Abstract
Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is an intracellular second messenger that controls the lifestyles of many bacteria. A high intracellular level of c-di-GMP induces a biofilm lifestyle, whereas a low intracellular level of c-di-GMP stimulates dispersal of biofilms and promotes a planktonic lifestyle. Here, we used the expression of different reporters to show that planktonic cells, biofilm cells, and cells dispersed from biofilms (DCells) had distinct intracellular c-di-GMP levels. Proteomics analysis showed that the low intracellular c-di-GMP level of DCells induced the expression of proteins required for the virulence and development of antimicrobial peptide resistance in Pseudomonas aeruginosa. In accordance with this, P. aeruginosa cells with low c-di-GMP levels were found to be more resistant to colistin than P. aeruginosa cells with high c-di-GMP levels. This finding contradicts the current dogma stating that dispersed cells are inevitably more susceptible to antibiotics than their sessile counterparts.
INTRODUCTION
It is now widely accepted that microbes are able to form surfaced-attached biofilm communities in the environment and during infection as an alternative to the planktonic or free-living style. Biofilm formation proceeds through several distinct steps, including initial attachment, with subsequent development of dense microcolonies embedded in self-generated extracellular matrix materials (1) and finally dispersal to seed new areas of biofilm formation (2).
Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is a global, intracellular second messenger that controls the lifestyles of many bacteria (3). The intracellular c-di-GMP concentration is controlled by diguanylate cyclases (DGCs) which catalyze the formation of c-di-GMP and phosphodiesterases (PDEs) which degrade c-di-GMP (4). Many bacteria contain multiple copies of DGCs and PDEs, which allow bacterial cells to sense and respond to diverse sets of environmental signals by adjusting the intracellular c-di-GMP content accordingly.
As a secondary messenger that binds to specific domains of regulatory proteins, high level of c-di-GMP stimulates bacteria to form biofilm by enhancing the synthesis of adhesive structures and biofilm matrix components and by reducing motility and chemotaxis (5, 6). In the aggregated biofilm mode, quorum sensing contributes to the production of matrix components that facilitate protection of the biofilm cells against cellular immunity attack and antimicrobial treatments (7–10). Recently, however, a low intracellular level of c-di-GMP has been shown to be necessary for the pathogenesis of bacteria (11, 12). The CheY-EAL-HTH domain protein VieA of Vibrio cholerae is required for the activation of certain virulence factors (13). Another EAL domain-containing protein, CdgR, has been shown to be required by Salmonella to resist phagocytosis and virulence during infection of mice (14).
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that can cause a wide range of infections, including those in cystic fibrosis, wounds, and the urinary tract (15). The success of P. aeruginosa as a human pathogen is largely dependent on its ability to form biofilms, produce virulence factors, and launch immune protective measures in an organized fashion, as well as its notorious resistance to antimicrobial agents (16, 17), all of which may allow infections to develop into chronic conditions (16, 17). Here, we studied the effects of modulating the intracellular content of c-di-GMP in P. aeruginosa in relation to biofilm dispersal and antimicrobial peptide resistance.
MATERIALS AND METHODS
Bacteria and growth conditions.
The bacterial strains, plasmids, and primers used in the present study are listed in Table 1. Escherichia coli DH5a strain was used for standard DNA manipulations. Luria-Bertani medium (18) was used to cultivate E. coli strains. Batch cultivation of P. aeruginosa was carried out at 37°C in ABT minimal medium (19) supplemented with 5 g of glucose liter−1 (ABTG) or 2 g of glucose liter−1 plus 2 g of Casamino Acids liter−1 (ABTGC). For plasmid maintenance in E. coli, the medium was supplemented with 100 μg of ampicillin ml−1, 15 μg of gentamicin (Gm) ml−1, 15 μg of tetracycline (Tc) ml−1, or 8 μg of chloramphenicol ml−1. For marker selection in P. aeruginosa, 30 μg of Gm ml−1, 50 μg of Tc ml−1, and 200 μg of carbenicillin ml−1 were used, as appropriate.
Table 1.
Strains,plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototypic nonmucoid wild-type strain | 20 |
| PAO1ΔwspF | wspF derivative of PAO1 constructed by allelic exchange | 21 |
| PAO1/plac-yhjH | Tcr; PAO1 containing the plac-yhjH vector | This study |
| PAO1/pBAD-yhjH | Gmr; PAO1 containing the pBAD-yhjH vector | This study |
| PAO1ΔwspF/plac-yhjH | Tcr; PAO1ΔwspF containing the plac-yhjH vector | This study |
| PAO1ΔwspF/pBAD-yhjH | Gmr; PAO1ΔwspF containing the pBAD-yhjH vector | This study |
| PAO1ΔpelAΔpslBCD/pcdrA-gfp | Gmr Cbr; low intracellular c-di-GMP content derivative of PAO1 | 21 |
| PAO1/pcdrA-gfp | Gmr; PAO1 containing the pcdrA-gfp vector | This study |
| PAO1ΔwspF/pcdrA-gfp | Gmr Cbr; PAO1ΔwspF containing the pcdrA-gfp vector | This study |
| PAO1/plac-yhjH/pcdrA-gfp | Tcr Cbr; PAO1/plac-yhjH containing the pcdrA-gfp vector | This study |
| PAO1/pBAD-yhjH/pcdrA-gfp | Gmr Cbr; PAO1/pBAD-yhjH containing the pcdrA-gfp vector | This study |
| PAO1-ppmr-gfp | Gmr; PAO1 tagged by miniTn7-ppmr-gfp | This study |
| PAO1ΔwspF/ppmr-gfp | Gmr; PAO1ΔwspF tagged by miniTn7-ppmr-gfp | This study |
| PAO1/plac-yhjH/ppmr-gfp | Tcr Gmr; PAO1/plac-yhjH tagged by miniTn7-ppmr-gfp | This study |
| PAO1/ppelA-lacZ | Tcr; PAO1 tagged by miniCTX-ppelA-lacZ | This study |
| PAO1ΔwspF/ppelA-lacZ | Tcr; PAO1ΔwspF tagged by miniCTX-ppelA-lacZ | This study |
| PAO1/pBAD-yhjH/ppelA-lacZ | Tcr Gmr; PAO1/pBAD-yhjH tagged by miniCTX-ppelA-lacZ | This study |
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Laboratory collection |
| Plasmids | ||
| pUCP22 | Apr Gmr; broad-host-range cloning vector | 22 |
| pJN105 | Gmr; broad-host-range vector carrying the araBAD promoter | 23 |
| pBBR1MCS3 | Tcr; broad-host-range ori from Bordetella bronchiseptica S87 | 24 |
| miniTn7-ppmr-gfp | Apr Gmr; miniTn7 vector carrying the ppmr-gfp fusion | 25 |
| miniCTX-ppelA-lacZ | Tcr; miniCTX vector carrying the ppelA-lacZ fusion | 26 |
| plac-yhjH | Tcr; pBBR1MCS3 carrying the yhjH gene | 27 |
| pBAD-yhjH | Gmr; pJN105 carrying the yhjH gene | This study |
| pcdrA-gfp | Apr Gmr; pUCP22 carrying the pcdrA-gfp fusion | 21 |
| pRK600 | Cmr; ori ColE1 RK2-Mob+ RK2-Tra+; helper vector for conjugation | 28 |
| Primers | ||
| yhjH-fwd | AAACTGCAGTAGTGGAGGAATTTGATGATAAGGCAGGTTATCCAGC | This study |
| yhjH-rev | AAATCTAGAGAAAATGAGGCAGCTTATAGCGC | This study |
Cmr, chloramphenicol resistance; Tcr,tetracycline resistance; Apr, ampicillin resistance; Gmr,gentamicin resistance; Cbr, carbenicillin resistance.
Construction of pBAD-yhjH vector.
Plasmid pJN105 contains an araC-PBAD promoter, which has been well studied and induced in the presence of l-arabinose (23). The yhjH gene of E. coli MG1655 was amplified by PCR using primers yhjH-rev and yhjH-fwd. The PCR product was cloned into the vector pJN105 by restriction with PstI and XbaI. DNA restriction enzyme digestions and modifications were performed according to the manufacturer's instructions (Fermentas and Invitrogen). The resulting plasmid pBAD-yhjH was transferred into E. coli DH5α by electroporation. Correct insertion of the yhjH gene into the vector pJN105 was verified by sequencing. The pBAD-yhjH plasmid was transformed into E. coli S17-1 by electroporation and thereafter conjugated into P. aeruginosa.
CdrA-gfp assay.
P. aeruginosa strains containing pcdrA-gfp reporter were cultivated in ABTGC medium at 37°C with shaking. Portions (200 μl) of overnight cultures were transferred into each of the wells of a 96-well microplate. The expression of pcdrA-gfp in P. aeruginosa was measured using a Tecan Infinite Pro2000 microplate reader. The optical density at 600 nm (OD600) and green fluorescent protein (GFP) fluorescence (in relative fluorescence units) were recorded for each well of the 96-well microplate.
For measuring pcdrA-gfp expression in biofilm cells of the PAO1 strain, the P. aeruginosa PAO1/pcdrA-gfp strain were cultivated in 50-ml BD Falcon tubes containing 15 ml of ABTGC medium. A sterile glass cover slide (24 by 60 mm) was inserted into each Falcon tube to support biofilm growth. After overnight incubation, PAO1 biofilms on the slides were washed twice with 1 ml of 0.9% NaCl and imaged using fluorescence microscopy (Carl Zeiss). The planktonically growing PAO1/pcdrA-gfp strain and strain PAO1ΔwspF/pcdrA-gfp were also imaged using fluorescence microscopy for comparison.
Pel-lacZ assay.
The mini-CTX-ppel-lacZ reporter fusion (26) was inserted into the chromosomes of P. aeruginosa PAO1, PAO1ΔwspF, and PAO1/pBAD-yhjH strains by triparental mating with the help of pRK600 vectors as previously described (29). PAO1/pBAD-yhjH biofilms were cultivated in ABTGC medium in a 24-well plate (Nunc) overnight at 37°C. The biofilms were washed twice with 1 ml of 0.9% NaCl and supplemented with ABTGC medium containing 0.25, 0.5, or 1% arabinose for 5 h to induce dispersal. Biofilms formed by the PAO1 and PAO1ΔwspF strains were dispersed by 5 μM NO donor sodium nitroprusside (SNP; Sigma). As controls, PAO1 and PAO1/pBAD-yhjH planktonic cultures were diluted 10 times to fresh ABTGC medium and incubated for 5 h at 37°C with shaking. The OD600 values of planktonic cells were measured and normalized to the same OD600 values of dispersed cells. A classical β-galactosidase assay was used to measure expression of the ppel-lacZ fusion in P. aeruginosa cells (30).
Intracellular c-di-GMP concentration in biofilm cells.
To assay c-di-GMP concentrations of biofilm cells, the glass slide biofilm assay was performed as previously reported (31). The pcdrA-gfp containing P. aeruginosa PAO1 and PAO1/pBAD-yhjH strain were cultivated in 50-ml BD Falcon tubes containing 15 ml of ABTGC medium. A sterile glass cover slide (24 by 60 mm) was inserted into each Falcon tube to support biofilm growth. After overnight incubation, slide biofilms were washed twice with 0.9% NaCl and imaged by using fluorescence microscopy (Carl Zeiss).
Microplate biofilm formation assay.
A microplate biofilm formation assay was carried out in ABTGC medium as previously described (32).
iTRAQ-based proteomics analyses.
P. aeruginosa cells were harvested after 48 h of cultivation in AB minimal medium supplemented with 5 g of glucose liter−1 and subjected to iTRAQ-based proteomics analyses (additional details for these analyses are provided in the supplemental material).
Pyoverdine quantification.
PAO1, PAO1ΔwspF, and PAO1/plac-yhjH strains were grown in ABTGC medium overnight. The pyoverdine fluorescence (excitation wavelength, 400 nm; emission wavelength, 450 nm) of each supernatant of P. aeruginosa overnight cultures was recorded by using the Tecan Infinite Pro2000 microplate reader as previously reported (33).
Pmr-gfp assay.
The miniTn7-Gm-ppmr-gfp fusion was inserted into the chromosomes of PAO1, PAO1ΔwspF, and PAO1/plac-yhjH strains by four-parental mating with the help of pBF13 and pRK600 vectors as previously described (29). P. aeruginosa PAO1, PAO1ΔwspF, and PAO1/plac-yhjH strains were grown in ABTGC medium overnight. The cultures were then diluted 10-fold into fresh ABTGC medium with or without 1 μg of colistin ml−1. Cultures, 3 μl for each condition, were spotted onto cover slides after 7 h growth for fluorescence microscopy imaging (Carl Zeiss). The level of fluorescence of 30 individual ppmr-gfp-tagged bacterial cells was measured for each sample by using ImageJ (http://rsbweb.nih.gov/ij/). The corrected total cell fluorescence of each cell was calculated as the sum of the fluorescence intensity within the region of interest minus the background intensity.
Antimicrobial peptide resistance assay of planktonic cells.
For comparison of the resistance of strains PAO1, PAO1ΔwspF, and PAO1/plac-yhjH to colistin, the growth curves of the three strains in the presence of 0, 0.125, and 2 μg of colistin ml−1 were produced in triplicate, as previously described (34). Colistin (0.25 μg ml−1) was selected to represent a concentration lower than the MIC of PAO1 (which is 1 μg ml−1), and 2 μg of colistin ml−1 was chosen for a concentration higher than the MIC. Overnight cultures were diluted to an OD600 of 0.15 with ABTGC minimal medium containing the appropriate concentrations of colistin. The OD600 was recorded every hour for 9 h using the Tecan Infinite Pro2000 microplate reader.
A time-kill kinetic assay was also performed to compare the resistance of PAO1, PAO1ΔwspF, and PAO1/plac-yhjH to colistin to concentrations of 2, 4, and 8 μg ml−1, respectively. Overnight cultures of PAO1, PAO1ΔwspF, and PAO1/plac-yhjH strains were diluted to an OD600 of ∼0.2 in fresh ABTGC medium containing 2, 4, and 8 μg of colistin ml−1, respectively. The absorbance of the surviving bacterial cells was monitored by using the Tecan Infinite Pro2000 microplate reader and Live/Dead BacLight bacterial viability kits (Invitrogen).
Colistin resistance assay of dispersed cells.
In order to compare the tolerance of cells that dispersed from biofilms with tube-cultivated planktonic cells to colistin, biofilms of PAO1 and PAO1/pBAD-yhjH were cultivated in ABTGC medium in a 24-well plate (Nunc) overnight at 37°C. The biofilms were washed twice with 1 ml of 0.9% NaCl and supplemented with ABTGC medium containing 5 μM SNP (PAO1 biofilms) or 0, 0.5, or 1% arabinose (PAO1/pBAD-yhjH biofilms) for 5 h to induce dispersal. Biofilms were stained with 0.01% crystal violet as previously described (32). As controls, biofilms of PAO1 and PAO1/pBAD-yhjH were washed twice with 1 ml of 0.9% NaCl and supplemented with ABTGC medium for 5 h. Planktonic cells were derived from both biofilms. The OD600 of dispersed cells and planktonic cells was measured and adjusted to an OD600 of 0.15. The growth curves of planktonic PAO1/pBAD-yhjH cells and dispersed biofilm cells from PAO1/pBAD-yhjH and PAO1 biofilms were measured in ABTGC medium containing 0, 0.125, and 2 μg of colistin ml−1. The OD600 was recorded every 15 min for 5 h using the Tecan Infinite Pro2000 microplate reader.
Biofilm colistin treatment assay.
The P. aeruginosa PAO1 strain was cultivated in 50-ml BD Falcon tubes containing 15 ml of ABTGC medium. A sterile glass cover slide (24 by 60 mm) was inserted into each Falcon tube to support biofilm growth. After overnight incubation, slide PAO1 biofilms were washed twice with 1 ml of 0.9% NaCl and supplemented with ABTGC medium containing 4 μg of colistin ml−1. PAO1 biofilms incubated in ABTGC medium without colistin were used as a control. After 2 h of treatment, the biofilms were washed twice with 0.9% NaCl, stained by using a Live/Dead bacterial viability kit (Invitrogen), and imaged using fluorescence microscopy (Carl Zeiss).
RESULTS
Construction of P. aeruginosa cells with different intracellular c-di-GMP levels.
We constructed P. aeruginosa strains with controllable intracellular levels of c-di-GMP so that we can mimic the three phases of the biofilm life cycle: planktonic cells (PCells), biofilm cells (BCells), and dispersed cells (DCells). The PAO1ΔwspF strain, which overexpresses the diguanylatecyclase WspR (21), is known to contain a high intracellular level of c-di-GMP (11) and can be used to mimic BCells. The PAO1/plac-yhjH strain contains a PBBRMCS-2 plasmid carrying the phosphodiesterase gene yhjH fused to a lac promoter, which is constitutively expressed in Pseudomonas (27), and can be used to mimic DCells. The PAO1/pBAD-yhjH strain contains a pJN105 plasmid carrying yhjH fused to an arabinose-inducible promoter. The intracellular content of c-di-GMP of the PAO1/pBAD-yhjH strain can be adjusted by arabinose and can therefore be used to mimic all of the three phases dependent on the conditions.
The cdrA and pel genes are both positively regulated by c-di-GMP in P. aeruginosa (35), and the fusions pcdrA-gfp and ppel-lacZ can therefore be used to monitor intracellular c-di-GMP levels in P. aeruginosa (21, 26). We measured the expression of the c-di-GMP biosensor pcdrA-gfp (21) in the PAO1 (PCells), PAO1ΔwspF strain (BCells), PAO1/plac-yhjH strain (DCells) and found that the intracellular level of c-di-GMP in the PAO1ΔwspF strain (BCells) was significantly higher than in the PAO1 strain (PCells) and the PAO1/plac-yhjH strain (DCells) (Fig. 1A). The wspF mutation was shown before to increase the intracellular content of c-di-GMP of P. aeruginosa up to 7-fold in planktonic growth (21). Expression of the plac-yhjH in PAO1ΔwspF strain was found to decrease the pcdrA-gfp fluorescence intensity (Fig. 1). PAO1 cells from biofilms had a high level of cdrA-gfp fluorescence intensity close to that of the PAO1ΔwspF cells (see Fig. S1 in the supplemental material). However, the pcdrA-gfp expression in PAO1 PCells and PAO1/plac-yhjH DCells was too low to indicate differences in the c-di-GMP level (Fig. 1). Nevertheless, the PAO1/plac-yhjH strain (DCells) was unable to form biofilms similar to the PAO1 (PCells) and PAO1ΔwspF strain during static cultivation (Fig. 1B), indicating that it had a low c-di-GMP level.
Fig 1.
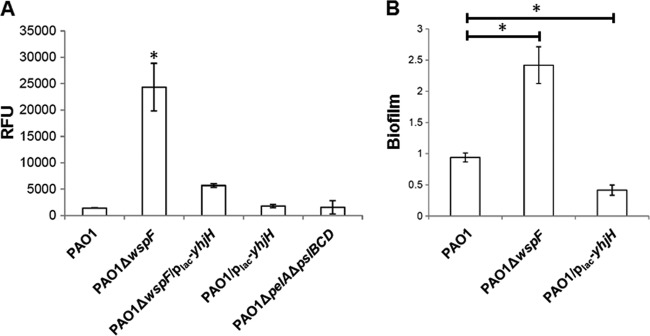
(A) Expression of pcdrA-gfp fusion in P. aeruginosa PAO1 (PCells), PAO1ΔwspF (BCells), PAO1ΔwspF/plac-yhjH, and PAO1/plac-yhjH (DCells) strains. Means and standard deviations (SD) in relative fluorescence units (RFU) from triplicate experiments are shown. *, P < 0.01. (B) Biofilm formation of P. aeruginosa PAO1 (PCells), PAO1ΔwspF (BCells), and PAO1/plac-yhjH (DCells) strains in microplates. Means and SD from triplicate experiments are shown. *, P < 0.01.
Due to the detection limit of the pcdrA-gfp biosensor, we then used the ppel-lacZ reporter gene (26) to compare the intracellular levels of c-di-GMP in SNP-dispersed biofilm cells (DCells*) and yhjH-dispersed biofilm cells (DCells). SNP reduces the intracellular c-di-GMP level in P. aeruginosa through activation of the DipA PDE (36), as an alternative to direct induction of the ectopically expressed YhjH PDE (27). The PAO1ΔwspF BCells showed a higher β-galactosidase activity than that of the PAO1 PCells (Fig. 2). SNP-dispersed PAO1 biofilm cells (DCells*) showed a level of β-galactosidase activity similar to that of PAO1/pBAD-yhjH cells dispersed by the addition of 0.5% arabinose (DCells) (Fig. 2). The dispersed cells (DCells* and DCells) expressed lower levels of β-galactosidase activity than did the planktonic PAO1 cells (PCells) (Fig. 2). This finding suggests that freshly dispersed P. aeruginosa cells (DCells) from the biofilms had lower levels of c-di-GMP than the planktonic cells (PCells).
Fig 2.

β-Galactosidase activity of P. aeruginosa strains grown as planktonic cells or biofilm cells containing the pel-lacZ biosensors. SNP was added to both PAO1 and the PAO1ΔwspF (BCells) at final concentration of 5 μM, whereas 0.25, 0.5, and 1% arabinose was added to the PAO1/pBAD-yhjH strain. For the biofilm cells, the β-galactosidase activity was measured in the dispersed cells. Means and SD in β-galactosidase activity from triplicate experiments are shown. *, P < 0.01.
Proteomics analysis of P. aeruginosa cells with different intracellular c-di-GMP levels.
Proteomics analysis of P. aeruginosa cells with different intracellular c-di-GMP levels was performed. Using a P value cutoff of 0.05, the abundances of 116 proteins were found to be significantly affected by low intracellular levels of c-di-GMP; the abundance of 44 proteins was upregulated, while the abundance of 72 proteins was downregulated (shown in Tables 2 and 3, respectively). As expected (3), extracellular matrix proteins were expressed more abundantly in PAO1ΔwspF strain (BCells) (Table 3), while motility and chemotaxis proteins were more abundant in PAO1/plac-yhjH (DCells) (Table 2).
Table 2.
Proteins whose abundance in P. aeruginosa PAO1 increased significantly in conditions of low intracellular levels of c-di-GMP
| PA no. | Gene | Description of product | Peptides (95%) | Coverage (95%) | Ratio (115:114) | Pa (115:114) |
|---|---|---|---|---|---|---|
| PA2452 | Enterochelin esterase | 43 | 53.42 | 99.08 | 5.22E–03 | |
| PA1092 | fliC | Flagellin type B | 146 | 54.3 | 99.08 | 3.19E–02 |
| PA3531 | bfrB | Bacterioferritin | 33 | 34.18 | 22.28 | 2.82E–04 |
| PA4777 | pmrB | PmrB: two-component regulator system signal sensor kinase PmrB | 2 | 5.451 | 11.91 | 3.28E–02 |
| PA1596 | htpG | Heat shock protein 90 | 30 | 22.4 | 10.57 | 1.58E–02 |
| PA2821 | gstA | Glutathione S-transferase | 5 | 20.91 | 10.28 | 3.68E–02 |
| PA4386 | groES | Cochaperonin GroES | 2 | 45.36 | 9.20 | 3.15E–02 |
| PA3126 | ibpA | Heat shock protein Hsp20 | 15 | 24.16 | 8.95 | 1.20E–02 |
| PA4228 | pchD | Pyochelin biosynthesis protein PchD | 14 | 13.71 | 8.24 | 3.35E–03 |
| PA1039 | ychJ | Hypothetical protein O1Q_07577 | 2 | 15.29 | 7.94 | 3.79E–02 |
| PA3552 | arnB | UDP-4-amino-4-deoxy-l-arabinose–oxoglutarate aminotransferase | 4 | 13.09 | 7.38 | 2.85E–02 |
| PA4942 | hflK | Protease subunit HflK | 6 | 17 | 7.31 | 2.24E–03 |
| PA1534 | recR | Recombination protein RecR | 2 | 21.21 | 7.31 | 3.72E–02 |
| PA4670 | prs | Ribose-phosphate pyrophosphokinase | 43 | 56.55 | 7.11 | 8.95E–05 |
| PA4710 | phuR | Heme/hemoglobin uptake outer membrane receptor PhuR | 20 | 25.52 | 6.55 | 1.57E–04 |
| PA4227 | pchR | Transcriptional regulator PchR | 3 | 15.54 | 6.31 | 2.34E–02 |
| PA4224 | pchG | Pyochelin biosynthetic protein PchG | 16 | 30.37 | 6.25 | 9.10E–03 |
| PA3158 | wbpB | UDP-2-acetamido-2-deoxy-d-glucuronic acid 3-dehydrogenase, WbpB | 10 | 22.78 | 6.08 | 3.03E–02 |
| PA0427 | oprM | Major intrinsic multiple antibiotic resistance efflux outer membrane protein OprM | 13 | 32.16 | 5.40 | 1.56E–03 |
| PA0018 | fmt | Bifunctional UDP-glucuronic acid decarboxylase/UDP-4-amino-4-deoxy-l-arabinose formyltransferase | 3 | 8.006 | 5.15 | 2.80E–02 |
| PA1803 | lon | Putative ATP-dependent protease | 23 | 25.28 | 4.79 | 1.21E–04 |
| PA3135 | Putative transcriptional regulator | 2 | 11.11 | 4.61 | 3.86E–02 | |
| PA4231 | pchA | Salicylate biosynthesis isochorismate synthase | 4 | 12.39 | 4.57 | 4.12E–02 |
| PA3114 | truA | tRNA pseudouridine synthase A | 3 | 17.54 | 3.98 | 3.39E–02 |
| PA3831 | pepA | Multifunctional aminopeptidase A | 42 | 38.59 | 3.80 | 2.66E–02 |
| PA3159 | wbpA | UDP-glucose/GDP-mannose dehydrogenase | 17 | 31.42 | 3.66 | 5.69E–03 |
| PA5054 | hslU | ATP-dependent protease ATP-binding subunit HslU | 6 | 13.87 | 3.44 | 4.85E–02 |
| PA4225 | pchF | pchF gene product | 24 | 16.8 | 3.25 | 5.80E–06 |
| PA0426 | mexB | RND multidrug efflux transporter MexB | 6 | 5.067 | 3.25 | 3.05E–03 |
| PA1288 | ompP1 | Putative outer membrane protein | 8 | 24.76 | 3.19 | 2.64E–02 |
| PA4749 | glmM | Phosphoglucosamine mutase | 5 | 8.09 | 3.16 | 3.36E–02 |
| PA5213 | gcvP1 | Glycine dehydrogenase | 3 | 3.967 | 2.91 | 8.00E–03 |
| PA3478 | rhlB | Rhamnosyltransferase chain B | 2 | 7.512 | 2.86 | 3.40E–03 |
| PA2086 | ephx | Epoxide hydrolase | 3 | 19.33 | 2.75 | 3.59E–02 |
| PA4336 | Hypothetical protein O1Q_03368 | 9 | 23.32 | 2.73 | 3.78E–03 | |
| PA4595 | yjjK | Putative ABC transporter ATP-binding protein | 15 | 24.37 | 2.68 | 9.43E–04 |
| PA4476 | Hypothetical protein O1Q_04078 | 6 | 5.956 | 2.51 | 9.67E–03 | |
| PA2290 | gcd | Glucose dehydrogenase | 9 | 9.34 | 2.42 | 3.72E–02 |
| PA5237 | yigC | 3-Octaprenyl-4-hydroxybenzoate carboxy-lyase | 3 | 6.967 | 2.42 | 4.15E–02 |
| PA2302 | ambE | Protein AmbE | 20 | 10.34 | 2.27 | 1.66E–02 |
| PA4307 | pctC | pctC gene product | 20 | 31.65 | 2.11 | 2.50E–03 |
| PA3707 | wspB | Hypothetical protein O1Q_02948 | 2 | 14.79 | 2.07 | 3.07E–02 |
| PA4588 | gdhA | Glutamate dehydrogenase | 30 | 41.57 | 2.05 | 3.19E–02 |
| PA4226 | pchE | pchE gene product | 78 | 30.6 | 2.03 | 1.85E–05 |
Significance was defined as having a 115:114 abundance score of >2.0 and a P value for 115:114 of <0.05. “115:114” refers to the ratio of the protein's abundance in the low c-di-GMP PAO1/plac-yhjH strain (strain 115) versus the high c-di-GMP PAO1ΔwspFstrain (strain 114).
Table 3.
Proteins whose abundance in P. aeruginosa PAO1 decreased significantly in conditions of low intracellular levels of c-di-GMP
| PA no. | Gene | Description of product | Peptides (95%) | Coverage (95%) | Ratio (115:114) | Pa (115:114) |
|---|---|---|---|---|---|---|
| PA1245 | aprX | Hypothetical protein O1Q_25902 | 21 | 33.82 | 0.03 | 2.74E–02 |
| PA3064 | pelA | PelA protein | 2 | 2.743 | 0.04 | 1.71E–02 |
| PA2395 | pvdO | Protein PvdO | 3 | 16.2 | 0.05 | 8.64E–03 |
| PA4554 | pilY1 | Type 4 fimbrial biogenesis protein PilY1 | 6 | 7.666 | 0.05 | 3.52E–02 |
| PA3613 | Hypothetical protein O1Q_02478 | 12 | 20.85 | 0.05 | 6.41E–03 | |
| PA2398 | fpvA | fpvA gene product | 40 | 25.64 | 0.06 | 8.02E–06 |
| PA2394 | pvdN | Protein PvdN | 57 | 50.82 | 0.06 | 1.24E–02 |
| PA0059 | osmC | Osmotically inducible protein OsmC | 2 | 26.24 | 0.07 | 3.22E–02 |
| PA5192 | pckA | Phosphoenolpyruvate carboxykinase | 8 | 18.13 | 0.07 | 7.83E–03 |
| PA0423 | pasP | Hypothetical protein O1Q_22583 | 17 | 59.16 | 0.08 | 1.49E–03 |
| PA0781 | Hypothetical protein O1Q_01027 | 4 | 6.259 | 0.08 | 1.13E–02 | |
| PA5171 | arcA | Arginine deiminase | 68 | 50.72 | 0.09 | 2.68E–05 |
| PA5427 | adhA | Alcohol dehydrogenase | 18 | 47.08 | 0.10 | 2.33E–02 |
| PA2397 | pvdE | Pyoverdine biosynthesis protein PvdE | 3 | 9.107 | 0.10 | 3.47E–02 |
| PA2392 | pvdP | pvdP gene product | 15 | 28.31 | 0.11 | 4.16E–04 |
| PA3117 | asd | Aspartate-semialdehyde dehydrogenase | 11 | 24.32 | 0.11 | 8.29E–03 |
| PA0764 | mucB | Sigma E regulatory protein, MucB/RseB | 4 | 21.52 | 0.13 | 2.89E–02 |
| PA3313 | Hypothetical protein | 17 | 44.48 | 0.13 | 6.00E–03 | |
| PA3190 | gltB | Putative binding protein component of ABC sugar transporter | 45 | 50 | 0.14 | 3.53E–03 |
| PA4624 | cdrB | Hypothetical protein O1Q_15965 | 5 | 12.16 | 0.14 | 3.22E–02 |
| PA5046 | maeB | Malic enzyme | 22 | 41.23 | 0.15 | 1.34E–03 |
| PA5312 | pauC | Putative aldehyde dehydrogenase | 15 | 16.1 | 0.15 | 4.37E–02 |
| PA3330 | Putative short-chain dehydrogenase | 21 | 36.18 | 0.15 | 1.05E–02 | |
| PA0895 | aruC | Bifunctional N-succinyldiaminopimelate-aminotransferase/acetylornithine transaminase protein | 23 | 49.26 | 0.15 | 1.40E–02 |
| PA4450 | murA | Bifunctional cyclohexadienyl dehydrogenase/3-phosphoshikimate 1-carboxyvinyltransferase | 32 | 35.12 | 0.15 | 9.69E–03 |
| PA3327 | Nonribosomal peptide synthetase | 31 | 15.48 | 0.15 | 2.35E–05 | |
| PA0482 | glcB | Malate synthase G | 33 | 24.69 | 0.16 | 3.22E–02 |
| PA3769 | guaA | GMP synthase | 31 | 35.62 | 0.17 | 1.49E–02 |
| PA3977 | hemL | Glutamate-1-semialdehyde aminotransferase | 27 | 37 | 0.18 | 6.48E–03 |
| PA2119 | adh | Alcohol dehydrogenase | 6 | 24.59 | 0.18 | 3.22E–02 |
| PA0552 | pgk | Phosphoglycerate kinase | 12 | 30.23 | 0.19 | 2.51E–02 |
| PA4687 | hitA | Ferric iron-binding periplasmic protein HitA | 10 | 29.55 | 0.19 | 2.03E–02 |
| PA2413 | pvdH | Diaminobutyrate–2-oxoglutarate aminotransferase | 22 | 41.36 | 0.19 | 7.34E–04 |
| PA4448 | hisD | Bifunctional histidinal dehydrogenase/histidinol dehydrogenase | 11 | 29.55 | 0.20 | 3.40E–02 |
| PA5172 | arcB | Ornithine carbamoyltransferase | 67 | 44.94 | 0.20 | 3.04E–02 |
| PA3686 | adk | Adenylate kinase | 4 | 25.58 | 0.20 | 3.45E–02 |
| PA3922 | Hypothetical protein O1Q_15760 | 7 | 21.54 | 0.21 | 4.15E–02 | |
| PA2385 | pvdQ | 3-Oxo-C12-homoserine lactone acylase PvdQ | 19 | 22.31 | 0.21 | 4.23E–04 |
| PA3729 | Hypothetical protein O1Q_03058 | 3 | 3.634 | 0.22 | 2.48E–02 | |
| PA0314 | fliY | l-Cysteine transporter of ABC system FliY | 7 | 30.86 | 0.24 | 1.02E–02 |
| PA2445 | gcvP2 | Glycine dehydrogenase | 21 | 24.19 | 0.24 | 2.85E–04 |
| PA0084 | tssC1 | Hypothetical protein O1Q_08024 | 8 | 19.48 | 0.24 | 2.89E–02 |
| PA3452 | mqoA | Malate:quinone oxidoreductase | 4 | 10.52 | 0.26 | 7.25E–03 |
| PA0400 | metB | Putative cystathionine gamma-lyase | 26 | 48.48 | 0.26 | 7.74E–03 |
| PA4138 | tyrS | Tyrosyl-tRNA synthetase | 7 | 19.8 | 0.26 | 4.93E–02 |
| PA4236 | katA | Catalase | 14 | 28.42 | 0.27 | 8.08E–03 |
| PA0139 | ahpC | Alkyl hydroperoxide reductase subunit C | 16 | 40.64 | 0.27 | 2.34E–02 |
| PA5322 | algC | Phosphomannomutase | 12 | 16.94 | 0.27 | 1.63E–02 |
| PA2944 | cobN | Cobaltochelatase subunit CobN | 8 | 9.936 | 0.29 | 1.96E–02 |
| PA3186 | oprB | Glucose-sensitive porin, partial | 7 | 7.432 | 0.29 | 3.49E–02 |
| PA0036 | trpB | Tryptophan synthase subunit beta | 12 | 25.87 | 0.30 | 2.98E–02 |
| PA4560 | ileS | Isoleucyl-tRNA synthetase | 20 | 16.65 | 0.32 | 4.14E–03 |
| PA4938 | purA | Adenylosuccinate synthetase | 17 | 41.86 | 0.34 | 5.56E–03 |
| PA0077 | tssM1 | Hypothetical protein O1Q_28197 | 5 | 6.378 | 0.34 | 4.85E–02 |
| PA4266 | fusA1 | Elongation factor G | 75 | 44.62 | 0.34 | 8.93E–04 |
| PA4829 | lpd3 | Dihydrolipoamide dehydrogenase | 20 | 45.91 | 0.35 | 6.32E–03 |
| PA3328 | Putative FAD-dependent monooxygenase | 12 | 19.85 | 0.35 | 1.96E–02 | |
| PA3666 | dapD | 2,3,4,5-Tetrahydropyridine-2-carboxylate N-succinyltransferase | 8 | 28.49 | 0.36 | 1.20E–02 |
| PA2391 | opmQ | Hypothetical protein O1Q_22777 | 7 | 23.94 | 0.37 | 7.47E–03 |
| PA0956 | proS | Prolyl-tRNA synthetase | 14 | 26.8 | 0.37 | 8.13E–03 |
| PA5131 | pgm | Phosphoglyceromutase | 5 | 11.46 | 0.37 | 3.54E–02 |
| PA3213 | Hypothetical protein O1Q_21511 | 7 | 46.4 | 0.41 | 2.14E–02 | |
| PA1833 | yhfP | Putative oxidoreductase | 13 | 34.24 | 0.42 | 4.81E–03 |
| PA0548 | tktA | Transketolase | 10 | 17.74 | 0.42 | 4.74E–03 |
| PA5497 | nrdJa | nrdJa gene product | 9 | 13.08 | 0.43 | 1.94E–02 |
| PA1342 | Putative binding protein component of ABC transporter | 10 | 31.79 | 0.44 | 1.39E–02 | |
| PA0067 | prlC | Oligopeptidase A | 13 | 19.82 | 0.44 | 4.51E–02 |
| PA1588 | sucC | Succinyl coenzyme A synthetase subunit beta | 52 | 42.53 | 0.45 | 6.66E–03 |
| PA1010 | dapA | Dihydrodipicolinate synthase | 21 | 45.89 | 0.47 | 6.41E–03 |
| PA3790 | oprC | Outer membrane copper receptor OprC | 15 | 24.76 | 0.48 | 7.21E–03 |
| PA3194 | edd | Phosphogluconate dehydratase | 36 | 36.84 | 0.49 | 3.74E–03 |
| PA1583 | sdhA | Succinate dehydrogenase flavoprotein subunit | 25 | 22.88 | 0.50 | 3.35E–02 |
Significance was defined as having a 115:114 abundance score of <0.5 and a P value for 115:114 of <0.05. “115:114” refers to the ratio of the protein's abundance in the low c-di-GMP PAO1/plac-yhjH strain (strain 115) versus the high c-di-GMP PAO1ΔwspF strain (train 114).
High intracellular levels of c-di-GMP were correlated with the increased expression of proteins for synthesis of the major iron siderophore, pyoverdine (Table 3). The data were corroborated by pyoverdine fluorescence measurements showing that the production of pyoverdine in the P. aeruginosa PAO1, PAO1ΔwspF, and PAO1/plac-yhjH strains was in accordance with the proteomics analysis (Fig. 3).
Fig 3.
Pyoverdine production by P. aeruginosa PAO1 (PCells), PAO1ΔwspF (BCells), and PAO1/plac-yhjH (DCells). The pyoverdine fluorescence levels (excitation wavelength, 400 nm; emission wavelength, 450 nm) of supernatants of P. aeruginosa overnight cultures were recorded using the Tecan Infinite Pro2000 microplate reader.
Low intracellular levels of c-di-GMP were found to favor the expression of a set of virulence-associated proteins (Table 2). Surprisingly, we found that dispersal correlated with the expression of proteins that contributes to the antimicrobial peptide resistance of P. aeruginosa. Antimicrobial peptides (AMPs; e.g., defensins) are secreted by a wide-range of host cells as a response to microbial infections and act by disrupting the bacterial cell (37). Bacteria have evolved a set of inducible AMP-sensing systems (38, 39). In P. aeruginosa, the PhoP/PhoQ system and the PmrA/PmrB two-component systems can sense the presence of AMPs and upregulate genes involved in AMP resistance, including lipopolysaccharide modification (40). The arn operon (PA3552-PA3559) can also be induced by AMPs, and its expression is partially regulated by the PmrA/PmrB two-component system (40). PmrB and ArnB, typically induced by antimicrobial peptides (41, 42), were observed to be induced here by low intracellular levels of c-di-GMP (Table 2).
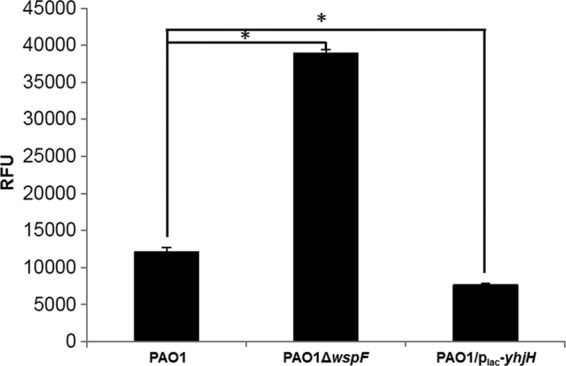
To examine whether the c-di-GMP effect found by proteomic analysis is on the level of transcription, we analyzed the expression of a ppmrA-gfp transcriptional fusion (25) in PAO1 (PCells), PAO1ΔwspF (BCells), and PAO1/plac-yhjH (DCells). The ppmrA-gfp fusion was expressed in all three strains in the presence of sublethal concentrations of colistin, but in the absence of any colistin, the fusion was only expressed in the PAO1/plac-yhjH (DCells) (Fig. 4 and see Fig. S2 in the supplemental material).
Fig 4.

Ppmr-gfp expression in P. aeruginosa PAO1 (PCells), PAO1ΔwspF (BCells), and PAO1/plac-yhjH (DCells) strains. Overnight cultures were diluted 10-fold into fresh ABTGC medium with or without 1 μg of colistin ml−1. Portions (3 μl) of cultures representing each condition were spotted onto cover slides after 7 h of growth for imaging by fluorescence microscopy. The level of fluorescence of 30 individual ppmr-gfp tagged bacterial cells was measured for each sample by using ImageJ. Means and SD in relative fluorescence intensity units (RFU) from 30 individual cells are shown. *, P < 0.01.
Antimicrobial peptide resistance of P. aeruginosa cells with different intracellular c-di-GMP levels.
Growth monitored in the presence of different concentrations of colistin revealed that the PAO1/plac-yhjH DCells were more resistant to colistin than PAO1 PCells and PAO1ΔwspF BCells during planktonic growth (Fig. 5A, B, and C). Colistin is a fast-killing bactericidal agent, and we thus measured the killing kinetics of 4 μg of colistin ml−1 in PAO1 PCells and PAO1/plac-yhjH DCells. PAO1 PCells and PAO1ΔwspF BCells were killed rapidly by 4 μg of colistin ml−1, whereas PAO1/plac-yhjH DCells were able to survive in the presence of 4 μg of colistin ml−1 (Fig. 5D and see Fig. S3 in the supplemental material).
Fig 5.
Colistin resistance assay. P. aeruginosa PAO1 (PCells) (A), PAO1ΔwspF (BCells) (B), and PAO1/plac-yhjH (DCells) (C) were cultivated at 37°C in ABTGC medium with 0, 0.25, or 2 μg of colistin ml−1. The OD600 was monitored for 10 h. Means and SD from triplicate experiments are shown. (D) Fast-kill assay of P. aeruginosa PAO1 (PCells), PAO1ΔwspF (BCells), and PAO1/plac-yhjH (DCells) by 4 μg of colistin ml−1. The proportion of dead bacterial cells was monitored by using the Live/Dead BacLight bacterial viability kits (Invitrogen) after 10 min of treatment. *, P < 0.01.
To examine whether cells that dispersed from biofilms were also more resistant to colistin than planktonic cells, the dispersal cells from biofilms of PAO1 and PAO1/pBAD-yhjH were tested for colistin resistance, and it was observed that PAO1/pBAD-yhjH cells dispersed from biofilms by the expression of yhjH were more resistant to colistin based on differences in growth rates (Fig. 6A and B). It was also observed that P. aeruginosa biofilms treated with dispersing agents (either arabinose or SNP) were more resistant to colistin than planktonic cells (Fig. 6C and D). In contrast, exposure of biofilms to colistin resulted in the killing of most biofilm cells and showed that a large fraction of the biofilm cells remained sensitive to colistin (Fig. 6E and F).
Fig 6.
Colistin resistance of planktonic cells (PCells), biofilm cells (BCells), and dispersed cells (DCells). Planktonic cells (PCells) of PAO1 (A), biofilm-dispersed cells (BCells) from PAO1 biofilm by 5 μM SNP (B), planktonic cells of (PCells) PAO1/pBAD-yhjH (C), and biofilm-dispersed cells (DCells) from PAO1/pBAD-yhjH biofilms by 1% arabinose (D) were cultivated at 37°C in ABTGC medium with 0 or 4 μg of colistin/ml. The OD600 was monitored for 300 min. Means of three replicates are shown. (E and F) Biofilms formed by PAO1 strain on glass slides were submerged into ABTGC medium with 0 (E) and 4 (F) μg of colistin ml−1 for 2 h. Live and dead cells in treated biofilms were stained by using Live/Dead BacLight bacterial viability kits, followed by confocal laser scanning microscopy imaging.
DISCUSSION
In this work, P. aeruginosa strains were constructed with a controllable intracellular c-di-GMP content to mimic the natural biofilm cells (BCells) and dispersed cells (DCells) from biofilms. Unlike the natural biofilm cells with a high level of physiological heterogeneity (43), our cells are cultivated as homogeneous planktonic cultures and are easy to manipulate. These P. aeruginosa strains thus enable us to study the overall impact of c-di-GMP on P. aeruginosa cells. Of course, the P. aeruginosa PAO1ΔwspF cells in planktonic cultures cannot functionally mimic the late stage biofilm cells since cells from mature biofilms have a slow growth rate, oxygen limitation, and a large amount of extracellular matrix material around them. Nevertheless, we showed here that the PAO1/plac-yhjH cells (DCells) have an intracellular c-di-GMP content similar to that of chemically dispersed cells (DCells*), which have a distinct physiology compared to planktonic cells (PCells). In fact, the PAO1/plac-yhjH cells were unable to form normal amounts of biofilms compared to the PAO1 cells (Fig. 1B).
It was also observed that the intracellular c-di-GMP level plays an important role in production of pyoverdine by P. aeruginosa. Pyoverdine is the major siderophore of P. aeruginosa and is required for subpopulation interactions and biofilm maturation (44). Previous work showed that pyoverdine is mainly produced by the nonmotile subpopulation at the bottom part of mature P. aeruginosa biofilms (44). The present result suggests that the nonmotile subpopulation might have a higher intracellular level of c-di-GMP compared to the motile subpopulation at the top part of mature P. aeruginosa biofilms. Further studies will be carried out to study the detailed regulation mechanism exerted by c-di-GMP on pyoverdine production.
Induction of expression of arnB and PA4773 (from the pmr operon) with 2 μg of polymyxin B ml−1 but not with 0.125 μg of polymyxin B ml−1 was reported to increase the polymyxin B resistance of P. aeruginosa clinical isolates from cystic fibrosis patients (34). A number of unique mutations in the pmrAB and phoPQ operons enable these clinical isolates to show an adaptive growth in medium containing 2 μg of polymyxin B ml−1 after a long lag phase (34). Our study has shown for the first time that c-di-GMP signaling plays a role in AMP resistance in P. aeruginosa. Reduced c-di-GMP levels were found to induce the expression of PmrB and AnrB even without the presence of AMPs. PhoP was recently found to be able to bind c-di-GMP (45); thus, it might be an effector of c-di-GMP in regulation of AMP resistance. However, further studies are needed to elucidate the mechanistic basis of induction of the pmr and arn genes by low levels of c-di-GMP. The reported induced resistance thus confers a “protection in advance” mechanism to protect dispersed cells from the otherwise detrimental action of antibiotics on planktonic cells and may be the first finding to contradict the current dogma stating that dispersed cells are inevitably more susceptible than their sessile counterparts.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme and by startup grant M4330002.C70 from Nanyang Technological University, Singapore.
We acknowledge Shu Sin Chng (National University of Singapore) for valuable discussions.
Footnotes
Published ahead of print 12 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02499-12.
REFERENCES
- 1. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 2. Yang L, Liu Y, Wu H, Song Z, Hoiby N, Molin S, Givskov M. 2012. Combating biofilms. FEMS Immunol. Med. Microbiol. 65:146–157 [DOI] [PubMed] [Google Scholar]
- 3. Hengge R. 2009. Principles of c-di-GMP signaling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 4. Romling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 5. Nakhamchik A, Wilde C, Rowe-Magnus DA. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74:4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75:827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alhede M, Bjarnsholt T, Jensen PO, Phipps RK, Moser C, Christophersen L, Christensen LD, van Gennip M, Parsek M, Hoiby N, Rasmussen TB, Givskov M. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508 [DOI] [PubMed] [Google Scholar]
- 8. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 9. Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S. 2011. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13:1705–1717 [DOI] [PubMed] [Google Scholar]
- 10. Yang L, Hengzhuang W, Wu H, Damkiaer S, Jochumsen N, Song Z, Givskov M, Hoiby N, Molin S. 2012. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 65:366–376 [DOI] [PubMed] [Google Scholar]
- 11. Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, Ehrt S, Zhang Z, Gaffney BL, Gandotra S, Holden DW, Murray D, Nathan C. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defense and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234–1245 [DOI] [PubMed] [Google Scholar]
- 15. Bodey GP, Bolivar R, Fainstein V, Jadeja L. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279–313 [DOI] [PubMed] [Google Scholar]
- 16. Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558 [DOI] [PubMed] [Google Scholar]
- 17. Fazli M, Bjarnsholt T, Kirketerp-Moller K, Jorgensen A, Andersen CB, Givskov M, Tolker-Nielsen T. 2011. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Repair Regen. 19:387–391 [DOI] [PubMed] [Google Scholar]
- 18. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99–112 [Google Scholar]
- 20. Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40:79–105 [DOI] [PubMed] [Google Scholar]
- 21. Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:5060–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 23. Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203 [DOI] [PubMed] [Google Scholar]
- 24. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 25. Haagensen JA, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakuragi Y, Kolter R. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189:5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75:815–826 [DOI] [PubMed] [Google Scholar]
- 28. Kessler B, de Lorenzo V, Timmis KN. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293–301 [DOI] [PubMed] [Google Scholar]
- 29. Koch B, Jensen LE, Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187–195 [DOI] [PubMed] [Google Scholar]
- 30. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [Google Scholar]
- 31. Liu Y, Yang L, Molin S. 2010. Synergistic activities of an efflux pump inhibitor and iron chelators against Pseudomonas aeruginosa growth and biofilm formation. Antimicrob. Agents Chemother. 54:3960–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 33. Greenwald J, Hoegy F, Nader M, Journet L, Mislin GL, Graumann PL, Schalk IJ. 2007. Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in Pseudomonas aeruginosa: a role for ferrous iron. J. Biol. Chem. 282:2987–2995 [DOI] [PubMed] [Google Scholar]
- 34. Schurek KN, Sampaio JL, Kiffer CR, Sinto S, Mendes CM, Hancock RE. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720 [DOI] [PubMed] [Google Scholar]
- 38. Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104:9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Otto M. 2009. Bacterial sensing of antimicrobial peptides. Contrib. Microbiol. 16:136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 41. Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macfarlane EL, Kwasnicka A, Hancock RE. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543–2554 [DOI] [PubMed] [Google Scholar]
- 43. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 44. Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74:1380–1392 [DOI] [PubMed] [Google Scholar]
- 45. Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jansch L, Herberg FW, Haussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J. Microbiol. Methods 88:229–236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




