to PCR amplify DNA upstream and downstream of the lasR and rhlR genes. We are grateful to Lars Christophersen for providing lysed PMNs.
This work was supported by grants from the Danish Strategic Research Council (to M.G.), the Danish Council for Independent Research (to T.T.-N.), the Novo Nordisk Foundation (to M.G.), and the Lundbeck Foundation (to T.T.-N.).
Footnotes
Published ahead of print 11 March 2013
REFERENCES
- 1. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 2. Ciofu O, Tolker-Nielsen T. 2010. Antibiotic tolerance and resistance in biofilms, p 215–230 In Bjarnsholt T, Jensen PØ, Moser C, Høiby N. (ed), Biofilm infections. Springer Publishing Company, New York, NY [Google Scholar]
- 3. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 4. Friedman L, Kolter R. 2003. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 5. Matsukawa M, Greenberg EP. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 8. Eckhart L, Fischer H, Barken KB, Tolker-Nielsen T, Tschachler E. 2007. DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Brit. J. Dermatol. 156:1342–1345 [DOI] [PubMed] [Google Scholar]
- 9. Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. 2010. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59:253–268 [DOI] [PubMed] [Google Scholar]
- 10. Nilsson M, Chiang WC, Fazli M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2011. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 13:1357–1369 [DOI] [PubMed] [Google Scholar]
- 11. Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318–1328 [DOI] [PubMed] [Google Scholar]
- 12. Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331–2343 [DOI] [PubMed] [Google Scholar]
- 13. Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74:1380–1392 [DOI] [PubMed] [Google Scholar]
- 14. Petersen FC, Pecharki D, Scheie AA. 2004. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 186:6327–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092 [DOI] [PubMed] [Google Scholar]
- 17. Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA. 2005. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 73:3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alhede M, Bjarnsholt T, Jensen PO, Phipps RK, Moser C, Christophersen L, Christensen LD, van Gennip M, Parsek M, Hoiby N, Rasmussen TB, Givskov M. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508 [DOI] [PubMed] [Google Scholar]
- 19. Fazli M, Bjarnsholt T, Kirketerp-Moller K, Jorgensen A, Andersen CB, Givskov M, Tolker-Nielsen T. 2011. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Repair Regen. 19:387–391 [DOI] [PubMed] [Google Scholar]
- 20. Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213 doi:10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramphal R, Lhermitte M, Filliat M, Roussel P. 1988. The binding of anti-pseudomonal antibiotics to macromolecules from cystic fibrosis sputum. J. Antimicrob. Chemother. 22:483–490 [DOI] [PubMed] [Google Scholar]
- 22. Hunt BE, Weber A, Berger A, Ramsey B, Smith AL. 1995. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob. Agents Chemother. 39:34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drew KRP, Sanders LK, Culumber ZW, Zribi O, Wong GCL. 2009. Cationic amphiphiles increase activity of aminoglycoside antibiotic tobramycin in the presence of airway polyelectrolytes. J. Am. Chem. Soc. 131:486–493 [DOI] [PubMed] [Google Scholar]
- 24. Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40:79–105 [DOI] [PubMed] [Google Scholar]
- 25. Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511–1524 [DOI] [PubMed] [Google Scholar]
- 26. Haagensen JA, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kessler B, de Lorenzo V, Timmis KN. 1992. A general system to integrate lacZ fusions into the chromosomes of Gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293–301 [DOI] [PubMed] [Google Scholar]
- 28. Simon R, Priefer UB, Puhler A. 1982. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 29. Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 30. Koch B, Jensen LE, Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187–195 [DOI] [PubMed] [Google Scholar]
- 31. Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 33. Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99–112 [Google Scholar]
- 34. Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68:223–240 [DOI] [PubMed] [Google Scholar]
- 35. Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 36. Crusz SA, Popat R, Rybtke MT, Camara M, Givskov M, Tolker-Nielsen T, Diggle SP, Williams P. 2012. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling 28:835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Hoiby N, Givskov M. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383 [DOI] [PubMed] [Google Scholar]
- 38. Shah PL, Scott SF, Knight RA, Marriott C, Ranasinha C, Hodson ME. 1996. In vivo effects of recombinant human DNase I on sputum in patients with cystic fibrosis. Thorax 51:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Potter JL, Spector S, Matthews LW, Lemm J. 1969. Studies on pulmonary secretions. 3. The nucleic acids in whole pulmonary secretions from patients with cystic fibrosis, bronchiectasis, and laryngectomy. Am. Rev. Respir. Dis. 99:909–916 [DOI] [PubMed] [Google Scholar]
- 40. Kolpen M, Hansen CR, Bjarnsholt T, Moser C, Christensen LD, van Gennip M, Ciofu O, Mandsberg L, Kharazmi A, Doring G, Givskov M, Hoiby N, Jensen PO. 2010. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65:57–62 [DOI] [PubMed] [Google Scholar]
- 41. Döring G, Knight R, Bellon G. 2000. Immunology of cystic fibrosis, p 109–141 In Hodson M, Geddes D. (ed), Cystic fibrosis. Arnold, London, United Kingdom [Google Scholar]
- 42. Macfarlane EL, Kwasnicka A, Hancock RE. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146(Part 10):2543–2554 [DOI] [PubMed] [Google Scholar]
- 43. Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305–316 [DOI] [PubMed] [Google Scholar]
- 44. McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 45. Jensen PO, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, Moser C, Williams P, Pressler T, Givskov M, Hoiby N. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329–1338 [DOI] [PubMed] [Google Scholar]
- 46. Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker-Nielsen T, Hoiby N, Givskov M, Bjarnsholt T. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558 [DOI] [PubMed] [Google Scholar]
- 48. Weiner DJ, Bucki R, Janmey PA. 2003. The antimicrobial activity of the cathelicidin LL37 is inhibited by F-actin bundles and restored by gelsolin. Am. J. Respir. Cell Mol. Biol. 28:738–745 [DOI] [PubMed] [Google Scholar]
- 49. Bucki R, Byfield FJ, Janmey PA. 2007. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur. Respir. J. 29:624–632 [DOI] [PubMed] [Google Scholar]
- 50. Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264 doi:10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khan W, Bernier SP, Kuchma SL, Hammond JH, Hasan F, O'Toole GA. 2010. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 13:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]