In 2006, the National Institutes of Health (NIH) created the Clinical and Translational Science Award (CTSA) program to “transform the local, regional and national environment for clinical and translational science, thereby increasing the efficiency and speed of clinical and translational research.”1 Since then, 61 CTSA‐funded sites have reengineered their local research enterprises to develop cadres of highly trained translational scientists and provide them with a robust array of research resources. Now under the direction of the National Center for Advancing Translational Sciences (NCATS), the maturing CTSA program is poised to enter its second stage of development to provide “institutions with greater flexibility and more opportunities to build on institutional strengths.”2 This approach is supported by a new CTSA Funding Opportunity Announcement (FOA) that will enable CTSA sites to develop programs to address the specific needs of their investigators and local communities. As a result, institutional CTSA programs will likely evolve from a model that provides a set of common research resources locally to unique programs with a mix of core infrastructure (e.g., training, informatics) that is supplemented by institution‐specific resources (e.g., drug discovery, comparative effectiveness research, and research implementation and dissemination cores).
Leveraging complementary institutional capabilities through closer collaborations between CTSA sites is the logical next step for the National CTSA Consortium. During the formative years of the CTSA program, several sites forged collaborations by creating regional consortia based on geographic proximity to enable sharing of local resources and meetings of trainees.3 For example, the Chicago Consortium for Community Engagement represents a collaboration among the three Chicago CTSAs to advance coordinated and synergistic approaches to developing community‐partnered research, disseminate research results, and provide training in community engagement methods to collaborators in academic and community‐based organizations. The Ohio Consortium of three CTSA sites implemented a statewide central institutional review board model. In Texas, several institutions meet to exchange best practices. However, collaborative efforts that are constrained by geographic location may limit the benefits that can be derived from linking scientists, institutions, resources, and innovative tools across an array of institutions with complementary capabilities. Accordingly, six geographically dispersed CTSA sites (Johns Hopkins University, University of Chicago, University of Pennsylvania, University of Pittsburgh, Washington University at St. Louis, Yale University) formed a “virtual CTSA consortium” based on a shared vision for data sharing and prior collaborations focused on informatics initiatives.
The initial goals of the six‐site Sharing Partnership for Innovative Research in Translation (SPIRiT) Consortium were (1) to develop a data sharing infrastructure for biospecimen‐based research and de‐identified clinical research data, and (2) to establish interinstitutional collaborations related to pilot studies, regulatory support, core laboratory facilities, and education and career development. Over the past 2 years, the SPIRiT Consortium embarked on several initiatives to achieve these goals, including the deployment of an informatics software and regulatory infrastructure to enable discovery and sharing of banked pathology specimens, development of an interinstitutional pilot study program to link investigators, and validation of a novel technology to improve the diagnosis of malignant melanoma. These programs are described below.
Enabling Tissue Specimen Discovery across Institutions
The Tissue Information Extraction System (TIES) is an open‐source end‐to‐end natural language processing system that was initially developed with support from the National Cancer Institute. TIES provides researchers with the ability to search information extracted from text‐based surgical pathology reports and to identify and access banked annotated tissue specimens across a network of federated sources.4 As its first initiative to share data, the SPIRiT Consortium is coordinating the implementation and use of TIES across three SPIRiT sites. This effort requires collaboration related to the TIES informatics platform and development of administrative and regulatory processes to implement an overall governance plan, common policies, trust documents, institutional review board protocols, materials transfer agreements, and security requirements. When this program is fully deployed over the next year, researchers will be able to directly access de‐identified pathology report data and acquire banked paraffin‐embedded and frozen tissues that are linked to pathology reports at participating SPIRiT sites. This resource will enable biomarker discovery and other translational research, including research related to rare diseases, that requires access to biospecimens.
Linking Investigators through a Pilot Study Funding Opportunity
In order to leverage complementary investigator expertise and accelerate collaborative research across our institutions, the SPIRiT Consortium issued a joint funding announcement for a pilot studies program. Research proposals addressing any aspect of clinical and translational science were required to represent a new collaboration between investigators from at least two SPIRiT sites, or a new project conducted by existing collaborators across sites. The lead scientist from each institution could request up to $25,000 of support from her/his institution. For example, a project proposed by a team of investigators from three SPIRiT sites could request a total of $75,000 in direct costs. Fifty‐six submitted applications underwent peer review and the six SPIRiT CTSA principal investigators collectively selected ten studies for funding.
To illustrate the impact of this program on cross‐institutional collaboration, we conducted social network analysis (SNA). SNA is used in sociology, psychology, organizational behavior, and more recently in health care research to identify interdependent relationships and patterns of relationships that promote or prevent performance.5, 6 After tabulating the number of pilot applications by institution, we examined two standard SNA measures to characterize structural patterns of collaboration: network density and centrality. Figure 1 is a sociogram of density and weighted point‐degree centrality that demonstrates that every SPIRiT site had investigators who collaborated with colleagues at other sites. Calculations of network density showed high levels of inter‐institutional interaction within the context of the 56 applications. Collaborative teams were formed in rich and varied ways between the six institutions, suggesting that the SPIRiT consortium is cohesive and capable of effective and efficient coordination.7 Point degree centrality refers to the concentration of collaborative activities between institutions.8 In our analysis, centrality was calculated by summing the number of dyadic connections of each institution and weighting those connections by the number of applications between the institutions. For the SPIRiT pilot studies program, centrality was highest for the University of Pennsylvania (43), which shared the most connections with other institutions on the submitted pilot grant applications. As shown in the sociogram, there was an overall high level of collaboration between all six institutions, with each having at least one shared pilot grant submission with every other consortium members.
Figure 1.
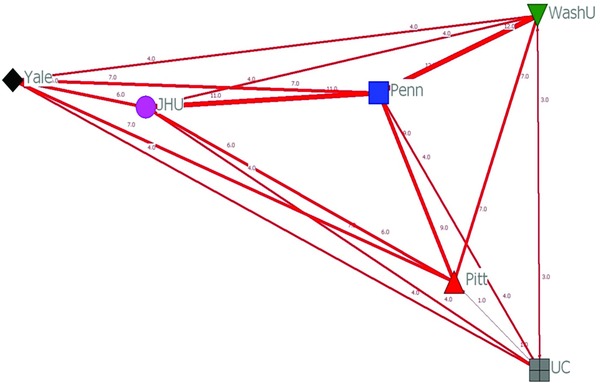
Sociogram of network density and weighted point‐degree centrality determined by social network analysis of collaborative pilot study applications submitted by investigators across SPIRiT sites. (JHU = Johns Hopkins University, Penn = University of Pennsylvania, Pitt = University of Pittsburgh, UC = University of Chicago, WashU = Washington University at St. Louis, Yale = Yale University).
Pooling Clinical Expertise to Validate a Novel Technology
Scientists at Carnegie Mellon University and Intel Research jointly created an open‐source software architecture, Diamond, for rapid scanning and filtering of large volumes of distributed loosely‐structured data.9, 10, 11 With support from the University of Pittsburgh CTSA, Diamond technology is being applied to medical images to provide clinicians with an interactive mechanism for comparing clinically acquired images (e.g., pathology slides) with large sets of archived images that are annotated with diagnoses. For example, a pathologist can view a whole slide image of a biopsied pigmented skin lesion, identify regions of interest with suspected pagetoid spread or cytologic atypia, and perform a Diamond search of a reference library of annotated whole slide images of biopsied skin lesions (Figure 2). The results of this Diamond search would provide images of known malignant or benign lesions that most closely resemble the clinical sample and can be used to guide the pathologist's diagnosis.
Figure 2.
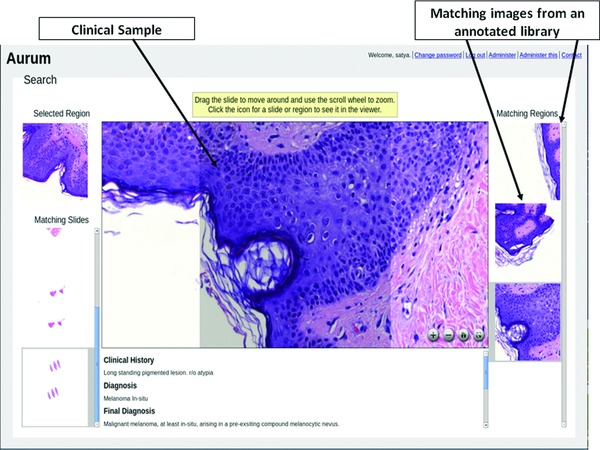
Screenshot of results from a Diamond search illustrating a whole slide image of a biopsied pigmented skin lesion and images of known malignant or benign lesions from an annotated library that most closely resemble the clinical sample.
The translational goal of this project is to validate and implement Diamond as a tool that can be incorporated into the workflow of general pathologists to improve diagnostic accuracy for melanoma on whole slide biopsy images. However, the incremental predictive value of Diamond over standard pathology methods to diagnose melanoma is not known. Through a CTSA supplement grant, Diamond's clinical utility is being tested by general pathologists at three SPIRiT sites (University of Pennsylvania, University of Pittsburgh, Yale University) who have been randomized to use of Diamond as a diagnostic aid versus standard diagnosis of the same slides. Gold standard diagnoses of these test slides were made by consensus of dermatopathologists from each of the participating SPIRiT sites. We anticipate that validation of Diamond as a diagnostic aid will lead to a subsequent implementation study in regions surrounding SPIRiT sites.
Future Directions
These three examples of interinstitutional collaboration represent the SPIRiT Consortium's early initiatives to leverage infrastructure (e.g., informatics programs, banked biospecimens), link investigators, and capitalize on clinical expertise across six CTSA sites. Future planned initiatives include the development of interinstitutional mechanisms to review pilot funding applications, educational initiatives and trainee tracking, sharing models for community engagement, broadening access to translational core labs, and provision of regulatory support (e.g., a SPIRiT institutional data and safety monitoring board program). The founding of the SPIRiT Consortium on the basis of a common priority for data sharing, as opposed to geographic proximity, can serve as a paradigm for the development of other consortia of CTSA sites with common areas of focus.
Acknowledgments
Supported by NIH grants UL1 TR000003, UL1 TR000005, UL1 TR000142, UL1 TR000430, UL1 TR000448, UL1 RR025005.
References
- 1. Institutional Clinical and Translational Science Award RFA‐RM‐06‐002 . Available at: http://grants.nih.gov/grants/guide/rfa‐files/RFA‐RM‐06‐002.html#SectionI. Accessed November 7, 2012.
- 2. Institutional Clinical and Translational Science Award (U54) RFA‐TR‐12‐006 . Available at: http://grants.nih.gov/grants/guide/rfa‐files/RFA‐TR‐12‐006.html. Accessed November 7, 2012.
- 3.Clinical and Translational Science Awards. Regional consortia. Available at: https://www.ctsacentral.org/regional‐consortia. Accessed November 7, 2012.
- 4. Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc. 2010; 17: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wasserman S, Faust K. Social Network Analysis: Methods and Applications. New York: Cambridge University Press; 1994. [Google Scholar]
- 6. Meltzer D, Chung J, Khalili P, Meltzer D, Chung J, Khalili P, Marlow E, Arora V, Schumock G, Burt R. Exploring the use of social network methods in designing healthcare quality improvement teams. Soc Sci Med. 2010; 71: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cross R, Parker A. The Hidden Power of Social Networks: Understanding How Work Really Gets Done in Organizations. Boston, MA: Harvard Business School Publishing Corporation; 2004. [Google Scholar]
- 8. Knoke D, Yang S. Social Network Analysis. 2nd ed Thousand Oaks, CA: Sage Publications; 2008. [Google Scholar]
- 9.What is Diamond? Available at: http://diamond.cs.cmu.edu/whatisdiamond.html Accessed November 7, 2012.
- 10. Huston L, Sukthankar R, Wickremesinghe R, Satyanarayanan M, Ganger GR, Riedel E, Ailamaki A. Diamond: a storage architecture for early discard in interactive search Paper presented at: 3rd USENIX Conference on File Storage Technologies 2004; San Francisco, CA. [Google Scholar]
- 11. Park SC, Sukthankar R, Mummert L, Satyanarayanan M, Zheng B. Optimization of reference library used in content‐based medical image retrieval scheme. Med Phys. 2007; 34: 4331–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]


