SUMMARY
p11, through unknown mechanisms, is required for behavioral and cellular responses to selective serotonin-reuptake inhibitors (SSRIs). Here we have identified SMARCA3, a chromatin-remodeling factor, as a novel target for the p11/annexin A2 heterotetrameric complex. Determination of the crystal structure indicates that SMARCA3 peptide binds to a hydrophobic pocket in the heterotetramer. Formation of this complex increases the DNA binding affinity of SMARCA3 and its localization to the nuclear matrix fraction. In the dentate gyrus, both p11 and SMARCA3 are highly enriched in hilar mossy cells and basket cells. In response to the SSRI, fluoxetine, the expression of p11 is induced in both cell types, and the amount of the ternary complex of p11/annexin A2/SMARCA3 is increased. SSRI-induced neurogenesis and behavioral responses are abolished by constitutive knockout of SMARCA3. Our studies indicate a central role for a chromatin-remodeling factor in the SSRI/p11 signaling pathway, and suggest a novel approach to the development of improved antidepressant therapies.
INTRODUCTION
Selective serotonin reuptake inhibitors (SSRIs) are currently the most widely used class of antidepressants (Berton and Nestler, 2006). SSRI medications generally take several weeks to show clinical efficacy, including mood elevation, in spite of their immediate effect on serotonergic neurotransmission. This therapeutic delay suggests the involvement of complicated downstream mechanisms, including long-term changes in gene expression and neuroplasticity. However, our knowledge of the molecular mechanisms underlying the efficacy of long-term treatment with SSRIs, and of the pathophysiology of depression, is still rudimentary.
p11 (S100A10) is a pivotal regulator of depression-like behaviors and a mediator of antidepressant responses (Svenningsson et al., 2006). Despite the importance of p11 in the actions of SSRIs, our knowledge about its underlying molecular mechanisms is limited (Svenningsson et al., 2006). Annexin A2 (AnxA2) is a well characterized binding partner for p11. AnxA2 together with p11 plays a role in trafficking of membranous/cytoplasmic proteins to plasma membrane or in providing them with firm anchorage at the plasma membrane and the cytoskeletal structure (Rescher and Gerke, 2008). p11 and AnxA2 were also found to localize in the nucleus and interact with nuclear proteins (Das et al., 2010; Liu and Vishwanatha, 2007), although the precise roles of p11 and AnxA2 in the nucleus have not been clearly defined. In this study, we have observed that chronic treatment with an SSRI, fluoxetine, increases the levels of the p11/AnxA2 complex. We have identified SMARCA3, a chromatin-remodeling factor, as a downstream target of the p11/AnxA2 complex. Our data indicate that the p11/AnxA2/SMARCA3 pathway mediates both neurogenic and behavioral responses to SSRIs.
RESULTS
Identification of SMARCA3 as a Specific Binding Partner of p11/AnxA2 Complex
p11, together with AnxA2, form a heterotetramer in cells. However, it is not yet established that p11 exists in a protein complex with AnxA2 in brain tissue. Here we show that the protein level of AnxA2 is drastically down-regulated in the frontal cortex and hippocampus of p11 KO mice (Figure 1A), in spite of unchanged levels of AnxA2 transcript (Figure 1B). In contrast, the protein level of AnxA1, another annexin family member, and of S100B, another S100 family member, were not altered in the brain of p11 KO mice, indicating the specificity of the physiological interaction between p11 and AnxA2 in the brain (Figure 1A). We have also observed that p11 protein level in the hippocampal lysates from AnxA2 knockout mice is reduced (data not shown). Given that components of a protein complex often stabilize each other, the data strongly support the existence of a protein complex of p11 and AnxA2 in the brain. In addition, we were able to co-immunoprecipitate AnxA2 with p11 from lysates of the hippocampus as well as from lysates of N2a neuroblastoma cells (Figure 1C).
Figure 1. p11/AnxA2 as an Antidepressant-Regulated Protein Complex.
(A) Brain lysates from frontal cortex and hippocampus of WT (+/+) and p11 KO (−/−) mice were immunoblotted for p11, AnxA2, S100B, and AnxA1.
(B) mRNA levels of p11 and AnxA2 in the hippocampus were measured using Q-PCR in p11 WT (+/+) and KO (−/−) mice. Data represent mean± SEM.
(C) Co-immunoprecipitation of p11 and AnxA2 from lysates of N2a neuroblastoma cells and mouse hippocampus using anti-p11 antibody.
(D) WT (+/+) or p11 KO (−/−) mice were administered vehicle (VEH) or fluoxetine (FLX) for 2 weeks. Hippocampal lysates were immunoblotted for p11, AnxA2, and β-actin.
(E) Quantitation of the immunoblot shown in (D) using infrared imaging system (Odyssey, LI-COR). Data represent mean± SEM. *p<0.05, **p<0.01, ns (non-significant), n.d. (not detectable), t-test.
Previous studies showed that p11 was induced in the frontal cortex (Svenningsson et al., 2006) and hippocampus (Warner-Schmidt et al., 2010) by chronic administration of antidepressants. In the present study, we observed concomitant up-regulation of p11(170.4±7.3% of p11(+/+)-VEH group, p=0.004) and of AnxA2 (167.1±20.8%, p=0.042) (Figure 1D and 1E). This AnxA2 increase was not observed in p11 KO mice (Figure 1E). Collectively, these results suggest that p11 and AnxA2 exist as a protein complex, which can be induced by antidepressant administration.
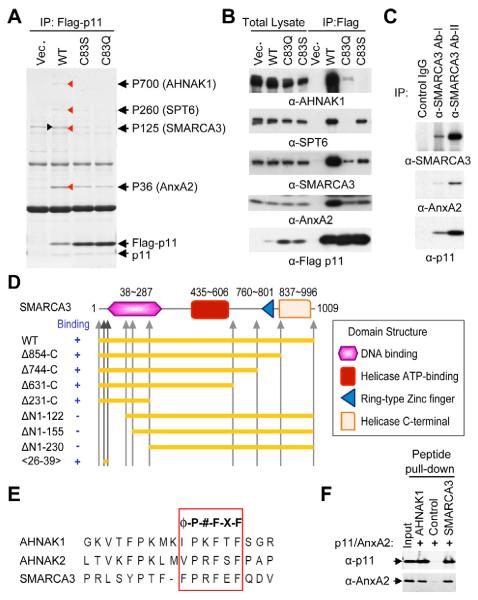
Next, we undertook a search for binding partners for p11/AnxA2. To ensure the specificity of the interaction with the p11/AnxA2 heterotetramer, we compared wild-type (WT) versus C83S and C83Q mutants of p11, which prevent the interaction between p11 and AnxA2 (Kube et al., 1992). C83S and C83Q mutations in p11 significantly decreased the interaction with AnxA2, without altering the interaction with endogenous p11 to form a p11 dimer, suggesting that C83 mutations selectively interfere with the heterotetramer formation, but not the homodimerization of p11 molecules (Figure S1A). After transfection of p11 WT and C83 mutant plasmids into HEK293 cells, we isolated the protein complex of p11 using immunoprecipitation (Figure 2A). Four proteins with relative molecular mass of 700, 260, 125 and 36 kDa were co-precipitated with WT p11, and were identified by mass spectrometry as AHNAK1 (AHNAK nucleoprotein), SPT6 (suppressor of Ty 6 homolog S. cerevisiae), SMARCA3 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 3), and AnxA2 respectively (Figure 2A). The identity of each protein was further confirmed by immunoblotting with specific antibodies (Figure 2B). AHNAK1, SPT6 and SMARCA3 were co-precipitated with WT p11 and the interaction was greatly reduced or abolished by either C83S or C83Q mutation of p11, indicating that the interaction likely needs AnxA2 binding to p11. AHNAK1 has been reported as a binding protein of p11/AnxA2 (Benaud et al., 2004). We focused on SMARCA3 in the following studies, because of the potential physiological importance of this chromatin-remodeling factor. To evaluate the role of AnxA2 in the intermolecular interaction, an in vitro pull-down assay using GST-p11, AnxA2, and 35S-labeled SMARCA3 was used. The SMARCA3 interaction was significantly increased by the addition of AnxA2 to the pull-down assay mixture (Figure S1B). The interaction of SMARCA3 with p11/AnxA2 was further confirmed by the inverse immunoprecipitaton using anti-SMARCA3 antibodies (Figure 2C). Collectively, these results identified SMARCA3 as a novel binding partner of p11/AnxA2.
Figure 2. Identification of the Binding Proteins of p11/AnxA2 Complex.
(A) Control vector (Vec.), and vectors containing WT or interaction-defective mutants (C83S and C83Q) of p11 were transfected into HEK293 cells. Proteins which co-precipitated with WT are marked with arrow heads (red). A non-specific band is indicated by a black arrow head. The proteins were identified by tandem mass spectrometry (black arrows).
(B) Immunoblots for AHNAK1, SPT6, SMARCA3, AnxA2 and Flag-p11, as indicated.
(C) Co-immunoprecipitation of p11/AnxA2/SMARCA3 complex from brain lysates by two different anti-SMARCA3 antibodies.
(D) WT, deletion mutants and a peptide were tested for their interaction with p11/AnxA2.
(E) Putative p11-binding sequences of AHNAK1 (aa5654-5671), AHNAK2 (aa5382-5399) and SMARCA3 (aa26-42).
(F) Interaction of the AHNAK1 or SMARCA3 peptides with p11/AnxA2 was confirmed by the pull down assay of biotinylated peptides. Scrambled peptide was used as control.
See also Figure S1.
SMARCA3 belongs to the family of SWI/SNF proteins that use the energy of ATP hydrolysis to remodel chromatin in a variety of nuclear processes, such as transcriptional regulation, and DNA replication and repair (Debauve et al., 2008). SMARCA3 contains multiple domain structures including DNA-binding, helicase ATP-binding, RING-type Zinc finger, and helicase C-terminal domains (Figure 2D). We next performed in vitro pull-down assay with a series of deletion constructs of SMARCA3 to determine the binding region for p11/AnxA2 (Figures 2D, S1C and S1D). While the serial deletion from the SMARCA3 C-terminus had no effect on the interaction with p11 (Figure S1C), the deletion of the N-terminus of SMARCA3 abolished the interaction (Figure S1D), localizing a binding region close to the N-terminus of SMARCA3 (Figure 2D). Through sequence alignment between AHNAK family proteins and the N-terminus of SMARCA3, we found a highly conserved putative binding motif, represented by φ-P-#-F-X-F (φ: hydrophobic amino acid, P: proline, #: basic amino acid, F: phenylalanine, X: any amino acid) (Figure 2E). Next, we performed a peptide pull-down assay to validate p11/AnxA2 binding to the putative binding motif. Indeed, the short synthetic peptides from AHNAK1, AHNAK2 and SMARCA3 covering the putative motif were sufficient to bind to p11/AnxA2 (Figure 2F; data not shown for AHNAK2 peptide).
Crystal Structure of p11/AnxA2 Bound to Its Ternary Target, SMARCA3
To understand the molecular interaction of SMARCA3 and AHNAK1 with p11/AnxA2 in detail, we conducted structural studies of p11 and AnxA2 complexed with SMARCA3 or AHNAK1 peptides. We have used a p11-AnxA2 fusion protein (henceforth named p11-AnxA2 peptide cassette) in which full-length p11 is connected to AnxA2 peptide with a nine amino acid linker (QENLYFQGD) (Rezvanpour et al., 2009) to generate complexes with added SMARCA3 (P26-F39) and AHNAK1 (G5654-F5668) peptides. Crystals of the complexes with SMARCA3 peptide (Figure 3) and AHNAK1 peptide (Figure S2) diffracted to 3.0 and 2.0 Å resolution, respectively. The structures of the two complexes exhibited similar recognition principles (although the peptide sequences are different, the backbones of bound SMARCA3 and AHNAK1 peptides superimpose well with an rmsd = 0.4 Å). The higher resolution structure of the AHNAK1 peptide complex (Figure S2A to S2F) proved useful in resolving ambiguities in the lower resolution structure of the SMARCA3 peptide complex (Figure 3A to 3F).
Figure 3. Crystal Structure of SMARCA3 Peptide Bound to p11-AnxA2 Peptide Cassette as Binary Complex, and to p11 and Full-Length AnxA2 as Ternary Complex.
(A) Ribbon view of crystal structure of p11-AnxA2 peptide cassette in complex with SMARCA3 peptide. p11 (blue and green) and AnxA2 peptide (salmon) are shown in ribbon, with the bound SMARCA3 peptides (yellow) in stick representations.
(B) Space-filling (peptide) and electrostatic surface (p11-AnxA2 peptide cassette) view of the complex illustrating the positioning of the pair of SMARCA3 peptides within the binding groove of the dimeric complex.
(C) Omit 2Fo-Fc electron density maps contoured at 1σ level of bound SMARCA3 peptide in one monomer of the complex. AnxA2 peptide (salmon) and p11 (blue) are represented in ribbon views, SMARCA3 peptide (yellow) is represented in a stick view, and electron density contours are in green.
(D) Intermolecular contacts within one monomer of the complex between the SMARCA3 peptide (yellow) and p11 (blue)-AnxA2 peptide (salmon) cassette, with hydrogen bonds depicted as red dashed lines.
(E) Positioning of the conserved residues Pro35 and Phe37 of the bound SMARCA3 peptide within a hydrophobic pocket formed by residues from AnxA2 peptide (salmon) and p11 (blue).
(F) A view of the superimposed structures of the p11-AnxA2 peptide cassette in the free (p11 in yellow and AnxA2 peptide in cyan) and SMARCA3-bound (p11 in blue and AnxA2 peptide in salmon) states. Black arrows highlight the conformational changes in p11 and AnxA2 peptide upon complex formation.
(G) Two views of 2.8 Å structure of SMARCA3 peptide (yellow) in stick representation bound to full-length AnxA2 (salmon) and p11 (blue and green) in ribbon representation shown for two different angles.
See also Figures S2 and S3, and Tables S1 and S2.
We have solved the 3.0 Å crystal structure of the complex between SMARCA3 (P26-F39) peptide and the p11-AnxA2 peptide cassette (Figure 3A and 3B; crystallization statistics in Table S1). The fusion linker does not affect the binding between p11 and AnxA2 peptide, as we observe the same intermolecular contacts in this complex as those reported previously for non-linked components (Rety et al., 1999). The p11-AnxA2 peptide cassette forms a symmetrical dimer, with individual SMARCA3 peptides (electron density for bound peptide shown in Figure 3C), binding each monomer in the dimer, while retaining two-fold symmetry (Figure 3A and 3B). Complex formation between SMARCA3 and p11-AnxA2 peptide cassette is mediated by van der Waals contacts and hydrogen bond interactions, whereby the SMARCA3 peptide interacts with elements of both p11 and AnxA2 peptide (Figure 3D). Two highly conserved amino acids (Pro35 and Phe37) in the bound SMARCA3 peptide are directed inwards and anchored within a hydrophobic pocket formed by residues from both p11 (Phe42, Ile55, Leu59, Leu75, Leu79, Ala82) and AnxA2 peptide (Leu8, Leu11, Leu13) in the complex (Figure 3E), with similar interactions observed for Pro5664 and Phe5666 in the AHNAK1 complex (Figure S2E). The complex is stabilized by additional intermolecular hydrogen-bonding interactions involving Phe33(SMARCA3)-Asp60(p11) and Arg36/Glu38(SMARCA3)-Ser12/Glu14(AnxA2 peptide) pairs (Figure 3D), with similar intermolecular contacts observed in the AHNAK1 complex (Figure S2D). In addition, the two bound SMARCA3 peptides are aligned in a head-to-head arrangement that is stabilized through intermolecular hydrogen bond formation between their N-terminal residues (Figure 3A). As expected, two point mutations (Pro35A, and Phe37Y) in the binding consensus motif of SMARCA3 diminished the interaction with p11/AnxA2 (Figure S3A). In addition, Leu13A mutation of AnxA2 abolished the interaction with SMARCA3 with no effect on p11 interaction, whereas Leu8A and Leu11A mutation blunted the interaction to p11 as well as to SMARCA3 (Figure S3B), showing a unique role of the Leu13 residue in creating a binding pocket for the ternary targets. These structural and mutagenesis studies account for the molecular basis of the preferential binding of SMARCA3 to p11/AnxA2.
We compared the structures of p11-AnxA2 peptide cassette in the free (PDB code: 1BT6) and SMARCA3 peptide-bound states following structural superimposition (Figure 3F). We observe that Asp60 in p11 is looped out from the peptide-binding groove in the free state but flips inwards by 4.8 Å on complex formation with the bound SMARCA3 peptide, in the process forming additional intermolecular hydrogen bonds (Figure 3D). In addition to this change, the Asp58-Asp64 loop segment also undergoes a conformational change by shifting towards and completes the peptide-binding groove on complex formation (Figure 3F). A similar conformational transition is observed on addition of AHNAK1 peptide to the p11-AnxA2 peptide cassette (Figure S2F). Indeed, the mutation on the p11 Asp60 residue abolished the interaction with SMARCA3 while maintaining homodimerization of p11, as well as heterotetramer formation with AnxA2, although the D60A mutant of p11 displayed a decrease in protein stability (Figure S3C). In our structures of complexes with bound SMARCA3 and AHNAK1 peptides, the disulfide bridges observed for both p11 alone and the p11-AnxA2 peptide cassette (Rety et al., 1999) are disrupted due to the movement of the Asp58-Asp64 loop on complex formation. As to the conformational change in AnxA2, we observe an inward flip of Ser12 towards the peptide-binding site by 4.6 Å for the SMARCA3 bound complex, thereby forming intermolecular hydrogen bonds with bound peptide in the complex (Figure 3D and 3F). In addition, due to this movement of Ser12, adjacent Leu13 in AnxA2 contributes to the closing of an additional face of the hydrophobic pocket, thereby anchoring the conserved residues Pro35 and Phe37 of bound SMARCA3 within the binding channel.
We have also successfully grown 2.8 Å crystals of SMARCA3 peptide bound to p11 and full-length AnxA2, with two views of the structure of the ternary complex shown in Figure 3G (x-ray statistics summarized in Table S2). Importantly, the crystal structure of p11/full-length AnxA2/SMARCA3 peptide illuminated the three dimensional organization of each component within the ternary complex (Figure 3G). Full-length AnxA2 is composed of an N-terminal p11 binding region and a C-terminal annexin repeat region with opposing convex and concave sides (Gerke et al., 2005). The convex side of AnxA2 faces the cellular membrane to mediate PIP2 binding, and the concave side faces away from the membrane. While two N-terminal peptides of AnxA2 contact the lateral/bottom side of the inverted p11 homodimer, two N-terminal peptides of SMARCA3 are anchored on the top position of the inverted p11 dimer while crossing each other. On the other hand, C-terminal annexin repeat regions of full-length AnxA2 are not involved in intermolecular recognition in the ternary complex. Furthermore, the C-terminal ends of the SMARCA3 peptides point downward, suggesting that the rest of the C-terminal parts of SMARCA3 including DNA binding-, ATP-dependent helicase-, and Zinc finger domains are likely positioned opposite to the convex side of the annexin repeat region of AnxA2. This mode of ternary complex assembly may leave C-terminal annexin repeat regions of AnxA2 free to execute the binding to PIP2 and actin, without steric hindrance. Taken together, our structural data on the complex with full-length AnxA2 implicate additional regulatory mechanisms through extended molecular interactions via C-terminal annexin repeat region.
Regulation of SMARCA3 Activity through Direct Interaction with p11/AnxA2
SMARCA3 was initially cloned as a regulatory factor that binds to the target DNA motifs of several gene promoters and enhancers, and was shown to regulate the transcription of tissue-specific target genes such as PAI-1, β-globin, and prolactin (Debauve et al., 2008). It is noteworthy that the p11-binding region (aa34-39) of SMARCA3 is located adjacent to the DNA binding domain (aa38-287) and seems to be partially overlapped. This observation prompted us to examine whether p11/AnxA2 interaction may regulate the DNA-binding affinity of SMARCA3. SMARCA3 was known to interact with the B-box element in the promoter of the PAI-1 gene (Ding et al., 1996). We carried out an in vitro reconstitution assay using purified recombinant proteins and the B-box oligonucleotide immobilized on beads. The oligonucleotide pull-down assay showed that SMARCA3 can form a quaternary complex together with p11/AnxA2 and the B-box oligonucleotide (Figure 4A). Furthermore, p11/AnxA2 interaction increases the DNA binding affinity of SMARCA3 by up to 2.5 fold, whereas the equivalent amount of AnxA2 alone didn’t show any effect (Figure 4B).
Figure 4. SMARCA3 Regulation by p11/AnxA2 Complex.
(A) The B-box oligonucleotide was incubated with the N-terminal domain of SMARCA3 (aa1-350) and p11 and/or AnxA2. Bound proteins were immunoblotted.
(B) Quantitation of SMARCA3 bound to the B-box oligonucleotide. Mean±SEM. *p<0.05, **p<0.01, t-test
(C) Whole cells (WC) and the nuclear matrix (NM) were prepared from control (Control) or p11-knockdown (KD) COS-7 cells and immunostained with the indicated antibodies. Arrows: knockdown cells; open arrowheads: non-knockdown cells. Scale Bar, 20 μm
(D) WC lysates and NM were prepared from COS-7 cells transfected as indicated, and immunoblotted for SMARCA3-V5 (α-V5), p11, AnxA2, Lamin-B (nuclear matrix marker).
(E) Transcriptional activity of SMARCA3 in N2a cells after co-transfection of luciferase reporter gene conjugated to PAI-1 promoter, together with indicated plasmids. Immunoblots of cell lysates and luciferase activity are shown. Mean±SEM. **p<0.01, ***p<0.001, t-test
(F) Transcriptional activity of SMARCA3 in Control or p11-knockdown (KD) COS-7 cells transfected with indicated SMARCA3 plasmids. Mean±SEM. **p<0.01, ***p<0.001, t-test
The distinct subnuclear localizations of nuclear factors are essential to conduct chromatin remodeling, transcription, replication, and mRNA processing (Zaidi et al., 2007). In fact, among SWI/SNF family chromatin remodelers, the hBAF (Brg1-associated factors) complex is known to be targeted to the nuclear matrix/chromatin through direct interaction with nuclear PIP2 and actin/actin-related protein (Rando et al., 2002; Zhao et al., 1998). It is well established that p11/AnxA2 binds to PIP2 as well as actin (Rescher and Gerke, 2008). Thus, we examined the possibility that p11/AnxA2 may regulate the subnuclear localization of SMARCA3. In the whole cell preparation, p11 is predominantly cytoplasmic, and a much less but significant level of p11 is in the nucleus, whereas SMARCA3 is mainly localized inside the nucleus. Our unpublished studies with primary cultured neurons indicate that p11 together with AnxA2 shuttles between cytoplasm and nucleus (data not shown). In the nuclear matrix, which is prepared after cell permeabilization followed by chromatin digestion, the presence and the colocalization of p11 and SMARCA3 are evident (Figure 4C). Importantly, the retention of SMARCA3 in the nuclear matrix is dramatically reduced by the silencing of p11 expression with siRNA. Consistent with that, biochemical fractionation of the nuclear matrix revealed that WT SMARCA3 is in the nuclear matrix preparation, whereas the P35A mutant, defective in p11/AnxA2 interaction, is not (Figure 4D), confirming the importance of the p11/AnxA2 complex in the subnuclear localization of SMARCA3. Taken together, p11/AnxA2 not only increases the DNA binding affinity of SMARCA3, but also anchors SMARCA3 to the nuclear matrix presumably via the interaction of AnxA2 with actin and PIP2.
To assess the functional consequence of p11-AnxA2 interaction in the transcriptional activity of SMARCA3, we utilized a PAI-1 promoter-driven luciferase reporter assay. Firstly, we examined the effect of overexpression of the p11/AnxA2 complex in N2a cells, which express low endogenous levels of p11 and AnxA2. Co-transfection of SMARCA3 with p11 and AnxA2 potentiated luciferase activity (Figure 4E). In addition, we examined the effect of p11 gene silencing in COS-7 cells, which express relatively higher endogenous levels of p11 and AnxA2. The silencing of the p11 gene leads to the down-regulation of SMARCA3-mediated luciferase activity (Figure 4F). In the same experimental set, we also compared WT SMARCA3 with its mutants (P35A, F37Y) defective in the interaction with the p11/AnxA2 complex. As expected, SMARCA3-mediated luciferase activity was abolished in the two mutants (Figure 4F). Collectively, these results suggest that p11/AnxA2 regulates transcriptional activity of SMARCA3 by controlling the DNA binding affinity of SMARCA3, as well as its localization.
Regulation of the p11/AnxA2/SMARCA3 Complex in Hilar Mossy Cells and Basket Cells in the Dentate Gyrus
p11 mediates the actions of antidepressants (Svenningsson et al., 2006). Chronic antidepressant administration increased the level of p11 in the hippocampus (Figure 1D and 1E). Antidepressant actions including neurogenesis require several weeks to show therapeutic effects. SMARCA3-mediated regulation of transcription may be associated with the therapeutic delay. Our previous study showed that p11 is expressed in GABAergic basket cells in the dentate gyrus and might play a critical role in antidepressant-induced hippocampal neurogenesis (Egeland et al., 2010). We thus examined the neuronal types expressing p11 and SMARCA3 in the dentate gyrus.
We took advantage of BAC (bacterial artificial chromosome) transgenic mice (Heintz, 2001), in which the expression of eGFP reporter is driven by p11 promoter activity ([p11]-eGFP). The excellence of immunostaining analysis enabled us to visualize the entire neuronal processes of the neurons expressing p11. Because eGFP-positive neurons in the BAC-[p11]-eGFP mice are doubly positive for the immunostaining of endogenous p11, eGFP-positive neurons represent p11-expressing neurons (Figure S4A). p11-expressing cells assessed with [p11]-eGFP signal localize in the hilus region of the dentate gyrus (Figure 5A). To identify neuronal types for the p11-expressing neurons, hippocampal sections from [p11]-eGFP BAC transgenic mice were doubly stained with eGFP and neuronal type markers. Notably, [p11]-eGFP reporter signal was found to be enriched both in CRT-positive mossy cells and in PV-positive basket cells, whereas the signal was negligible in granule cells, and is rarely (< 5%) observed in HIPP cells (Figure 5A). Furthermore, p11 expression is not observed in astrocytes or neural stem cells (Figure 5A).
Figure 5. SSRI Regulates p11 Expression in Mossy Cells and Basket Cells in the Dentate Gyrus.
(A) Cell types expressing [p11]-eGFP in the dentate gyrus. Calretinin (CRT, hillar mossy cells (MC)), parvalbumin (PV, subpopulation of basket cells (BC(PV+))), calbindin (CBD, mature granule cells (GC)) neuropeptide Y (NPY, hilar interneurons perforant path (HIPP)), glial fibrillary acidic protein (GFAP, astrocytes) and nestin (neural stem cells (SC)) were used to identify cell types. Solid arrow heads: representative doubly labeled cells. Open arrow heads: cells labeled only with markers. Scale bars, 20 m.
(B) Distinct laminar projections of mossy cells and parvalbumin-positive basket cells in the dentate gyrus. GCL: granule cell layer, IML: inner molecular layer, OML, outer molecular layer. Scale Bars, 100 m.
(C-E) Induction of [p11]-eGFP in dentate gyrus by chronic SSRI. The dentate gyrus slices were co-stained with anti-eGFP antibody (C and D) and either anti-CRT (C) or anti-PV (D) antibodies. Scale Bars, 100 m. eGFP intensity was quantitated in the indicated subregions (E). Values were normalized to fluorescence intensity in OML. Data represent mean±SEM (n=5-6 mice per group). *p<0.05, **p<0.01, t-test.
See also Figure S4.
We next examined if p11 expression in hippocampal neuronal subpopulations is altered in response to antidepressant administration. We took advantage of the distinct laminar segregation of the two types of p11-expressing neurons. The mossy cells assessed by CRT expression locate their soma and dendrites within the deep hilus and project their axonal arbor to the inner molecular layer (IML), whereas a subpopulation of basket cells visualized by PV staining locate their somas in the hilar border and project their axons to the granule cells within the granular cell layer (GCL) (Figure 5B). Since p11 is mainly enriched in mossy cells and basket cells, the [p11]-eGFP signal in the hilus and IML represents p11 promoter activity in mossy cells and the signal of eGFP in GCL does for basket cell activity. BAC-[p11]-eGFP mice were treated with fluoxetine for 2 weeks and [p11]-eGFP signal was examined in the areas of hilus, IML and GCL (Figure 5C-5E). Since a high density of serotonergic innervation prevails in the caudal part of the hippocampal formation, whereas the rostral part receives only a moderate to weak serotonergic innervation (Bjarkam et al., 2003; Gage and Thompson, 1980), the caudal part (bregma −2.5~−4.0) of the dentate gyrus was used for the analysis. Fluoxetine treatment increased [p11]-eGFP in the dentate gyrus (Figure 5C and 5D). Quantitative analysis revealed that fluoxetine increased [p11]-eGFP intensity in hilus and IML as well as in GCL, suggesting p11 induction in mossy cells and basket cells (Figure 5E). p11 is also induced in the rostral part of the hippocampus, but with less potency (Figure S4B). AnxA2 together with p11 is induced by fluoxetine and requires p11 for its protein stability (Figure 1D and 1E). Conversely, p11 protein requires AnxA2 for its protein stability (data not shown). Thus, AnxA2 is also likely to be induced in the same neurons.
We examined the expression of SMARCA3 in the dentate gyrus by immunohistochemistry. The specificity of the immunostaining was tested using SMARCA3 KO mice. SMARCA3 KO mice were generated by targeting exons 11-13 of the SMARCA3 gene (Figure S5A). Immunoblotting (Figure S5B and S5C) and immunohistochemistry (Figure 6A) confirmed the absence of SMARCA3. SMARCA3 is expressed primarily in the hilar area of the dentate gyrus (Figure 6A), where it is enriched in mossy cells and parvalbumin-positive basket cells (Figure 6B), and is expressed in [p11]-eGFP-positive cells (Figure 6C).
Figure 6. SMARCA3 Expression in Mossy Cells and Basket Cells in the Dentate Gyrus.
(A) Dentate gyrus slices from wild-type (top) or SMARCA3 KO (bottom) mice stained with anti-SMARCA3 antibody (Red) and nuclear dye DraQ5 (Blue). Scale Bars, 100 m.
(B) Neuronal cell types expressing SMARCA3 in the dentate gyrus. Scale Bars, 20 m. Representative cells doubly labeled are indicated by arrow heads.
(C) Co-expression of [p11]-eGFP and SMARCA3. BAC-[p11]-eGFP mice were stained with anti-eGFP and anti-SMARCA3 antibodies. Representative cells doubly labeled are indicated by arrow heads. Scale Bar, 20 μm.
(D and E) Level of SMARCA3 is not altered by fluoxetine administration for 2 weeks. Hippocampal lysates were immunoblotted for SMARCA3, and β-actin (D). mRNA level of SMARCA3 in the hippocampus was measured using Q-PCR (E). ns (non-significant).
(F) Formation of p11/AnxA2/SMARCA3 complex is increased after treatment with FLX for 2 weeks. SMARCA3 was immunoprecipitated from hippocampal lysates. Total lysates and immunoprecipitates (IPs) were immunoblotted for SMARCA3, p11, and AnxA2.
See also Figure S5.
In contrast to p11 and AnxA2 (Figure 1D and 1E), the levels of SMARCA3 protein (Figure 6D) and mRNA (Figure 6E) were not altered after treatment with fluoxetine. Analysis of hippocampal lysates, following immunoprecipitation of SMARCA3, revealed that chronic fluoxetine administration increased the ternary complex of p11/AnxA2/SMARCA3 by about 2.3 fold (Figure 6F). Thus, p11 and AnxA2 induction facilitates the assembly of the p11/AnxA2/SMARCA3 complex. Taken together, these results identify the mossy cells and the basket cells in the dentate gyrus as primary neuronal types for SSRI/p11/SMARCA3 signaling.
SMARCA3 is Required for Neurogenic and Behavioral Response to Chronic SSRI Administration
p11 KO results in the loss of enhanced hippocampal neurogenesis and behavioral change in response to chronic antidepressant treatment (Egeland et al., 2010), suggesting a crucial role for p11 and its downstream signaling pathways in those antidepressant actions. We examined here whether SMARCA3, as a downstream signaling molecule of p11, might mediate chronic antidepressant-induced hippocampal neurogenesis and behaviors.
Adult neurogenesis is controlled by multi-step processes including the proliferation of neural progenitors, differentiation, and maturation into functional granule neurons (Ming and Song, 2011). We examined proliferation using in vivo BrdU labeling of the neural progenitors in the S-phase in WT and SMARCA3 KO mice. A significant increase was observed in the number of BrdU-labeled neural progenitors in WT mice treated with fluoxetine (160.6±13.7% of VEH group, p<0.01), but not in SMARCA3 KO mice (Figure 7A and 7B). We also carried out an alternative assay measuring an endogenous mitotic marker Ki-67. Ki-67-positive cells were significantly increased in response to fluoxetine in WT mice (143.8±8.6%, p<0.01), but not in SMARCA3 KO mice (Figure S6A and S6B), confirming the results of the BrdU assay.
Figure 7. SMARCA3 Is Required for SSRI-Induced Neurogenesis and Behavioral Changes.
(A and B) Fluoxetine-induced cell proliferation in WT and SMARCA3 KO mice. WT (+/+) and SMARCA3 KO (−/−) mice were administered VEH or FLX for 14 days and labeled with BrdU for the last 2 hrs prior to perfusion. (A) Immunostaining with anti-BrdU. Scale Bars, 100 μm. (B) Quantitation of BrdU-positive cells in the subgranular zone (n=6-8 mice per group).
(C and D) Fluoxetine-induced increase of doublecortin (DCX)-positive cells in WT and SMARCA3 KO mice. (C) Immunostaining with anti-DCX. Scale Bars, 20 μm. (D) Quantitation of DCX-positive cells in the subgranular and granular zone. (n=6-8 per group).
(E and F) Survival of newborn cells in WT and SMARCA3 KO mice treated with VEH or FLX. BrdU was injected for three consecutive days prior to fluoxetine administration for 28 days. (E) Immunostaining with anti-BrdU and anti-NeuN. Scale Bars, 100 μm. (F) Quantitation of BrdU-positive cells.
(G and H) Behavior was assayed using (G) the novelty suppressed feeding (NSF) paradigm after chronic administration of VEH or FLX (4 weeks, n=14-16 per group), or (H) the sucrose preference test (SPT) in non-stressed (NS) or restraint-stressed (RS) mice after chronic administration of VEH or escitalopram (eCIT) (4 weeks, n=8-11 per group).
All data represent mean±SEM. *p<0.05, **p<0.01, ***p<0.001, ns (non-significant), two-way ANOVA followed by the post-hoc Bonferroni test
We next analyzed the expression level of doublecortin (DCX), a marker for immature neurons, which represents a snapshot of newborn cells undergoing neuronal maturation and differentiation (Couillard-Despres et al., 2005). DCX immunofluorescence signal was increased by chronic fluoxetine administration in WT mice (218.4±18.0%, p<0.001), but not in SMARCA3 KO mice (Figure 7C and 7D). Chronic fluoxetine administration promotes the newborn cells to survive at post-mitotic stages and also to become mature neurons (Wang et al., 2008). Chronic fluoxetine administration greatly increased the survival of the BrdU-labeled newborn cells in WT mice (288.2±21.1%, p<0.01), but the effect of fluoxetine was attenuated in SMARCA3 KO mice (196.0±20.3%, p<0.05) (Figure 7E and 7F). Taken together, these results indicate that SMARCA3 contributes to multiple stages of antidepressant-stimulated neurogenesis, and this phenotype of SMARCA3 KO mice is reminiscent of that observed in p11 KO mice (Egeland et al., 2010).
Next, we investigated the functional significance of the p11/AnxA2/SMARCA3 complex for SSRI-induced behavioral changes. WT and SMARCA3 KO mice did not display a baseline difference in locomotor activity (open field test, Figure S6C), depressive behaviors (tail suspension test (TST) and sucrose preference test (SPT) Figures S6D and S6E), or anxiety behaviors (light/dark and elevated plus maze tests, Figures S6F and S6G). Novelty-suppressed feeding (NSF) is believed to represent depression-like and anxiety behavior, and this test is commonly used to measure the chronic effect of antidepressants (David et al., 2009; Surget et al., 2011). p11 KO mice are refractory to behavioral change in response to chronic fluoxetine administration in the NSF test (Egeland et al., 2010; Schmidt et al., 2012). In the same model, we analyzed the behavior of SMARCA3 KO mice. Chronic treatment with fluoxetine shortened latency to feed compared to vehicle treatment in WT mice (VEH vs FLX: 279±13 vs 187±20, p<0.01), but not in SMARCA3 KO mice (Figure 7G), as with p11 KO mice (Egeland et al., 2010). Neither fluoxetine nor SMARCA3 KO caused any significant effect on home cage feeding (Figure S6H) or body weight (data not shown).
Anhedonia is a core symptom of human depression (Berton and Nestler, 2006) and a chronic stress-induced decrease in sucrose preference in rodents is regarded as a sign of an hedonic deficit (Katz, 1982), which can be treated with chronic SSRI. We examined the effect of chronic administration of escitalopram (eCIT, another SSRI) on the behaviors of non-stressed (NS) or restraint-stressed (RS) mice in the SPT. Daily restraint for 14 days induced a reduction of sucrose consumption in both WT (NS+VEH vs RS+VEH: 100±12% vs 68±8%) and SMARCA3 KO mice (NS+VEH vs RS+VEH: 97±6% vs 61±8%). Post-stress treatment with eCIT recovered sucrose consumption to the normal level in WT mice (RS-VEH vs RS+eCIT: 68±8% vs 99±13%), but not in SMARCA3 KO mice (Figure 7H). No differences were observed between WT and SMARCA3 KO mice as well as among the experimental groups used for the sucrose preference test with regard to their locomotor activities (Figure S6I). Notably, the behavioral despair in TST is known to be treated by acute SSRI, while the therapeutic effect of SSRI in the NSF and SPT requires chronic treatment. SMARCA3 KO did not alter the effect of acute administration of eCIT in the TST (Figure S6J). Taken together, our results suggest a crucial role for SMARCA3 in p11-dependent neurogenic and behavioral response to chronic antidepressant administration.
DISCUSSION
Molecular Interaction of p11/AnxA2 with SMARCA3
p11 and AnxA2 cooperate to create a unique binding pocket, but the optimal binding condition is not achieved without conformational changes associated with target binding. Upon interaction with the SMARCA3 peptide, both Asp60 in p11 and Ser12 in AnxA2 flip inward toward the peptide-binding groove, forming additional intermolecular hydrogen bonds (Figure 3D-3F). Similar intermolecular contacts were also observed in the AHNAK1 complex (Figure S2D-S2F). These findings support an induced-fit model for the assembly of the p11/AnxA2/SMARCA3 complex. Among the key residues found in the binding pocket, the role of Ser12 (AnxA2) is of particular interest, because this residue is regulated by phosphorylation (Jost and Gerke, 1996), which may interfere with high-affinity binding of ternary targets.
The current study identifies a binding motif, which is represented as ϕ-P-#-F-X-F and can be used for in silico analysis to identify new binding targets of p11/AnxA2. Our binding motif is different from the p11-binding sequences previously reported for TRPV5/6 (VATTV) (van de Graaf et al., 2003), and TASK1 (RRSSV) (Girard et al., 2002). It is possible that the p11/AnxA2 heterotetramer has additional binding pocket(s) on the surface.
Regulation of SMARCA3 Function by Interaction with the p11/AnxA2 Complex
Most chromatin remodelers, including four well-characterized subfamily members (SWI/SNF, INO80/SWR1, ISWI and CHD), form a large multi-subunit complex with a core ATPase motor subunit and unique accessory/regulatory subunits (Hargreaves and Crabtree, 2011). The core subunit displays DNA- and nucleosome-dependent ATPase activity, and the accessory/regulatory subunits are essential for the function of the core ATPase subunit, by facilitating the interaction with the transcriptional regulatory factors, mediating the indirect binding to DNA and/or modified histones, and targeting the complex to subnuclear locations (Mohrmann and Verrijzer, 2005). SMARCA3, a relatively uncharacterized member of the SWI/SNF protein family, is composed of multiple functional domains, including a DNA binding domain, a SWI2/SNF2 ATPase-domain, a RING-type Zinc finger domain for the binding to RFBP, a Type IV P-type ATPase, and a C-terminal domain for the binding of transcription factors such as Sp1, Sp3, Egr1, and cRel (Debauve et al., 2008) (Figure 2D). While p11 and AnxA2 stabilize each other as structural components of a protein complex (Figure 1), the levels of p11 and AnxA2 are not altered in SMARCA3 KO mice (Figure S5B and S5C) and the level of SMARCA3 is not altered in p11 KO mice (data not shown). Thus, p11 and AnxA2 act as regulatory proteins but are not likely structural core components of SMARCA3. Notably, the DNA binding domain at the N-terminal side of the ATPase domain mediates the sequence-dependent binding of SMARCA3 to the target DNA (Debauve et al., 2008). p11/AnxA2 facilitates the DNA binding affinity of SMARCA3 (Figure 4A and 4B). Most chromatin-remodeling complexes, including SMARCA3, display DNA- and/or nucleosome-dependent ATPase activity (Hargreaves and Crabtree, 2011). Therefore, it is conceivable that the enhanced DNA binding of SMARCA3, upon the interaction with p11/AnxA2, may lead to the activation of SMARCA3 to initiate ATP-dependent chromatin remodeling of the target genes. Thus, p11/AnxA2 binding to SMARCA3 would open the chromatin structure and recruit specific transcription factors bound to the C-terminal domain of SMARCA3 to the specific locus of the genomic DNA.
Critical nuclear events such as gene expression, replication and repair processing occur at a distinct subnuclear region, the nuclear matrix, which is composed of nuclear lamins, nuclear actin/actin-related proteins, and phospholipids (Barlow et al., 2010; Zaidi et al., 2007). Indeed, key gene regulatory machineries such as transcription factors, chromatin-remodeling complexes, RNA polymerase II and processing factor SC35 are associated with nuclear matrix structures (Zaidi et al., 2007). p11/AnxA2 mediates the subnuclear targeting of SMARCA3 (Figure 4C and 4D). The subnuclear targeting of SMARCA3 is likely controlled by intrinsic phospholipid- and actin-binding properties of AnxA2 (Gerke et al., 2005). Our crystal structure of p11/full length AnxA2/SMARCA3 peptide visualized the spatial organization of each component in the ternary complex, in which the p11 dimer is ideally positioned in the core of the complex to link SMARCA3 to AnxA2 (Figure 3G). Thus, our current model is reminiscent of the role of p11/AnxA2 in mediating the membrane translocation of AHNAK1 (Benaud et al., 2004). Both SMARCA3 and AHNAK1 use the interaction with p11/AnxA2 to localize properly to the distinct subcellular sites where they become functionally active.
p11/AnxA2/SMARCA3 Complex in Antidepressant Action
p11/AnxA2/SMARCA3 complex-mediated hippocampal neurogenesis may contribute to the behavioral response to SSRIs. However, the involvement of hippocampal neurogenesis in depression and antidepressant-induced behavioral change is still controversial (Hanson et al., 2011). Such neurogenesis has been associated with mood, stress responses, and antidepressant effects in some studies (Santarelli et al., 2003; Snyder et al., 2011; Surget et al., 2011), but not in others (Bessa et al., 2009).
p11 and SMARCA3 are enriched in mossy cells and basket cells but not detectable in neural progenitor cells. Thus, the regulation of SSRI-induced neurogenesis by the p11/AnxA2/SMARCA3 signalling pathway is non-cell autonomous. In fact, GABAergic interneurons play an important role in the differentiation, development, and integration of newborn neurons (Ge et al., 2006; Tozuka et al., 2005). Specifically, interneuronal GABA release is thought to act directly on neural progenitor cells and, due to an age-specific increase in intracellular chloride levels in these young cells, causes an atypical depolarization that subsequently affects neurogenic processes (Tozuka et al., 2005). The synaptic regulation of granule cells by basket and mossy cells may contribute to the neurogenic and behavioral responses to SSRIs. It has been suggested that, upon activation by local inputs from the granule cells in the dentate gyrus, glutamatergic mossy cells primarily provide excitatory feedback to the granule cells through their axonal projections to the IML, and also provide excitatory drive to local GABAergic HIPPs in the hilus (Henze and Buzsaki, 2007; Scharfman, 1995). The basket cells primarily provide feedforward inhibition to the granule cells in response to excitatory inputs from the entorhinal cortex, and feedback inhibition to the granule cells in response to excitatory inputs from the granule cells (Houser, 2007). Thus, despite their relatively low abundance (approximately 3×104 mossy cells and 0.5×104 basket cells versus 1×106 granule cells in rat), the mossy cells and basket cells regulate the flow of extrinsic input to the dentate gyrus through modulation of the activity of the granule cells and/or the HIPPs, and such regulation is important to generate the distinct oscillation patterns of neuronal excitation in the dentate gyrus (Amaral et al., 2007; Henze and Buzsaki, 2007).
In this study, we have identified and characterized a novel nuclear protein complex mediating chronic actions of SSRIs. Many interesting questions have been raised as a result of this study: what are the roles of p11 and SMARCA3 in the excitability and synaptic transmission of basket cells and mossy cells?; which genes are regulated by the p11/AnxA2/SMARCA3 complex in basket cells and mossy cells?; which of these genes contribute to the neurogenic and behavioral responses to antidepressants? Current and future studies of the p11/AnxA2/SMARCA3 pathway should contribute not only to our understanding of SSRI actions, but also provide molecular and cellular targets for the development of advanced therapeutics for mood and anxiety disorders.
EXPERIMENTAL PROCEDURES
Generation of Transgenic Mice
All procedures involving animals were approved by the Rockefeller University Institutional Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines. The constitutive SMARCA3 KO mice were generated in Taconic-Artemis (Germany) and maintained at The Rockefeller University. The BAC-[p11]-eGFP transgenic line (GENSAT; Clone No. HC85) was provided by GENSAT (Gong et al., 2002). The mouse breeding and drug treatment methods are in the Extended Experimental Procedures.
Crystallization and Structure Determination of p11/AnxA2/SMARCA3 and p11/AnxA2/AHNAK1 Complexes
Coordinates and structure factors for p11-AnxA2 peptide cassette in complex with AHNAK1 peptide (PDB code 4HRG) and SMARCA3 peptide (PDB code 4HRH), and full-length p11/AnxA2 heterotetramer in complex with SMARCA3 peptide (PDB code 4HRE), have been deposited in the RCSB Protein Data Bank. Details of protein preparations, protein expression, purification, crystallization conditions, data collection, and refinement are included in Extended Experimental Procedures.
Preparation of Nuclear Matrix Fraction
Cells were incubated with cell-permeable crosslinker, DSP (1 mM), and extracted with CSK buffer containing 0.5% NP40. After chromatin digestion with DNase I and then elution with 0.25 M (NH4)2SO4, the nuclear matrix fraction was scraped for the biochemical assay or fixed with 2 % paraformaldehyde for immunocytochemistry.
Histological Methods
Immunostaining was carried out using the standard free floating method. Detailed description of antibody preparation, antigen retrieval, image acquisition and quantification are in Extended Experimental Procedures.
Behavioral Analysis
Mood and anxiety-related behaviors (NSF, SPT, TST, light-dark box test and elevated plus maze) and locomotor activity (open field test) were tested as described in Extended Experimental Procedures.
Data Analysis and Statistics
All data are presented as mean±SEM. Two group comparisons were done by two-tailed, unpaired Student′s t test or the non-parametric Mann-Whitney test. Multiple group comparisons were assessed using a one-way or two-way analysis of variance (ANOVA), followed by the post-hoc Newman-Keuls test or Bonferroni test respectively, when significant main effects or interactions were detected. Statistical significance was set at p<0.05 level. Summary for statistical analysis for animal experiments is included as Supplemental Table S3.
Supplementary Material
HIGHLIGHTS.
Identification of SMARCA3 as a novel binding protein of the p11/AnxA2 complex
Crystal structure of p11/AnxA2 heterotetramer bound to SMARCA3 peptide
Mechanism of SMARCA3 activation by interaction with p11/AnxA2 heterotetramer
SMARCA3 mediates chronic SSRI-induced neurogenesis and behaviors
ACKNOWLEDGEMENTS
This work was supported by DOD/USAMRAA Grant W81XWH-09-1-0392 to Y.K.; DOD/USAMRAA Grant W81XWH-09-1-0402 to P.G.; the JPB Foundation to P.G.; the Fisher Center Foundation to P.G.; NIH grants (MH090963, DA010044 and AG09464) to Y.K. and P.G.; the Maloris Foundation and the Abby Rockefeller Mauze Trust to D.J.P. We thank the staff at beamline 24ID-C of the Advanced Photon Source at the Argonne National Laboratory and beamline X29 of the National Synchrotron Light Source at the Brookhaven National Laboratory for assistance with data collection. We thank Daesoo Kim, Eric Schmidt, Jennifer Wagner-Schmidt and Yotam Sagi for their helpful advice and discussion, and Elisabeth Griggs for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information Supplemental Information includes Extended Experimental Procedures in article online at doi: xx.xxx.
REFERENCES
- Amaral DG, Scharfman HE, Lavenex P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies) Progress in brain research. 2007;163:3–22. doi: 10.1016/S0079-6123(07)63001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends in cell biology. 2010;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaud C, Gentil BJ, Assard N, Court M, Garin J, Delphin C, Baudier J. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. The Journal of cell biology. 2004;164:133–144. doi: 10.1083/jcb.200307098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton O, Nestler EJ. New approaches to antidepressant drug discovery: beyond monoamines. Nature reviews Neuroscience. 2006;7:137–151. doi: 10.1038/nrn1846. [DOI] [PubMed] [Google Scholar]
- Bessa JM, Ferreira D, Melo I, Marques F, Cerqueira JJ, Palha JA, Almeida OF, Sousa N. The mood-improving actions of antidepressants do not depend on neurogenesis but are associated with neuronal remodeling. Molecular psychiatry. 2009;14:764–773. 739. doi: 10.1038/mp.2008.119. [DOI] [PubMed] [Google Scholar]
- Bjarkam CR, Sorensen JC, Geneser FA. Distribution and morphology of serotonin-immunoreactive axons in the hippocampal region of the New Zealand white rabbit. I. Area dentata and hippocampus. Hippocampus. 2003;13:21–37. doi: 10.1002/hipo.10042. [DOI] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, Aigner L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci. 2005;21:1–14. doi: 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- Das S, Shetty P, Valapala M, Dasgupta S, Gryczynski Z, Vishwanatha JK. Signal Transducer and Activator of Transcription 6 (STAT6) Is a Novel Interactor of Annexin A2 in Prostate Cancer Cells. Biochemistry-Us. 2010;49:2216–2226. doi: 10.1021/bi9013038. [DOI] [PubMed] [Google Scholar]
- David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP, et al. Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron. 2009;62:479–493. doi: 10.1016/j.neuron.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debauve G, Capouillez A, Belayew A, Saussez S. The helicase-like transcription factor and its implication in cancer progression. Cellular and molecular life sciences : CMLS. 2008;65:591–604. doi: 10.1007/s00018-007-7392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Descheemaeker K, Marynen P, Nelles L, Carvalho T, CarmoFonseca M, Collen D, Belayew A. Characterization of a helicase-like transcription factor involved in the expression of the human plasminogen activator inhibitor-1 gene. DNA Cell Biol. 1996;15:429–442. doi: 10.1089/dna.1996.15.429. [DOI] [PubMed] [Google Scholar]
- Egeland M, Warner-Schmidt J, Greengard P, Svenningsson P. Neurogenic Effects of Fluoxetine Are Attenuated in p11 (S100A10) Knockout Mice. Biol Psychiatry. 2010;67:1048–1056. doi: 10.1016/j.biopsych.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Gage FH, Thompson RG. Differential distribution of norepinephrine and serotonin along the dorsal-ventral axis of the hippocampal formation. Brain research bulletin. 1980;5:771–773. doi: 10.1016/0361-9230(80)90220-8. [DOI] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Bio. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Girard C, Tinel N, Terrenoire C, Romey G, Lazdunski M, Borsotto M. p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. The EMBO journal. 2002;21:4439–4448. doi: 10.1093/emboj/cdf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome research. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson ND, Owens MJ, Nemeroff CB. Depression, antidepressants, and neurogenesis: a critical reappraisal. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2011;36:2589–2602. doi: 10.1038/npp.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Bac to the future: The use of bac transgenic mice for neuroscience research. Nature Reviews Neuroscience. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Henze DA, Buzsaki G. Hilar mossy cells: functional identification and activity in vivo. Progress in brain research. 2007;163:199–216. doi: 10.1016/S0079-6123(07)63012-X. [DOI] [PubMed] [Google Scholar]
- Houser CR. Interneurons of the dentate gyrus: an overview of cell types, terminal fields and neurochemical identity. Progress in brain research. 2007;163:217–232. doi: 10.1016/S0079-6123(07)63013-1. [DOI] [PubMed] [Google Scholar]
- Jost M, Gerke V. Mapping of a regulatory important site for protein kinase C phosphorylation in the N-terminal domain of annexin II. Biochimica et biophysica acta. 1996;1313:283–289. doi: 10.1016/0167-4889(96)00101-2. [DOI] [PubMed] [Google Scholar]
- Katz RJ. Animal model of depression: pharmacological sensitivity of a hedonic deficit. Pharmacology, biochemistry, and behavior. 1982;16:965–968. doi: 10.1016/0091-3057(82)90053-3. [DOI] [PubMed] [Google Scholar]
- Kube E, Becker T, Weber K, Gerke V. Protein-protein interaction studied by site-directed mutagenesis. Characterization of the annexin II-binding site on p11, a member of the S100 protein family. The Journal of biological chemistry. 1992;267:14175–14182. [PubMed] [Google Scholar]
- Liu J, Vishwanatha JK. Regulation of nucleo-cytoplasmic shuttling of human annexin A2: a proposed mechanism. Molecular and cellular biochemistry. 2007;303:211–220. doi: 10.1007/s11010-007-9477-7. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochimica et biophysica acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescher U, Gerke V. S100A10/p11: family, friends and functions. Pflugers Archiv : European journal of physiology. 2008;455:575–582. doi: 10.1007/s00424-007-0313-4. [DOI] [PubMed] [Google Scholar]
- Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, Russo-Marie F, Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- Rezvanpour A, Phillips JM, Shaw GS. Design of high-affinity S100-target hybrid proteins. Protein Sci. 2009;18:2528–2536. doi: 10.1002/pro.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Scharfman HE. Electrophysiological evidence that dentate hilar mossy cells are excitatory and innervate both granule cells and interneurons. Journal of neurophysiology. 1995;74:179–194. doi: 10.1152/jn.1995.74.1.179. [DOI] [PubMed] [Google Scholar]
- Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149:1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–U112. doi: 10.1038/nature10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surget A, Tanti A, Leonardo ED, Laugeray A, Rainer Q, Touma C, Palme R, Griebel G, Ibarguen-Vargas Y, Hen R, et al. Antidepressants recruit new neurons to improve stress response regulation. Molecular psychiatry. 2011;16:1177–1188. doi: 10.1038/mp.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, El Yacoubi M, Vaugeois JM, Nomikos GG, Greengard P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- Tozuka Y, Fukuda S, Namba T, Seki T, Hisatsune T. GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron. 2005;47:803–815. doi: 10.1016/j.neuron.2005.08.023. [DOI] [PubMed] [Google Scholar]
- van de Graaf SF, Hoenderop JG, Gkika D, Lamers D, Prenen J, Rescher U, Gerke V, Staub O, Nilius B, Bindels RJ. Functional expression of the epithelial Ca(2+) channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. The EMBO journal. 2003;22:1478–1487. doi: 10.1093/emboj/cdg162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, David DJ, Monckton JE, Battaglia F, Hen R. Chronic fluoxetine stimulates maturation and synaptic plasticity of adult-born hippocampal granule cells. J Neurosci. 2008;28:1374–1384. doi: 10.1523/JNEUROSCI.3632-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner-Schmidt JL, Chen EY, Zhang X, Marshall JJ, Morozov A, Svenningsson P, Greengard P. A role for p11 in the antidepressant action of brain-derived neurotrophic factor. Biological psychiatry. 2010;68:528–535. doi: 10.1016/j.biopsych.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nature reviews Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.