Summary
Epstein-Barr virus (EBV) attachment to primary B-cells initiates virus entry. While CD21 is the only known receptor for EBVgp350/220 a recent report documents EBV infects B-cells from a patient genetically deficient in CD21. On normal resting B-cells CD21 forms two membrane complexes, one with CD19 another with CD35. Whereas the CD21/CD19 complex is widely retained on immortalized and B-cell tumor lines, the related complement-regulatory protein CD35 is lost. To determine the role(s) of CD35 in initial infection, we transduced a CD21-negative pre-B-cell and myeloid leukemia line with CD35, CD21 or both. Cells expressing CD35 alone bound gp350/220 and became latently infected when the fusion receptor HLA II was co-expressed. Temporal, biophysical and structural characteristics of CD35-mediated infection were distinct from CD21. Identification of CD35 as an EBV receptor uncovers a salient role in primary infection, addresses unsettled questions of virus tropism, and underscores the importance of EBVgp350/220 for vaccine development.
INTRODUCTION
EBV is the first virus causally associated with human malignancy (Epstein et al., 1965). Like all herpesviruses, EBV persists lifelong through support of a complex life cycle consisting of alternate lytic and latent phases, each family member sequestered in a distinct cellular niche. EBV latency is maintained in the B-cell compartment (Miyashita et al., 1997). To date, CD21 is the only known B-cell receptor for EBV (Fingeroth et al., 1984; Frade et al., 1985; Nemerow et al., 1985; Tanner et al., 1987). Tethering of EBV to the B-cell membrane initiates efficient entry and is mediated by a high affinity (10−8M) interaction between envelope glycoprotein gp350/220 and CD21 (Tanner et al., 1988). Unique among herpesviruses gp350/220 dominates the outer membrane of EBV and is the major target of neutralization (Edson and Thorley-Lawson, 1981; Sashihara et al., 2009). Proximity to the B-cell surface promotes interaction between a second envelope protein gp42 in complex with gHgL and the virus co-receptor HLA II, activating fusion that in concert with other EBV proteins (Connolly et al., 2011; Hutt-Fletcher, 2007) delivers nucleocapsid to the cytoplasm. Whether initial contact with surface receptors influences the outcome of infection is unknown.
In vivo, latent infection typically begins with EBV entry into naïve tonsillar B-cells. In vitro the normal resting B-cell is most efficiently infected and immortalized to form lymphoblastoid cell lines (LCLs). EBV also infects oral epithelial cells though the pathways of infection and physiologic outcome, lytic infection, are distinct. As a tumor virus, EBV is primarily associated with a spectrum of B-lymphoid and nasopharyngeal epithelial malignancies (Rickinson and Kieff, 2007), reflecting tropism of EBV for these cells. Although most EBV+ tumors of hematopoietic lineage originate from B- and some T-cells (activated T-cells can express CD21 and HLA II), sporadic identification of EBV in blood cells not known to express CD21 as well as reports of gp350/220-induced signaling in CD21- cells (e.g. pre-B, monocyte, dendritic, NK cells, and activated neutrophils) (Busse et al., 2010; Calattini et al., 2010; Larochelle et al., 1998) has long proved puzzling. Importantly, the recent finding that B-cells from a patient with genetic deletion of CD21 can be readily immortalized with EBV indicates that even on B-cells an alternate attachment receptor must exist (Thiel et al., 2012).
Human CD21, consistently detected on B-cells and FDCs, is the receptor for C3d (complement (C’) receptor type 2, CR2), for CD23 (low affinity IgE receptor) and binds interferon (Asokan et al., 2006). An ectodomain (ED) of 15-16 short consensus repeat (SCR) modules is followed by a hydrophobic transmembrane (TM) and short cytoplasmic tail (CT)(Moore et al., 1987). All CD21-ligands bind within two flexible N-terminal SCRs (Prota et al., 2002). CD21’s role(s) is best understood in the context of humoral immunity though much investigation was conducted in mice. A related though clearly diverged murine protein (mCD21/CD35) serves the dual function of both a C3d and a C3b/C4b receptor (C’ receptor type 1, CR1)/CD35 based on alternative splicing of the ED (Roozendaal and Carroll, 2007). Studies mainly provide insight into the roles of CD21 as the CT of human CD21 is most similar to that of mCD21/CD35 (Fingeroth, 1990). On quiescent B-cells, CD21 associates non-covalently with other proteins forming two mutually exclusive membrane complexes along with free CD21. The CD21/CD19/CD81 complex is conserved in mouse, retained on human B-cell tumor lines, and has been extensively characterized, whereas the distinct complex between CD21 and CD35 is lost (Tuveson et al., 1991).
Human CD35 is an independent membrane glycoprotein best studied as an immune adherence and phagocyte receptor. A regulator of C’ activity (RCA) gene family member, CD35 also consists of a long ED, standard TM, and short CT. The common F allele contains thirty SCRs organized into four long homologous repeats (LHRs) A–D of seven SCRs each, followed by SCRs29-30 adjacent to the plasma membrane. Although highly expressed on B-cells and FDCs, CD35 is also present on RBCs, neutrophils, monocyte/macrophage, dendritic, subpopulations of T-cells and renal podocytes. Depending on cell type, CD35 may bind, transport and/or phagocytose particles or immune complexes (ICs) tethered by activated C’ proteins C3b, iC3b, C4b, C1q and mannan-binding lectin (Klickstein et al., 1988; Krych-Goldberg and Atkinson, 2001). Multiple SCRs from distinct LHRs may be utilized for the different interactions. Unlike CD21, but analogous to most RCA proteins, CD35 provides co-factor activity for Factor I modulating the C’ cascade to protect cells from membrane attack; and cleavage of C3b by CD35 generates C3d that may in turn bind nearby CD21 (Krych-Goldberg and Atkinson, 2001). RBC CD35 is also a pathogen receptor that binds P. falciparum merozoites via adhesin PfRH4, rosettes infected RBC via PfEMP1 (Spadafora et al., 2010) and indirectly tethers HIV and other infectious agents.
Of note, though consistently identified on primary B-cells, CD35 expression is minimal or undetectable on B-LCLs and most tumor lines (Schlossman et al., 1995). Thus, knowledge of its role in B-cell biology is more limited. On normal B-cells (tonsillar) nearly all CD35 associates with CD21 (Tuveson et al., 1991), though each receptor can cap independently (Tsokos et al., 1988). While crosslinking CD21 to mIgM enhances Ca2+ flux, promotes tyrosine phosphorylation of intracellular targets and augments B-cell proliferation, crosslinking of CD35 stimulates terminal B-cell differentiation (Weiss et al., 1987) and is anti-proliferative (Erdei et al., 2009), underscoring the complexity of signals integrated when B-cells engage ICs bearing diverse C’ fragments.
Herein we show CD35 is itself an attachment receptor for EBV. Infection of cells expressing CD35 alone was unaltered by monoclonal antibodies (mAbs) that directly block infection through CD21. Although CD35 is sufficient for EBV binding, HLA II co-expression is additionally required for entry and latent infection. The dominant virion ligand gp350/220 binds CD35, as mAb 72A1 directed to a glycan free region that interacts with CD21 inhibits CD35-mediated infection as well. Yet, structural, biophysical and temporal characteristics of CD35-based infection are distinct. Finally, we provide evidence that EBV downregulates CD35 upon B-cell infection, likely contributing to delayed recognition of its role. Identification of CD35 as an attachment receptor reconciles unsolved observations about how EBV signals to and infects CD21 negative hematopoietic lineage cells including immature B-cells and provides broad new understanding of viral pathogenesis with implications for cancer biology, autoimmune disease and vaccine development.
RESULTS
Nalm6/Pre-B and K562/Myeloid Cell Lines Lacking CD21 and CD35 Support Stable Expression after Gene Transfer
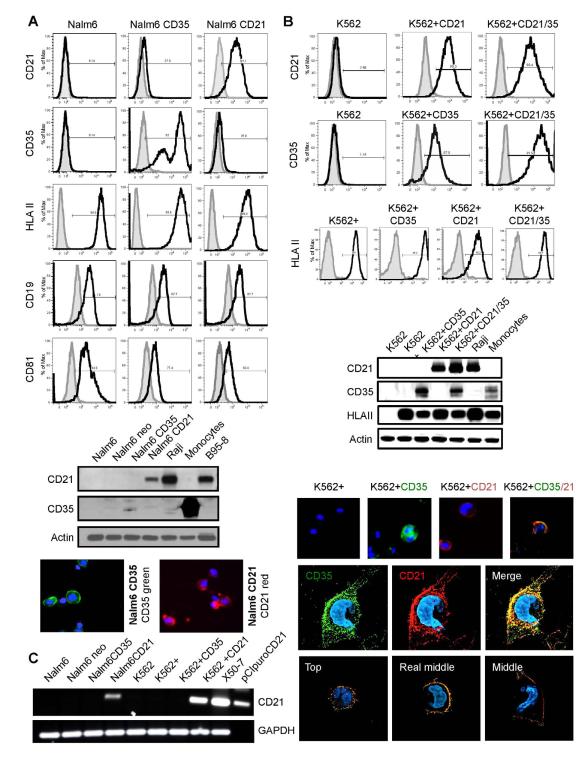
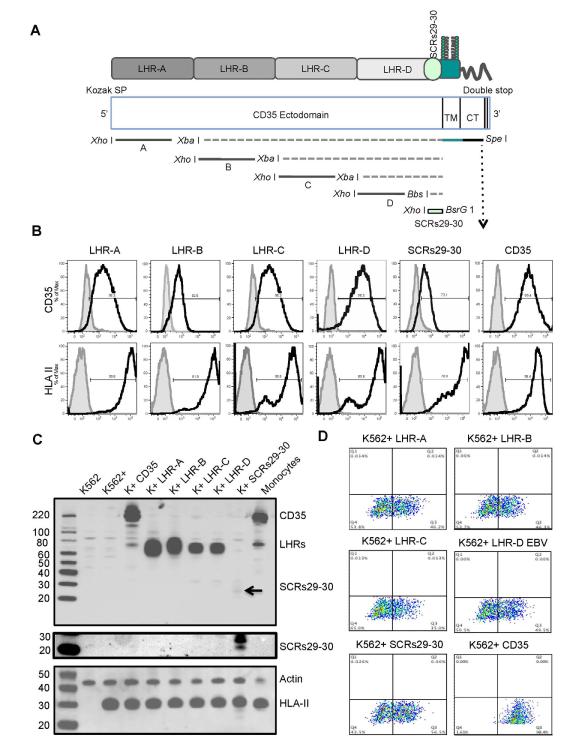
To analyze whether CD35 contributes to EBV infection, we developed a panel of informative cell lines that display different combinations of key surface proteins associated with CD21 on primary B-cells (Figure 1A,B). Antigen expression was documented by flow cytometry (FC), immunoblot (IB) and immunofluorescence analysis (IFA). We chose two background hematopoietic lineages, lymphoid and myeloid as EBV+ tumors from the latter have been reported (Larochelle, 1998) though normal precursors lack CD21. Whereas B-cell lines rarely express CD35 most express CD21 except certain pre-B cell lines such as Nalm6 (Table S1). Nalm6 expresses a pre-B-cell receptor, CD19 and HLA-II that arise earlier in ontogeny than CD35 or CD21 (Tedder et al., 1997). K562, an immature myeloid line, lacks a B-cell receptor, CD19, HLA II, CD21 and CD35 (Figure 1B), but is readily transduced with different components of the EBV entry pathway. Both lines express CD81, a tetraspan that can also complex with CD19/CD21. No CD21 expression was detected in lone CD35 or HLA II transfectants by IB (Figures 1A, B) and confirmed by PCR (Figure 1C). Cell line panels: Nalm6, Nalm6neo, Nalm6puro, Nalm6 CD35, Nalm6 CD21 and K562, K562neo, K562puro, K562-HLA II (denoted K562+), K562+CD35, K562+CD21, K562+CD21/CD35 were used for further investigation. Ab sources for all experiments are listed in Table S2.
Figure. 1. Stable Expression of Recombinant CD35 and CD21 in (A) Nalm6 (Pre-B) and (B) K562 (Myeloid) Cells +/− HLA II.
(A) Top, Flow Cytometry (FC). Nalm6 cells lack CD21 and CD35, but express HLA II, CD19 and CD81 as shown using specific anti-human mAbs CD35, CD21, HLA II, CD19, and CD81 (black lines). Isotype controls are filled. X axis=log fluorescence intensity, Y axis=relative cell number. (A) Middle, Immunoblot (IB). Neither CD21 nor CD35 were visualized in Nalm6 or Nalm6 neo (control) cells, though evident in the respective transfectants (Nalm6 CD35, CD21). Raji and B958 (B-cell lines) control for CD21 expression and human splenic monocytes/macrophages control for CD35 expression. Actin served as loading control. Bottom, Immunofluorescence assay (IFA). CD35 (green) and CD21 (red) were visualized at the surface of the respective Nalm6 transfectants. Nuclei were stained blue (DAPI). (B) Top, FC. K562 lacks CD21 and CD35. However, after transfection and selection CD35, CD21 and co-expressed CD21/CD35 were detected (black lines) using mAbs. Isotype controls are filled. Although K562 lacks HLA II, when CIITA was transfected HLA II was expressed at the cell surface (denoted K562+) as shown by FC in the bottom row. (B) Middle, IB. Expression patterns displayed left to right in K562 parent (CD21−, CD35−, HLA II−), K562+(CD21−, CD35−, HLA II+), K562+CD35 (CD21−, CD35+, HLAII+), K562+CD21 (CD21+, CD35−, HLA II+), K562+CD21/CD35 (CD21+, CD35+, HLA II+) were confirmed by IB. Raji and splenic mono/macrophages served as controls. (B) Bottom, IFA. Receptor expression in dual K562 transfectants demonstrated by microscopy. CD35 (green), CD21 (red), co-localized receptors (yellow). Nuclei are stained blue (DAPI). Sequential views of CD35 and CD21 in a single K562 cell. 3-D acquisition followed by deconvolution and reconstitution showed that in this representative cell 73.62% of CD21 molecules co-localized with CD35 whereas of 57.02% CD35 molecules co-localized with CD21. (C) PCR. Reverse transcription of cellular RNA followed by quantitative PCR was performed using described primers (Methods). CD21 cDNA was undetectable in Nalm6, Nalm6neo, Nalm6 CD35, K562, K562+, K562+ CD35, but present in Nalm6 CD21, K562+ CD21, X50-7 (control, Table S1) and reference plasmid pCI-puroCD21. GAPDH cDNA (control) was generated under identical conditions.
CD35 Binds EBV
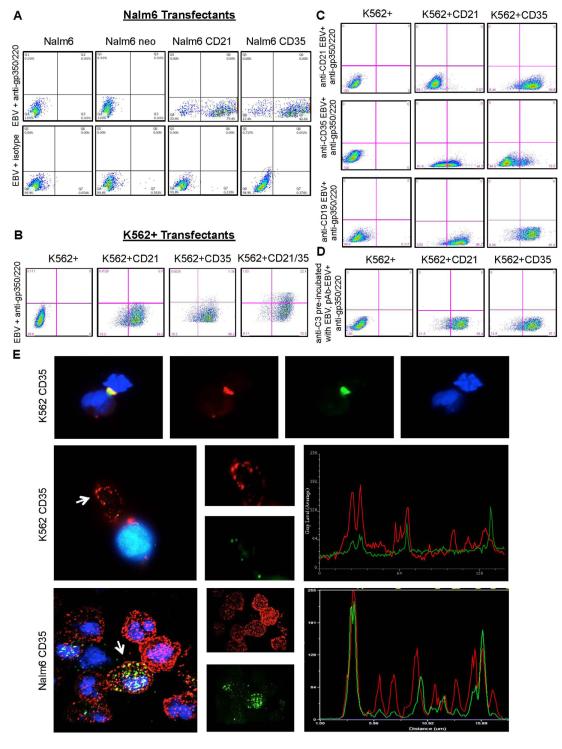
As a prelude to infection experiments, we sought to document EBV bound CD21 expressed on Nalm6 and K562 cells and that binding was specific. Cells were incubated with EBV, washed and attachment detected with 2L10, a mAb directed to a gp350/220 epitope distant from the CD21 contact site (Tanner et al., 1988). To our surprise EBV also bound to cells expressing CD35 on both Nalm6 and K562 transfectants, though neither parental nor vector control lines bound virus (Figure 2A, B). As there was no evidence of cross contamination (Figure 1), and neither 2L10 nor secondary Ab bound non-specifically to Nalm6, K562 or transfected sublines (data not shown) we investigated the specificity of binding. Multiple sources of EBV including standard B958, variants EBfaV-GFP and B958-Tet-BZLF1 and Akata virus all bound CD35 expressing cells, supporting specific interaction (data not shown). Although none of several mAbs (2B11, YZ1, E11, 543, 3D9) that recognize distinct epitopes on CD35 (Chen et al., 2007) blocked EBV binding to CD35+ cells, even upon addition of a 20 Ab or when pooled (data not shown), a rabbit polyclonal IgG directed to CD35 partially inhibited attachment (Figure 2C) while polyclonal anti-CD21 did not. Based on cell culture and experimental systems employed (absence of relevant C’ factors, heat inactivation of serum, washing of cells before EBV incubation, use of purified EBV), it seemed unlikely C’ activation occurred generating C3b on the virus envelope for interaction with CD35. Still, to rule out this possibility we pre-incubated EBV with polyclonal anti-C3, which did not alter EBV binding as shown for K562+CD35 (Figure 2D). Moreover, EBV co-localized with CD35 at the surface of K562CD35 (no HLA II) and Nalm6CD35 cells (Figure 2E). Thus, EBV directly binds CD35, an interaction independent of virus-associated C3b and CD21.
Figure 2. EBV Binds CD35 Expressed on Nalm6 and K562+.
(A) EBV indirectly detected with mAb 2L10 anti-gp350/220 and GAM FITC binds CD21, and also CD35+ Nalm6 cells, but not Nalm6 or Nalm6neo controls. (B) Identical results were obtained when EBV was incubated with K562+ clones and analyzed as described. K562+CD21, +CD35 and +CD21/35 binds EBV, whereas K562+ does not. (C) Polyclonal anti-CD35 partially blocks binding of CD35+, but not CD21+ K562+ cells (middle row). Polyclonal anti-CD21 does not prevent binding of CD35+ cells, though it blocks CD21+ cells (top row, control). Polyclonal anti-CD19 (irrelevant control) does not alter any EBV-receptor interaction (bottom row). (D) Polyclonal anti-C3 pre-incubated with EBV also does not alter any EBV-receptor interaction. (E) EBV co-localizes with CD35. Top, EBV (titer non-saturating) (green) co-localizes with CD35 (red) on the surface of K562CD35 (HLA II−) and Nalm6CD35 (HLA II+) cells. Nuclei are stained blue (Hoechst). Top, adjacent K562CD35 cells are joined by binding of aggregated EBV. Middle, co-localization of EBV with CD35 at the plasma membrane of a single cell is indicated (arrow) and graphically displayed by pixel analysis in overlapping channels (Methods). Bottom, EBV co-localizes with CD35 on Nalm6CD35 cells.
CD35 Mediates EBV Infection in the Presence of HLA II
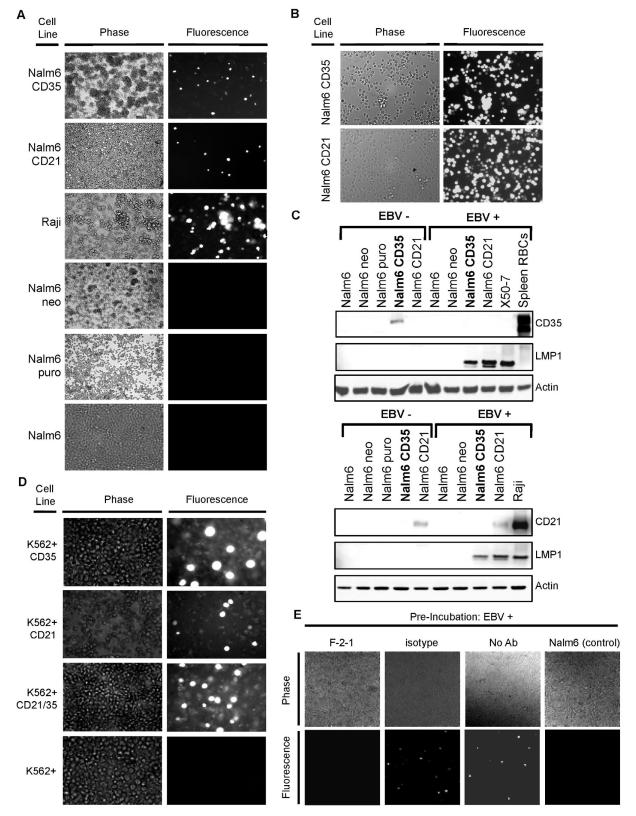
Next, we queried whether CD35 also mediated infection, or whether EBV might be irreversibly tethered to the cell surface by CD35 (a rigid protein), or be phagocytosed and destroyed. Nalm6 CD35, Nalm6 CD21, Nalm6 and control lines were incubated with EBfaV-GFP (a variant B958 strain that expresses EGFP from the viral genome after infection) and analyzed by microscopy at 18, 36, 72 and 96 hr post-infection. By 72 hr, Nalm6 CD21, Raji (CD21+ CD35−, EBV+ Burkitt lymphoma (BL) line susceptible to EBV “superinfection”) and also Nalm6 CD35 became infected with EBfaV-GFP as shown by fluorescence (Figure 3A). The time course of infection via CD21 compared with CD35 was distinct: fluorescent cells first appeared at ~18 hr in Nalm6 CD21, but were not visible until ~36 hr after Nalm6 CD35 was incubated with EBV (data not shown). Although the relative receptor density of CD35 and CD21 may have varied in some assays, delayed fluorescence of CD35 expressing cells was consistent in multiple experiments. Sorting EGFP expressing cells enabled establishment of stable lines that grew well and were further evaluated after six months (Figure 3B). We reliably detected EBV latent membrane protein 1 (LMP1) in these cells by IB (Figure 3C), whether they originated from Nalm6 CD21 or CD35 lines, documenting persistent infection. Interestingly, whereas CD21 expression was retained in infected Nalm6 CD21 cells, CD35 expression could not be detected after stable infection of Nalm6 CD35 lines, an observation we will return to (Figure 3C bottom).
Figure 3. EBV Infects CD35+ Cells that Express the Fusion Receptor HLA II.
(A) 72 hr after incubation of Nalm6 CD21 or CD35 with EBfaV-GFP, fluorescent cells were detected by microscopy. No fluorescence was detected in identically incubated vector control or parent Nalm6 cells. When Raji (CD21+, HLA II+, EBV+ BL line) was superinfected with EBfaV-GFP, fluorescence was also detected (control). B. EGFP-expressing Nalm6 CD35 and CD21 cells enriched by sorting maintained stable fluorescence after six months in culture. C. IB of stable Nalm6 CD35 and Nalm6 CD21 confirmed LMP1 expression in both lines. Nalm6 CD21 retained long-term receptor expression after EBV infection, whereas Nalm6 CD35 did not. D. K562 cells that expressed CD21, CD35 or both became fluorescent (susceptible to EBV infection) when the fusion receptor HLA II was co-expressed, unlike cells that expressed HLA II alone. E. When EBV was pre-incubated with mAb F-2-1 to gp42 infection of Nalm635 (HLA II+) was abolished, whereas pre-incubation of EBV with the isotype control UPC10 had no effect comparable to cells infected with EBV in the absence of any Ab. Nalm6 cells lacking CD35, but HLA II+ were not infected.
Despite EBV binding neither K562 CD21 nor CD35 expressed EGFP after incubation (data not shown). K562, unlike Nalm6, lacks HLA II, the lymphocyte co-receptor and only known cellular ligand for the EBV fusion protein gp42 extensively reviewed in (Hutt-Fletcher, 2007; Shaw et al., 2010). To assess whether HLA II could also serve as a co-receptor for CD35 expressing cells, dual transfectants were generated as previously reported using transcription factor CIITA (LeibundGut-Landmann et al., 2004) to upregulate HLA II (Li et al., 1997) (Figure 1B). Although neither K562, nor K562+ were susceptible to infection, K562+CD21 (control), K562+CD35 and K562+CD21/CD35 were all readily infected as shown by visualization of EGFP (Figure 3D). Moreover, pre-incubation of EBV with mAb F-2-1 directed to gp42, but not with an isotype control Ab blocked infection of HLA II+ Nalm6CD35 cells (Figure 3E).
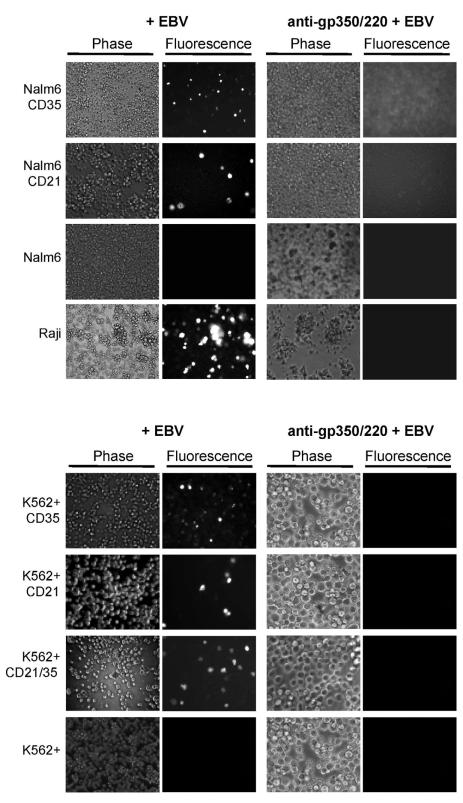
EBV Contacts CD35 Expressing Cells through Gp350/220
Gp350/220 is the most highly expressed protein on the EBV envelope. The prior observation that specific Abs to gp350/220 entirely blocked EBV absorption to the normal B-cell membrane (Tanner et al., 1987), suggested EBV gp350/220 might mediate attachment to CD35 as well. Mutant analyses and structural studies showed N-terminal gp350/220 was the major site of CD21 interaction (SCR1-linker-SCR2), and contacts were mapped (Kovacs et al., 2010). However, the ED of gp350/220 is long (863 amino acids) and highly N- and O-glycosylated, thus ample potential epitopes for further interaction with CD35 (perhaps other ligands) remained. MAb 72A1 binds the N-terminal domain of gp350/220, directly blocking absorption by CD21, whereas mAb 2L10, binds a C-terminal region (Tanner et al., 1988) that has no effect on attachment. Pre-incubation with 72A1 prevented EBV infection of Nalm6 CD35, K562+CD35 and K562+CD21/35 (Figure 4), though neither 2L10 nor 2L10 with secondary Ab were inhibitory. These results not only confirmed prior reports that 72A1 was potently neutralizing, but importantly revealed N-terminal gp350/220 was also utilized for CD35 attachment, thereby suggesting the docking site was near, possibly the same as that used for interaction with CD21, or that a major conformational change produced upon 72A1 binding, perhaps mimicking CD21, compromised subsequent CD35 interaction.
Figure 4. MAb 72A1, Directed to N-terminal Gp350/220 Blocks Infection of CD35 Expressing Nalm6 and K562+ lines.
Top, Nalm6 CD21, CD35 and Raji (control) are susceptible to EBV infection, whereas Nalm6 is not, as shown by EGFP visualization at 72 hr. Pre-incubation of EBV with anti-gp350/220 (mAb 72A1) blocks infection of CD21 and/or CD35+ cells. Bottom, Infection of all K562+ receptor-bearing lines is likewise blocked when EBV is pre-incubated with mAb 72A1.
Temperature and Structure Distinguish CD35-EBV Interaction
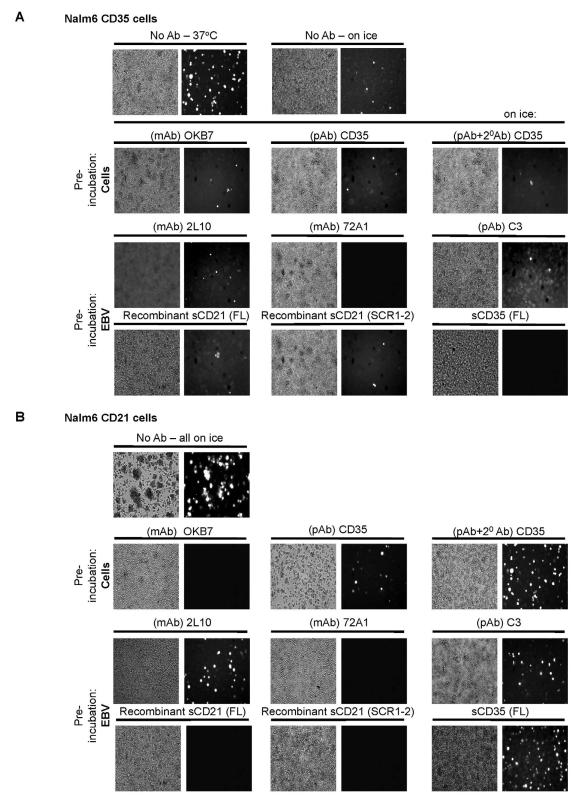
Virus infection blocking experiments are classically performed on ice to prevent capping, internalization and/or shedding of ligand-receptor pairs and probes. Nalm6 CD21 cells avidly bound EBV when incubated on ice or at 37°C in the presence of azide (personal observation). However, binding of EBV to Nalm6 CD35 was at least 10-fold more avid at 37°C than on ice (Figure 5A, top row). Nevertheless, a series of comparative infection inhibition assays was carried out on ice. When Nalm6 CD35 (Figure 5A) was compared to Nalm6 CD21 (Figure 5B) it was evident mAb 72A1, but not 2L10 anti-gp350/220 (control) blocked EBV infection of both C’ receptor expressing Nalm6 cells (Figures 5A, B third row). MAb OKB7, which recognizes the gp350/220 attachment site on CD21, only prevented infection of Nalm6 CD21 (Figure 5B second row), whereas polyclonal anti-C3 Ab had no effect on EBV infection of either line (Figures 5A, B third row). Polyclonal anti-CD35 IgG partially inhibited infection of Nalm6 CD35, but did not alter Nalm6 CD21 infection (Figures 5A, B second row). Soluble recombinant CD35 (full length ED), while decreasing Nalm6 CD35 infection, in three experiments appeared to enhance infection of Nalm6 CD21 cells, raising the possibility of trans interaction between soluble recombinant CD35 and cell surface CD21. These findings demonstrate for the first time that CD35 as well as CD21 can engage EBV on the cell surface and mediate infection in the presence of HLA II. However, temperature requirements for EBV absorption, domain structure of ligand-receptor binding, and the tempo of initial infection all differ.
Figure 5. Temperature and Structure Distinguish CD35 from CD21-mediated Infection.
Cells were infected with EBfaV-GFP in the absence or presence of a competing ligand. Experiments were performed on ice unless otherwise noted. (A) Top row, Infection of CD35+ cells is > ten-fold more efficient when virus is introduced, washed and incubated at 37°C, compared with initial incubation and wash on ice. Second row, Pre-incubation of Nalm6 CD35 with OKB7 anti-CD21 does not block virus infection, nor does polyclonal anti-CD35-though addition of 20 Ab confers modest inhibition. Third row, Pre-incubation of EBV with mAb 72A1 blocks infection of CD35+ lines, whereas mAb 2L10 and anti-C3 have no effect. Fourth row, Pre-incubation of EBV with soluble recombinant CD35 blocks most infection of CD35+ cells, whereas soluble recombinant forms of CD21 are ineffective. (B) Top row, Infection of CD21+ cells on ice. Second row, Pre-incubation with OKB7 entirely blocks EBV infection, whereas polyclonal anti-CD35 even with 20 Ab has no effect. Third row, Pre-incubation of EBV with mAb 72A1 blocks infection of CD21+ lines, whereas mAb 2L10 and anti-C3 have no effect. Fourth row, Pre-incubation of EBV with soluble recombinant CD21 SCR1-2, but not soluble recombinant CD35 blocks EBV infection of CD21+ cells. One representative of three experiments is displayed.
EBV Contacts Multiple LHRs and SCRs29-30 at the Plasma Membrane
The highly repetitive structure of the SCRs comprising each LHR has yielded complex patterns of ligand binding based on subtle differences in affinity. For example, LHR-A preferentially binds C4b, while both LHR-B and -C preferentially bind C3b and likely iC3b. LHR-D binds C1q and mannan binding lectin (Ghiran et al., 2000; Krych-Goldberg and Atkinson, 2001). Recently, Plasmodium falciparum PfRh4 was shown to bind CD35 with high affinity within three N-terminal SCRs (Tham et al., 2011). To begin to more precisely decipher the structural basis for EBV-CD35 interaction at the plasma membrane we constructed five truncation mutants consisting of one LHR (A-D) or SCRs29-30 fused to the CD35 TM-CT (Figure 6A). All mutants were expressed at the surface of K562+ cells as detected by FC and confirmed by IB (Figures 6B, C). Membrane adjacent SCRs29-30 was readily detected by FC, but poorly detected after denaturation by IB. EBV binding was promiscuous, similar to reports for certain C’ fragments. EBV bound each of the LHR mutants as well as mutant SCRs29-30 (Figure 6D). However, all mutants bound less well than full-length CD35. Somewhat unexpectedly, despite attachment, infection was absent based on lack of fluorescence in HLA II+ cells expressing CD35 mutants (data not shown) compared to full-length controls. These results indicate EBV binds CD35 differently than CD21 as contact with multiple LHRs is more similar to that of certain C’ ligands.
Figure 6. CD35 Truncation Mutants: Surface Expression and EBV Binding.
(A) Schematic representation of CD35, and truncation mutants (LHRs A-D, SCRs29-30). Five individual cDNA mutants were constructed ligated to the TM and CT of parental CD35 and incorporated into the vector pCIneo. The relative position of mutant fragments (LHRs A-D, SCRs29-30) and flanking restriction enzyme sites are indicated. (B) Each mutant was independently transfected into K562+ cells and surface expression documented by FC using polyclonal anti-CD35. Concurrent expression of co-receptor HLA II is displayed, as no mutant-bearing cells became infected. (C) IB confirmed each mutant was of the predicted MW. Controls (K562, K562+, K562+CD35, monocytes) were as expected. (D) EBV bound each mutant as shown by FC, but in no case was binding equal to that of full-length CD35. See also Figure 1B.
EBV Downmodulates CD35
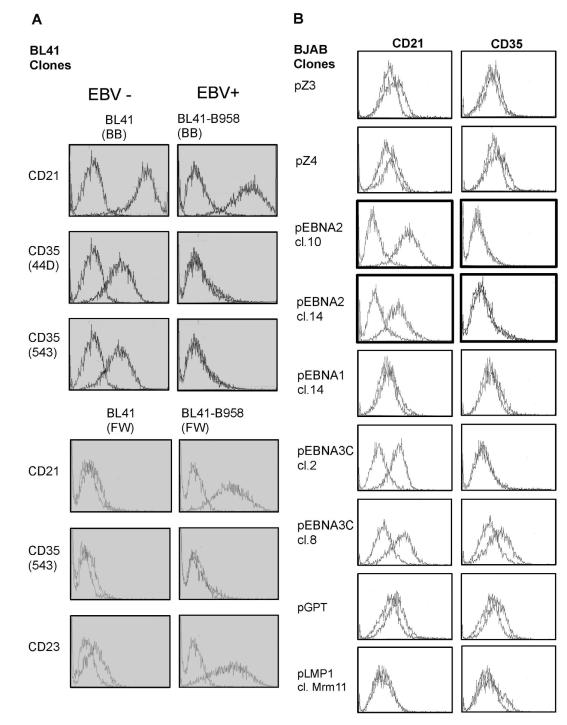
Loss of CD35 after stable infection of Nalm6 CD35 in contrast to CD21 upregulation, prompted re-inspection of their expression patterns on different B-cells. CD21 upregulation is unusual as viral binding proteins are typically downmodulated after infection. A multi-laboratory blinded evaluation of anti-CD35 mAbs (Schlossman et al., 1995) confirmed consistent CD35 expression on fetal and adult B-cells whereas no recorded B-cell lines (BJAB (EBV−), Daudi (EBV+), IM9 (EBV+), JY (EBV+), Nalm6 (EBV−), Raji (EBV+), U266 (EBV−)) expressed CD35. In contrast, all lines (except Nalm6, pre-B) expressed CD21. While several groups previously reported increased or stable CD21 expression on superinfected BL lines (Calender et al., 1990; Wang et al., 1990), a single account (using polyclonal Ab) described upregulation of CD35 (Cohen et al., 1987). BL41, an EBV-BL is frequently used to model “physiologic” EBV infection (Rickinson and Kieff, 2007) as its surface phenotype resembles that of a normal B-cell. Upon infection with strain B958, a stable type III latent (immortalizing) infection is established, similar to LCLs. Two sources (BB, FW) of BL41 expressed CD21 and variably CD35. However, after superinfection CD21 was upregulated, while CD35 was lost (Figure 7A, Table S1). EBNA-2, a transcription factor expressed in latency III, was previously shown to upregulate CD21 (Calender et al., 1990; Wang et al., 1990). Using a series of BJAB sublines (CD21 and CD35 are low but detectable in some BJAB clones each expressing a single EBV latent protein (gift of Fred Wang) we assessed whether EBNA-2 also regulates CD35. While CD21 expression uniformly increased in clones expressing EBNA-2, CD35 became undetectable (Figure 7B). There was no change in CD21 or CD35 expression in clones expressing vector (pZ-3, pZ-4), EBNA-1 or LMP1. In EBNA-3C expressing clones, CD21 upregulation was as described (Wang et al., 1990), but results for CD35 were equivocal. Normal human B-cells are short-lived in culture unless immortalized. Upon isolation B-cells express both CD21 and CD35. However, after outgrowth of oligo or monoclonal LCLs, no or minimal CD35 is detected by FC (personal observation, Table S1). In contrast, CD21 is consistently upregulated, though to varying degrees. Our results support published literature indicating most B-cell tumor lines (EBV− or EBV+) express no or low CD35 and suggest EBV infection of normal B-cells and superinfection of most CD21+ B-cell lines downmodulates CD35. Though EBNA-2 may mediate repression of CD35 as well as upregulate CD21, the background of the tumor line in which EBNA-2 is expressed may also be relevant (Cordier-Bussat et al., 1993).
Figure 7. Comparative Expression of CD35 and CD21 on EBV− and + converted BL lines by FC.
(A) BL41 (EBV−) from two sources (BB top, FW bottom) upregulates CD21, but downregulates CD35 after stable EBV (B958) infection. Irrespective of source and clonality, CD21 was up and CD35 downmodulated. As predicted CD23 expression increased upon superinfection (control). (B) Analysis of BJAB (EBV−) clones that express individual EBV latent cycle proteins (EBNA-2, EBNA-1, EBNA3C, LMP1) or vector controls (pZ3, pZ4, pGPT) confirmed EBNA-2 upregulates CD21 and suggested CD35 loss was also EBNA-2 dependent. Assays were repeated a minimum of three times. X axis=log fluorescence intensity, 3 log display. Y axis=cell number (50,000 events).
DISCUSSION
The B-cell tropism of EBV was established shortly after its discovery in 1964 and a high affinity B-cell receptor, CD21, identified two decades later. Still, unresolved questions about how EBV contacts, signals, variably enters, and at times persists in certain CD21− hematopoietic cells (some pre-B, plasmablast, monocyte/macrophage, dendritic, activated neutrophil, and NK cells) remained. On review, virtually all implicated cells regularly or sporadically expressed CD35 lending credence to the idea this structurally related protein might substitute for CD21. The recent demonstration that B-cells from a patient with genetic deletion of CD21 (compound heterozygote) are immortalized by EBV provide clear evidence an alternate receptor exists. Herein we demonstrate that CD35 mediates EBV attachment to the cell surface and supports entry into a CD21− pre-B and myeloid cell line when the fusion receptor HLA II is co-expressed. Whether CD35 plays the key role in establishing or alters the course or pattern of primary B-cell infection is under review.
Significantly, although CD35 like CD21 bound gp350/220, and absorption was blocked by an identical mAb (72A1) each EBV-C’ receptor interaction was unique. CD35 binding was not restricted to N-terminal SCRs (a feature of several pathogen-RCA protein interactions). Rather, EBV bound each LHR as well as SCRs29-30, a feature analogous to certain C’-type contacts and that may explain why individual anti-CD35 mAbs were ineffective in preventing attachment. CD35 is adjacent to CD21 on the RCA locus chromosome 1q32 and evidence supports development of CD35 as an independent type 1 TM protein by homologous recombination late in primate evolution (Birmingham and Hebert, 2001). Indeed, a study of homology relationships among all known RCA and other SCR containing proteins showed CD21 SCRs1-2 were most similar to SCRs3-4 (LHR A), 10-11(LHR B), 17-18 (LHR C), 24-25 (LHR D) and 29-30 of CD35 (Krushkal et al., 2000), signifying gp350/220 may contact related SCRs. Unlike full length CD35, none of the truncation mutants conferred infectivity implying (among other scenarios) that multiple contacts were needed to retain EBV on CD35 during membrane transit, a longer span or a flexible spacer domain(s) was required for delivery to the fusion receptor or signaling was modified.
EBV infection of CD35+ cells was highly sensitive to temperature. An approximate ten-fold decrease in infection was observed when cells were infected on ice vs 37°C. Such variance often occurs in warm-blooded animals and typically reflects thermodynamic effects (Haywood, 1994) on affinity or avidity (e.g. bond formation or dissolution), movement in the plasma membrane (e.g. capping vs patching – a notable characteristic distinguishing EBV entry into normal B-cells from cell lines (Nemerow and Cooper, 1984)), and/or rate of internalization of ligand-receptor complexes (Holland and Mc, 1959). One or all noted properties may differentiate CD35-EBV interactions. Time to infection (at 37°C) scored by initial visualization of EGFP EBV was consistently slower (~18 hr delay) in CD35 compared with CD21 or co-expressing cells suggesting the respective receptor-EBV complexes may form distinct cytoskeletal associations and/or deliver independent intracellular signals that separately impact the competence of recipient cells to support infection. On resting B-cells, where both receptors are expressed and associate, signals may be integrated in ways that ultimately reveal how differences in transformation efficiency of individual B-cells arise, a phenomenon not well understood.
What does CD35-mediated infection imply for EBV biology?
Among the barriers to the study of EBV biology is the inherent diversity of the primary human B-cell. Human B-cells require isolation from mixed specimens, cannot be cultured long-term, and short-term culture changes their behavior. They are extremely difficult to transfect and lentivirus infection alters gene expression. Even B-LCLs are suboptimal for many studies due to high biological noise and in vitro artefacts (Choy et al., 2008). This, coupled with absence of a robust animal model (both EBV infection and CD35 as a type 1 TM protein are restricted to humans) has impeded decisive work. Experiments of nature, that uncover and resolve important biological questions are often most informative. For example, discovery of EBV-lymphoproliferative disease upon introduction of T-cell suppressive transplant therapy and of SAP mutation in Duncan’s syndrome, each provided essential evidence that EBV infection was regulated by the cellular immune system (Rickinson and Kieff, 2007). Likewise the observation agammaglobulinemic patients with no or immature (typically CD35−, CD21−) B-cells are not infected by EBV (even in the epithelial compartment) revealed an essential role of B-cells in initiation of primary infection (Faulkner et al., 1999). As CD35 arises earlier than CD21 in B-cell ontogeny (Tedder et al., 1997) virtually all naive B-cells express both. Thus the recent finding that B-cells from a patient lacking CD21 (compound heterozygote) can be readily immortalized with EBV in vitro and serological evidence of infection highlights the existence of an alternative EBV receptor. To date genetic absence of CD35 has not been reported.
Is CD35 a physiologically relevant EBV receptor?
Several lines of evidence suggest CD35 is a physiologically relevant EBV receptor on normal human B-cells. For example, human pro and early pre B-cells that express only CD35 (precedes CD21 in B-ontogeny) can be infected and immortalized (Ernberg et al., 1987). Although transfection of CD21 and HLA II may mediate restricted (latency I or II) infection of a spectrum of transformed cell lines – including mouse B-lymphoma cells, neither primary murine B-cells expressing CD21 and HLA II nor murine stem cells electroporated with EBV and B-differentiated can be immortalized and expanded in vitro (Zychlinska et al., 2008). As CD35 in the form of a type 1 TM protein first appears late in primate evolution, it is unclear if expression in mouse B-cells will be informative. While superinfection of human CD21+ cell lines results in restricted latency, infection of primary B-cells (and certain model lines such as BL41 that are CD21+ and CD35+) yields latency III. When primary B-cells are mitogen activated, CD21 is upregulated, CD35 downregulated and susceptibility to immortalization rapidly decreases (Einhorn and Klein, 1981; Roome and Reading, 1987). Further, the observation that CD35 but not CD21 is downregulated after EBV infection replicates behavior of other major virus binding and entry proteins that thereby avoid superinfection and inhibition of virus release. When mature tonsillar B-cells are first exposed to salivary EBV, CD35 and CD21 are closely associated and interact through their ED (Tuveson et al., 1991) and possibly other domains. Thus it is most likely they work in concert to optimize primary infection and establish a latent state.
EBV, CD35, Lupus, Cancer – and Malaria?
The current results also raise provocative new questions relevant to EBV-associated disease. For example, active lupus patients (often females of African heritage) have high rates of EBV+ B-cells in their blood (Gross et al., 2005) and CD35 expression on memory B-cells is depressed (Isaak et al., 2008). Does EBV enforce CD35 downregulation and does it contribute to ongoing polyclonal B-cell activation and lupus flares, particularly among children in whom EBV seroprevalence is high (James et al., 1997)? Does the sustained B-cell activation predispose to lymphoproliferative disease? CD35 on RBCs is a receptor for P. falciparum, a driving force in human evolution. Does EBV modulate RBC CD35 – in lupus, in malaria? Might ongoing EBV production, while potentially predisposing to lupus and lymphoma (rare outcomes), paradoxically afford protection from severe malaria?
Gp350/220 as a vaccine
The observation that CD35 contacts an identical EBV ligand coupled with a slower tempo of infection, distinct protein interactions and temperature dependence in addition to frequent absence of CD35 on B-cell lines likely confounded interpretation of past experiments. Our results establish that both CD21 and CD35 can mediate EBV attachment and support entry when a fusion co-receptor is present (Nalm6CD35) or introduced (K562+CD35). As CD35 also binds gp350/220, reports attributing signaling to gp350/220 contact with CD21 alone on normal B-cells or to innate immune receptors in other leukocytes may need to be re-examined in light of possible CD35 interaction. Identification of this additional gp350/220 receptor further suggests vaccination might protect from a wider spectrum of disease than is currently appreciated as some of the rare EBV+ tumors and autoimmune phenomena not well explained by a CD21-mediated path might result from CD35-mediated entry in a relevant microenvironment.
Significantly, unlike most herpesviruses, gp350/220 alone dominates the EBV envelope. Certain Abs to gp350/220 completely neutralize in vitro EBV infection, gp350/220 is a target of Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) and maternal serum with a high content of anti-gp350/220 Ab is protective. Small vaccine trials of soluble recombinant gp350/220 have shown partial efficacy (Sashihara et al., 2009). Demonstration that gp350/220 binds CD35 as well as CD21 highlights the potential of gp350/220 as an immunogen, particularly if delivered in an innovative vehicle free of viral nucleic acid to mucosal sites that robustly harness humoral and cellular immune responses – a vaccine poised to prevent infection, cancer and autoimmune disease.
EXPERIMENTAL PROCEDURES
Cell Lines
Nalm6, EBV+/− BL lines, EBV+ LCLs; EBV producer lines: Akata, Akata-GFP, B958, EBfaV-GFP, Tet-BZLF1 B958 and the myeloid leukemia line K562 were obtained and cultured as detailed in Extended Experimental Procedures. Monocyte/macrophages were isolated as described (Fingeroth et al., 1989) from discarded de-identified spleens consented for research and approved by both New England Organ Bank and BIDMC Institutional Review Boards.
Plasmids
Full-length CD35 and CD21 cDNAs were cloned into pCIneo (Promega) and pCIpuro, respectively. pCIpuro was constructed from pCIneo and pPUR (Clontech) backbones as detailed in Extended Experimental Procedures. Plasmid containing CIITA (pZeoCIITA) was a gift of Dr. Jeremy Boss (Riley et al., 1995). Truncated forms of CD35 ED encoding LHR-A, LHR-B, LHR-C, LHR-D and SCRs29-30 were synthesised in vitro (Genewiz) based on the genomic sequence of CD35 (GenBank NM_000573.3) and cloned into pCIneo (Figure 6).
Transfection/Selection
Plasmids containing relevant cDNAs were transfected into parental Nalm6 and K562 cells using Lipofectamine™ (Life Technologies) according to the manufacturer’s directions. After 48 hr cells were selected in media containing the indicated antibiotic (Life Technologies): Zeocin (CIITA), puromycin (CD21) and Geneticin (CD35, truncation mutants).
Antibodies
Abs used for all experiments are listed in Table S2.
Cytometric Analysis and Sorting
Single cell suspensions were prepared in PBS containing 2% doubly heat inactivated FCS. Abs used for staining are listed in Table S2. FC was performed on a FACScan or LRSII benchtop FC (Becton-Dickinson, B-D) and data analyzed using CellQuest Pro Version 4.0.1 (B-D) and/or FlowJo Cytometry Analysis software (Tree Star Inc). A minimum of 10,000 events was recorded for each analysis. Sorting of antibiotic selected cells was performed on a FACSAria (B-D Biosciences).
Imaging of C’ Receptors
Cells were stained with specific mAbs for 30 min on ice. Nuclei were stained with DAPI 33342 (Sigma) for 5 min at RT. Stained cells were washed, mounted (Mounting Medium, DakoCytomation) and imaged using an UPlanApo 60×1.42 NA objective on an Olympus BX62 microscope fitted with a cooled Hamamatsu Orca AG CCD camera. The microscope, filters, camera were controlled as outlined (Ghiran et al., 2008). Deconvolution is detailed in Extended Experimental Methods.
Co-localization of EBV and CD35
Cells were fixed in 4% formaldehyde for 10 min at RT, washed 2x in buffer containing 0.5% IgG free BSA and incubated with Alexafluor 594 Zenon-labeled anti-CD35 rabbit polyclonal Ab and Alexafluor 488 labeled mouse anti-EBV IgG for 20 min at RT, washed 2x in the presence of 0.03% Hoechst 33342, mounted on slides using Fluoromount (DakoCytomation) and visualized with a UplanApo 60×1.42 objective fitted on an Olympus BX62 motorized microscope. Images were recorded with a Qimaging Emc2 cooled CCD camera controlled by Slidebook v. 5.0 (Intelligent Imaging Innovations, 3i). For co-localization studies, the microscope was fitted with Zero-Shift DAPI, Texas Red and GFP-green filters (Semrock) providing exact registration between pixels upon channel overlap.
Imaging of EGFP-EBV
EGFP-EBV expressing cells were imaged using filters set for FITC with an excitation wavelength of 488 nm resulting in emission at 507 nm as described (Speck and Longnecker, 1999).
Immunoblot
IBs were performed by standard methods (Extended Experimental Procedures) using non-reducing Laemmli SDS-PAGE sample buffer (Boston Bioproducts). Protein was visualized with an ECL kit (GE-Life Sciences) and images obtained using a LAS-4000 luminescent image analyzer (Fujifilm).
PCR
Cellular RNA was extracted with a RNeasy Total RNA kit (QIAGEN), treated with DNase (Promega), repurified on RNeasy spin columns and reverse transcribed with the RNA to cDNA EcoDry Premix™ (Oligo dT) kit (Clontech) using an iCycler™ thermal cycler (BioRad). PCR amplification of cDNA was performed using High Yield PCR EcoDry Premix™ (Clontech) with specific primers spanning the CT of CD21: sense 5′ GTGCCAATCGGATCACC 3′ and anti-sense 5′ TCCGCTGAATTCCAAGC 3′. Control GAPDH primers were sense 5′ GAGTCAACGGATTGTGTCGT 3′, antisense 5′ TTGATTTTGGAGGGATCTCG 3′. Amplification conditions: 95°C for 5 min for denaturation, 35 cycles at 95°C for 1 min, 55°C for 2 min, and 72°C for 2 min using an iCycler™.
Virus Production
B958 cells harboring Tet-BZLF1 or EBfaV-GFP were induced to release EBV into the supernatant as respectively described (Kudoh et al., 2003; Speck and Longnecker, 1999). Supernatants were passed through a 0.45μm cellulose acetate filter and centrifuged at 10,000 rpm (SLA-1500 rotor, Sorvall RC-5B Centrifuge, Du Pont) for 90 min. Concentrated pellets were diluted 1:50 with RPMI, aliquoted and stored at −80°C until use. Other EBV strains were prepared as described (Fingeroth et al., 1984).
Virus Binding and Infection Assays
Cells (1 × 106) seeded in triplicate were incubated with standard amounts of EBV under different conditions, as detailed in Extended Experimental Methods. EBV binding was assessed by FC, infection by microscopic visualization of EGFP EBV in cells and detection of LMP1 by IB (Extended Experimental Procedures).
Supplementary Material
Epstein-Barr virus (EBV) infection begins upon attachment of EBVgp350/220 to CD21 on normal B-cells where CD21 forms two complexes, one with CD19 another with CD35. The CD21/CD19 complex persists on immortalized and B-cell tumor lines, whereas CD35 is lost. To investigate CD35’s role, cells were transduced with CD35, CD21, or both. Strikingly, CD35+ cells bound gp350/220 and were latently infected when HLA II was co-expressed. These findings identify a novel receptor and underscore the importance of EBVgp350/220 for vaccine development.
ACKNOWLEDGMENTS
We apologize to the many individuals whose contributions were not directly cited due to space restriction. JDF was supported by NIH R01AI063571, AHA Grant-in-Aid, St Baldrick’s Foundation and JGO by a Cancer Research Institute fellowship. We thank the BIDMC Cytometry Core, Anastasia Hyrina for technical assistance and Dr. David Isenman for soluble CD21 SCR1-2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asokan R, Hua J, Young KA, Gould HJ, Hannan JP, Kraus DM, Szakonyi G, Grundy GJ, Chen XS, Crow MK, et al. Characterization of human complement receptor type 2 (CR2/CD21) as a receptor for IFN-alpha: a potential role in systemic lupus erythematosus. J Immunol. 2006;177:383–394. doi: 10.4049/jimmunol.177.1.383. [DOI] [PubMed] [Google Scholar]
- Birmingham DJ, Hebert LA. CR1 and CR1-like: the primate immune adherence receptors. Immunol Rev. 2001;180:100–111. doi: 10.1034/j.1600-065x.2001.1800109.x. [DOI] [PubMed] [Google Scholar]
- Busse C, Feederle R, Schnolzer M, Behrends U, Mautner J, Delecluse HJ. Epstein-Barr viruses that express a CD21 antibody provide evidence that gp350’s functions extend beyond B-cell surface binding. J Virol. 2010;84:1139–1147. doi: 10.1128/JVI.01953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S, Sereti I, Scheinberg P, Kimura H, Childs RW, Cohen JI. Detection of EBV genomes in plasmablasts/plasma cells and non-B cells in the blood of most patients with EBV lymphoproliferative disorders by using Immuno-FISH. Blood. 2010;116:4546–4559. doi: 10.1182/blood-2010-05-285452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calender A, Cordier M, Billaud M, Lenoir GM. Modulation of cellular gene expression in B lymphoma cells following in vitro infection by Epstein-Barr virus (EBV) Int J Cancer. 1990;46:658–663. doi: 10.1002/ijc.2910460418. [DOI] [PubMed] [Google Scholar]
- Chen CH, Ghiran I, Beurskens FJ, Weaver G, Vincent JA, Nicholson-Weller A, Klickstein LB. Antibody CR1-2B11 recognizes a non-polymorphic epitope of human CR1 (CD35) Clin Exp Immunol. 2007;148:546–554. doi: 10.1111/j.1365-2249.2007.03355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JH, Fischer E, Kazatchkine MD, Lenoir GM, Lefevre-Delvincourt C, Revillard JP. Expression of CR1 and CR2 complement receptors following Epstein-Barr virus infection of Burkitt’s lymphoma cell lines. Scand J Immunol. 1987;25:587–598. doi: 10.1111/j.1365-3083.1987.tb01085.x. [DOI] [PubMed] [Google Scholar]
- Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier-Bussat M, Billaud M, Calender A, Lenoir GM. Epstein-Barr virus (EBV) nuclear-antigen-2-induced up-regulation of CD21 and CD23 molecules is dependent on a permissive cellular context. Int J Cancer. 1993;53:153–160. doi: 10.1002/ijc.2910530128. [DOI] [PubMed] [Google Scholar]
- Edson CM, Thorley-Lawson DA. Epstein-Barr virus membrane antigens: characterization, distribution, and strain differences. J Virol. 1981;39:172–184. doi: 10.1128/jvi.39.1.172-184.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhorn L, Klein E. EBV infection of mitogen-stimulated human B lymphocytes. Int J Cancer. 1981;27:181–183. doi: 10.1002/ijc.2910270209. [DOI] [PubMed] [Google Scholar]
- Epstein MA, Henle G, Achong BG, Barr YM. Morphological and Biological Studies on a Virus in Cultured Lymphoblasts from Burkitt’s Lymphoma. J Exp Med. 1965;121:761–770. doi: 10.1084/jem.121.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdei A, Isaak A, Torok K, Sandor N, Kremlitzka M, Prechl J, Bajtay Z. Expression and role of CR1 and CR2 on B and T lymphocytes under physiological and autoimmune conditions. Mol Immunol. 2009;46:2767–2773. doi: 10.1016/j.molimm.2009.05.181. [DOI] [PubMed] [Google Scholar]
- Ernberg I, Falk K, Hansson M. Progenitor and pre-B lymphocytes transformed by Epstein-Barr virus. Int J Cancer. 1987;39:190–197. doi: 10.1002/ijc.2910390212. [DOI] [PubMed] [Google Scholar]
- Faulkner GC, Burrows SR, Khanna R, Moss DJ, Bird AG, Crawford DH. X-Linked agammaglobulinemia patients are not infected with Epstein-Barr virus: implications for the biology of the virus. J Virol. 1999;73:1555–1564. doi: 10.1128/jvi.73.2.1555-1564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingeroth JD. Comparative structure and evolution of murine CR2. The homolog of the human C3d/EBV receptor (CD21) J Immunol. 1990;144:3458–3467. [PubMed] [Google Scholar]
- Fingeroth JD, Benedict MA, Levy DN, Strominger JL. Identification of murine complement receptor type 2. Proc Natl Acad Sci U S A. 1989;86:242–246. doi: 10.1073/pnas.86.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade R, Barel M, Ehlin-Henriksson B, Klein G. gp140, the C3d receptor of human B lymphocytes, is also the Epstein-Barr virus receptor. Proc Natl Acad Sci U S A. 1985;82:1490–1493. doi: 10.1073/pnas.82.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiran I, Barbashov SF, Klickstein LB, Tas SW, Jensenius JC, Nicholson-Weller A. Complement receptor 1/CD35 is a receptor for mannan-binding lectin. J Exp Med. 2000;192:1797–1808. doi: 10.1084/jem.192.12.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiran I, Glodek AM, Weaver G, Klickstein LB, Nicholson-Weller A. Ligation of erythrocyte CR1 induces its clustering in complex with scaffolding protein FAP-1. Blood. 2008;112:3465–3473. doi: 10.1182/blood-2008-04-151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. EBV and systemic lupus erythematosus: a new perspective. J Immunol. 2005;174:6599–6607. doi: 10.4049/jimmunol.174.11.6599. [DOI] [PubMed] [Google Scholar]
- Haywood AM. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J Virol. 1994;68:1–5. doi: 10.1128/jvi.68.1.1-5.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, Mc LL. The mammalian cell-virus relationship. II. Adsorption, reception, and eclipse of poliovirus by HeLa cells. J Exp Med. 1959;109:487–504. doi: 10.1084/jem.109.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt-Fletcher LM. Epstein-Barr virus entry. J Virol. 2007;81:7825–7832. doi: 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaak A, Gergely P, Jr., Szekeres Z, Prechl J, Poor G, Erdei A, Gergely J. Physiological up-regulation of inhibitory receptors Fc gamma RII and CR1 on memory B cells is lacking in SLE patients. Int Immunol. 2008;20:185–192. doi: 10.1093/intimm/dxm132. [DOI] [PubMed] [Google Scholar]
- James JA, Kaufman KM, Farris AD, Taylor-Albert E, Lehman TJ, Harley JB. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100:3019–3026. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klickstein LB, Bartow TJ, Miletic V, Rabson LD, Smith JA, Fearon DT. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J Exp Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JM, Hannan JP, Eisenmesser EZ, Holers VM. Biophysical investigations of complement receptor 2 (CD21 and CR2)-ligand interactions reveal amino acid contacts unique to each receptor-ligand pair. J Biol Chem. 2010;285:27251–27258. doi: 10.1074/jbc.M110.106617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krushkal J, Bat O, Gigli I. Evolutionary relationships among proteins encoded by the regulator of complement activation gene cluster. Mol Biol Evol. 2000;17:1718–1730. doi: 10.1093/oxfordjournals.molbev.a026270. [DOI] [PubMed] [Google Scholar]
- Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. Immunol Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- Kudoh A, Fujita M, Kiyono T, Kuzushima K, Sugaya Y, Izuta S, Nishiyama Y, Tsurumi T. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J Virol. 2003;77:851–861. doi: 10.1128/JVI.77.2.851-861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle B, Flamand L, Gourde P, Beauchamp D, Gosselin J. Epstein-Barr virus infects and induces apoptosis in human neutrophils. Blood. 1998;92:291–299. [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- Li Q, Spriggs MK, Kovats S, Turk SM, Comeau MR, Nepom B, Hutt-Fletcher LM. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Cooper NR, Tack BF, Nemerow GR. Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes. Proc Natl Acad Sci U S A. 1987;84:9194–9198. doi: 10.1073/pnas.84.24.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow GR, Cooper NR. Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology. 1984;132:186–198. doi: 10.1016/0042-6822(84)90102-8. [DOI] [PubMed] [Google Scholar]
- Nemerow GR, Wolfert R, McNaughton ME, Cooper NR. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Sage DR, Stehle T, Fingeroth JD. The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding. Proc Natl Acad Sci U S A. 2002;99:10641–10646. doi: 10.1073/pnas.162360499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. Lippiincott, Williams & Wilkins; Philadelphia: 2007. pp. 2655–2700. [Google Scholar]
- Riley JL, Westerheide SD, Price JA, Brown JA, Boss JM. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- Roome AJ, Reading CL. Frequency of B-lymphocyte transformation by Epstein-Barr virus decreases with entry into the cell cycle. Immunology. 1987;60:195–201. [PMC free article] [PubMed] [Google Scholar]
- Roozendaal R, Carroll MC. Complement receptors CD21 and CD35 in humoral immunity. Immunol Rev. 2007;219:157–166. doi: 10.1111/j.1600-065X.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- Sashihara J, Burbelo PD, Savoldo B, Pierson TC, Cohen JI. Human antibody titers to Epstein-Barr Virus (EBV) gp350 correlate with neutralization of infectivity better than antibody titers to EBV gp42 using a rapid flow cytometry-based EBV neutralization assay. Virology. 2009;391:249–256. doi: 10.1016/j.virol.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlossman SF, Boumsell L, Gilks W, Harlan JM, Kishimoto T, Morimoto C, Ritz J, Shaw S, Silverstrin R, Springer T, et al. Leukocyte Typing V. Vol 1. Oxford University Press; Oxford, New York, Tokyo: 1995. [Google Scholar]
- Shaw PL, Kirschner AN, Jardetzky TS, Longnecker R. Characteristics of Epstein-Barr virus envelope protein gp42. Virus Genes. 2010;40:307–319. doi: 10.1007/s11262-010-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C, Awandare GA, Kopydlowski KM, Czege J, Moch JK, Finberg RW, Tsokos GC, Stoute JA. Complement receptor 1 is a sialic acid-independent erythrocyte receptor of Plasmodium falciparum. PLoS Pathog. 2010;6:e1000968. doi: 10.1371/journal.ppat.1000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck P, Longnecker R. Epstein-Barr virus (EBV) infection visualized by EGFP expression demonstrates dependence on known mediators of EBV entry. Arch Virol. 1999;144:1123–1137. doi: 10.1007/s007050050574. [DOI] [PubMed] [Google Scholar]
- Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- Tanner J, Whang Y, Sample J, Sears A, Kieff E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J Virol. 1988;62:4452–4464. doi: 10.1128/jvi.62.12.4452-4464.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedder TF, Inaoki M, Sato S. The CD19-CD21 complex regulates signal transduction thresholds governing humoral immunity and autoimmunity. Immunity. 1997;6:107–118. doi: 10.1016/s1074-7613(00)80418-5. [DOI] [PubMed] [Google Scholar]
- Tham WH, Schmidt CQ, Hauhart RE, Guariento M, Tetteh-Quarcoo PB, Lopaticki S, Atkinson JP, Barlow PN, Cowman AF. Plasmodium falciparum uses a key functional site in complement receptor type-1 for invasion of human erythrocytes. Blood. 2011;118:1923–1933. doi: 10.1182/blood-2011-03-341305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J, Kimmig L, Salzer U, Grudzien M, Lebrecht D, Hagena T, Draeger R, Volxen N, Bergbreiter A, Jennings S, et al. Genetic CD21 deficiency is associated with hypogammaglobulinemia. J Allergy Clin Immunol. 2012;129:801–810. e806. doi: 10.1016/j.jaci.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Tsokos GC, Thyphronitis G, Jack RM, Finkelman FD. Ligand-loaded but not free complement receptors for C3b/C4b and C3d co-cap with cross-linked B cell surface IgM and IgD. J Immunol. 1988;141:1261–1266. [PubMed] [Google Scholar]
- Tuveson DA, Ahearn JM, Matsumoto AK, Fearon DT. Molecular interactions of complement receptors on B lymphocytes: a CR1/CR2 complex distinct from the CR2/CD19 complex. J Exp Med. 1991;173:1083–1089. doi: 10.1084/jem.173.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L, Delfraissy JF, Vazquez A, Wallon C, Galanaud P, Kazatchkine MD. Monoclonal antibodies to the human C3b/C4b receptor (CR1) enhance specific B cell differentiation. J Immunol. 1987;138:2988–2993. [PubMed] [Google Scholar]
- Zychlinska M, Herrmann H, Zimber-Strobl U, Hammerschmidt W. Restricted expression of Epstein-Barr virus latent genes in murine B cells derived from embryonic stem cells. PLoS One. 2008;3:e1996. doi: 10.1371/journal.pone.0001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.