Abstract
Circadian clocks are found in a wide variety of organisms from cyanobacteria to mammals. Many believe that the circadian clock system evolved as an adaption to the daily cycles in light and temperature driven by the rotation of the earth. Studies on the cyanobacterium, Synechococcus elongatus PCC 7942, have confirmed that the circadian clock in resonance with environmental cycles confers an adaptive advantage to cyanobacterial strains with different clock properties when grown in competition under light-dark cycles. The results thus far suggest that in a cyclic environment, the cyanobacterial strains whose free running periods are closest to the environmental period are the most fit and the strains lacking a functional circadian clock are at a competitive disadvantage relative to strains with a functional clock. In contrast, the circadian system provides little or no advantage to cyanobacteria grown in competition in constant light.
To explain the potential mechanism of this clock-mediated enhancement in fitness in cyanobacteria, several models have been proposed; these include the limiting resource model, the diffusible inhibitor model and the cell-to-cell communication model. None of these models have been excluded by the currently available experimental data and the mechanistic basis of clock-mediated fitness enhancement remains elusive.
Background
Circadian clocks are endogenous timing mechanisms that function to regulate a variety of cellular, metabolic and behavioral activities over the course of the day-night cycle. Circadian systems allow organisms to anticipate daily changes in environmental signals such as light and temperature. Regulated by circadian clocks, organisms sustain roughly 24-hour rhythms even in the absence of environmental timing cues, and these clock-driven rhythms sustain stable free-running periods (FRPs) within the physiologically tolerable temperature range [1, 2].
Circadian clocks have been found in a broad range of organisms from cyanobacteria to mammals. Given their ubiquity, circadian clocks are considered to be an evolutionary adaptation that enhances the fitness of organisms possessing them [3]. For instance, chipmunks with disrupted circadian clocks were more susceptible to predation in the wild than those with intact circadian systems. Ecological observations suggested that the nighttime restlessness of the arhythmic chipmunks resulted in elevated detection rates by predators [4]. Studies of Drosophila melanogaster showed that the life span of flies with altered circadian periods was significantly reduced by up to 15% [5], and that the disruption of the circadian clock also reduced sperm production in males [6]. Furthermore, Arabidopsis strains lacking a circadian clock showed lower viability, less carbon fixation and slower photosynthesis rates than wild-type strains [7, 8]. Moreover, Arabidopsis is more resistant to herbivory when plants were entrained in the same phase as the herbivores, indicating that the circadian system in Arabidopsis assists in defending against herbivory [9].
Although these studies demonstrate that circadian regulation of cellular, metabolic and behavioral events is beneficial, few studies have rigorously tested the adaptive value of circadian clocks in terms of their contribution to fitness and adaptation. Fitness primarily describes reproductive success [10], whereas longevity, growth and development are secondary factors affecting the fitness of an organism. An adaptation is an acquired feature as a result of natural selection that enhances the fitness of an organism under certain selective pressures [10]. An adaptation can only be presumed to be adaptive when it first emerges [3]. In the process of evolution, the adaptation may retain an “extrinsic” value only if the selective pressure remains. Alternatively, the adaptation may acquire an “intrinsic” value by becoming integrated with other processes. In this case, even if the original adaptation persists in the absence of the selective pressures, it is no longer considered to be an adaptation [10]. In order to fully test the adaptive value of circadian clocks, two questions must be addressed. Does the presence of a circadian clock (i) enhance the fitness and (ii) if so, is the adaptive value conferred by the circadian clock intrinsic or extrinsic? [3] To date, most studies have only partially or indirectly addressed these questions. Furthermore, little if any research has addressed the potential mechanisms by which circadian clocks mediate fitness enhancement.
The cyanobacterium, Synechococcus elongatus PCC 7942 (S.elongatus) is an ideal model system to address these questions for several reasons [3, 11, 12]. First, the central clock mechanism is relatively simple; the core circadian clock in S. elongatus is composed of three proteins (KaiA, KaiB, and KaiC) that are encoded by the genes, kaiA, kaiB and kaiC [13] and a number of clock mutants have been generated [14]. Among these mutants are those with short and long periods as well as arhythmic mutants. These mutant strains allow us to test the adaptive value of the cyanobacterial circadian clock by directly comparing them to the wild-type strain under different growth conditions. Second, S.elongatus reproduces asexually by binary fission and therefore growth rates are a direct measurement of fitness [15]. Third, the growth conditions of S. elongatus are relatively simple and therefore laboratory conditions can approximate the relevant features of natural conditions. Fourth, S.elongatus can grow in either constant light or in light/dark cycles, thus the extrinsic versus intrinsic adaptive value can be determined by artificially introducing or removing selective pressures [3]. Finally, S.elongatus represents one of the most evolutionarily ancient organisms possessing a circadian system; therefore, elucidating the mechanisms of clock-mediated adaptation in this species could help us understand the selective pressures that may have led to the evolution of circadian clocks.
In this chapter, the previous work that has been done to test the adaptive value of the circadian clock in S. elongatus will be described and the possible mechanisms that might explain how the cyanobacterial circadian system exerts its influence on overall fitness will be discussed. The models that have been considered to explain these mechanisms are “the limiting resource model”, the “diffusible inhibitor model” and the “cell-to-cell communication model.” [3]
Testing the Adaptive Value of the Circadian Clock in Cyanobacteria
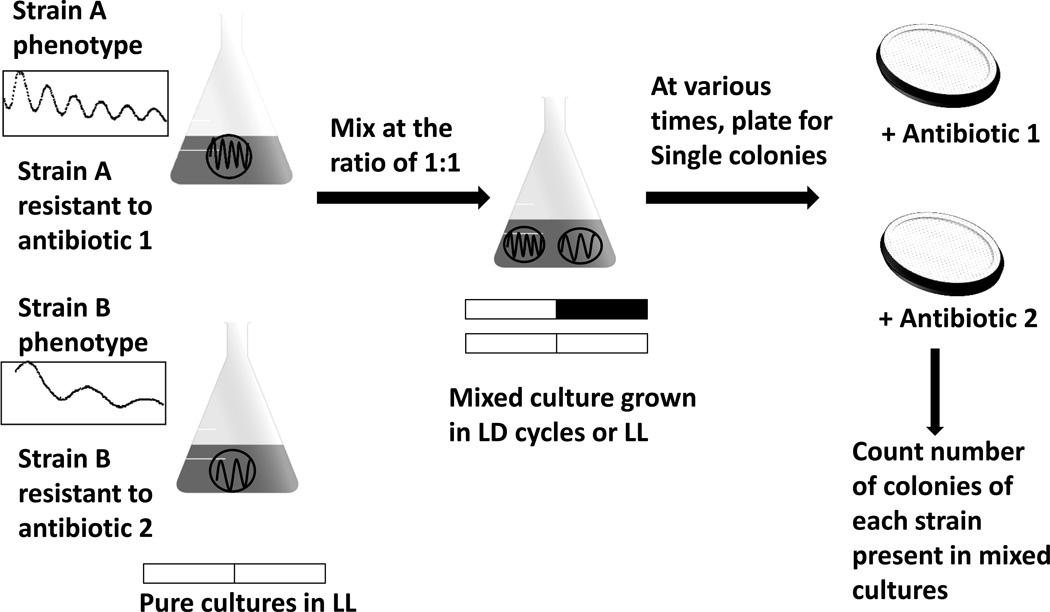
The adaptive value of circadian clocks in cyanobacteria was tested by using growth in competition between the wild-type S.elongatus and several different clock mutant strains [3, 11, 12]. These clock mutants are due to point mutations in the kaiA, kaiB or kaiC genes respectively, resulting in altered clock properties such as arhythmicity or rhythmicity that exhibits FRPs that are longer or shorter than 24 hours [14]. In pure cultures, neither these mutant strains nor the wild-type strain have growth rates that are significantly different from each other in constant light (LL) or in light/dark (LD) cycles [11, 12]. This observation does not exclude the possibility that the circadian clock system enhances fitness in cyanobacteria; however the adaptive value may only be detectable under some selective circumstances such as competition. For this reason, competition experiments were designed to assess the reproductive fitness of the wild-type strain (WT) and the clock mutant strains under controlled environmental conditions (Fig.1) [3, 11, 12]. In these experiments, two cyanobacterial strains with different clock phenotypes were mixed and grown together in either constant light or in light/dark cycles and the composition of these mixed cultures was assayed over time as a test of reproductive fitness.
Figure 1. Competition experiment between S.elongatus strains with different clock phenotypes [3, 11, 12].
The clock phenotypes of two strains, A and B, are shown as luminescence rhythms that report the promoter activity of psbAI. Strains A and B are resistant to different antibiotics such that their fractions in mixed cultures can be tracked by plating on selective media. Pure cultures of A and B were set up under LL, and when they reached log phase, equal numbers of A and B cells were mixed and cultured under different LD cycles or LL condition. Aliquots were taken from the mixed cultures every ~8 generations in LD and every ~16 generations in LL, and they were plated on selective media to count the number of colony-forming units (CFU) of each strain. Meanwhile, the mixed culture was diluted into fresh medium and grown for another ~8 generations (LD) and ~16 generations (LL). This process was repeated for 4 cycles to allow cells to grow for 40-50 generations. The fraction of each strain in the mixed culture was calculated by the number of colonies of each strain growing on selective media. Circadian phenotypes were confirmed by monitoring the luminescence rhythms of colonies of each strain at different sampling times. Figure modified from [3].
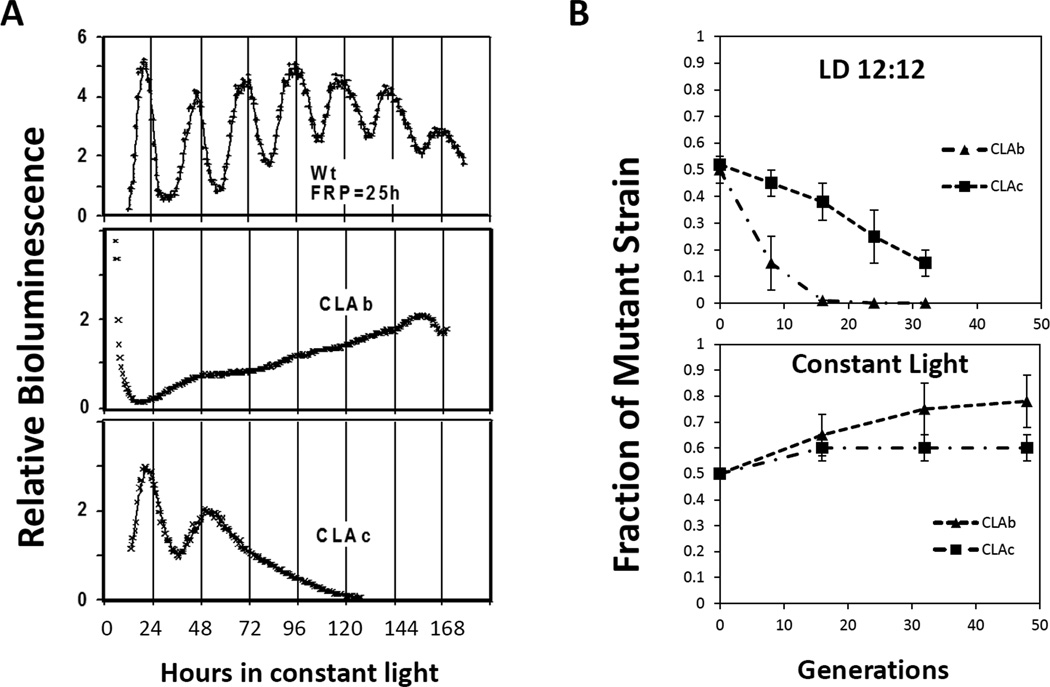
For example, to test whether the circadian clock enhances reproductive fitness in cyanobacteria, competition experiments were conducted between the WT strain with a FRP of approximately 24–25 hours and an arhythmic mutant (CLAb) whose circadian clock was disrupted by a point mutation (G460E) in the kaiC gene (Fig.2A) [11, 13, 14]. In LD 12:12 (12 hours of light followed by 12 hours of darkness) cycles, the WT strain quickly (within 20 generations) became the predominant strain in mixed cultures (Fig.2B). As a control, the point mutation in CLAb was rescued by introducing a wild-type copy of the kaiC gene into the genome. When the rescued CLAb strain was grown in competition with the WT strain, the proportions of the WT and mutant strain remained approximately equal in the mixed cultures over many generations indicating that the reduction in fitness of CLAb was due to altered clock properties rather than some other difference in the genetic background of the two strains in competition [11]. This experiment confirmed that the circadian clock in cyanobacteria confers adaptive value in light/dark cycles, but it does not reveal whether this adaptive value is an intrinsic or extrinsic property of the clock. If the value is an intrinsic property of the clock, one would expect that the WT strain would also defeat CLAb when grown in mixed cultures in constant conditions as well as when grown together in light/dark cycles. To address this question, the WT and arhythmic strains were co-cultured and grown in constant light condition removing the presumed selective pressure of the day-night cycles. Surprisingly (at least, to some chronobiologists who favor the intrinsic adaptiveness of circadian timekeeping!), the CLAb strain was not only able to successfully maintain itself in mixed cultures with WT, but the proportion of CLAb significantly increased in these mixed population (p-value=0.01; Fig.2B). Additionally, competition experiments using the WT strain and a second kaiC mutant CLAc (T495A; which expresses a rapidly damped circadian oscillation [13, 14]) yielded similar results. The WT strain once again became the predominant strain in mixed cultures in LD cycles, but when grown in constant light both strains maintained approximately equal proportions over many generations. Interestingly, the CLAc strain was able to remain as a small fraction of the mixed-culture even after 30 generations in light/dark cycles, while the fraction of the arhythmic CLAb mutant decreased rapidly within 20 generations (Fig.2B). Because CLAc is able to oscillate for one or two cycles in constant conditions (Fig.2A), the discrepancy in the competition kinetics may be due to the difference in their clock phenotypes, supporting the idea that even limited rhythmicity is of benefit to cyanobacteria in LD cycles. Based on these observations, it seems that the adaptive value of the circadian clock is of extrinsic value to S. elongatus cells rather than intrinsic, and additionally, the data in constant light conditions implies that having a functional clock may not always be adaptive under non-selective conditions (e.g., constant light condition).
Figure 2. Competition of the WT strain with arhythmic strains [11].
A, phenotypes of three strains used in the competition experiments. The WT strain (top) shows circadian rhythms with a ~25 h FRP. CLAb (middle), a clock-disrupted kaiC mutant, is arhythmic. Another kaiC mutant, CLAc (bottom), is also ultimately arhythmic but initially shows a rapidly damped oscillation. B, competitions between the WT strain and arhythmic mutants under LD 12:12 (upper) or LL conditions (lower). Data are plotted as the fraction of the mutant strain in mixed cultures (ordinate) versus the estimated number of generations (abscissa). Figure modified from [3, 11].
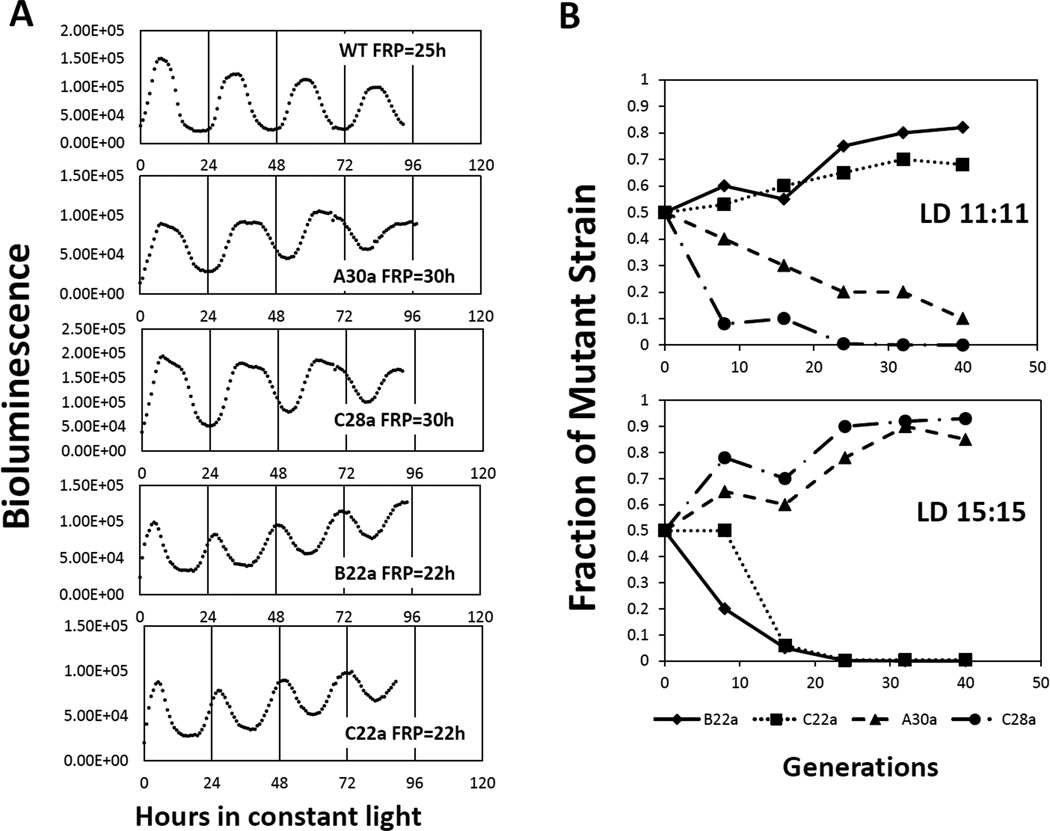
We wanted to determine if a circadian clock that is optimally resonating with the environmental cycle is of more value than a clock that is functional, but is entrained to the environmental cycle in a non-optimal phase relationship. The reproductive fitness for this scenario was tested by competition experiments between the WT strain and several mutants with altered FRPs [12]. In one set of competition experiments, either a kaiB (B22a; R74W) or a kaiC mutant (C22a; A87V), both with a FRP of approximately 22 hours [13, 14], was grown in mixed cultures with the WT strain. In a second set of competition experiments, either a kaiA (A30a; R249H) or a different kaiC mutant (C28a; P236S), both with a FRP of 28–30 hours [13, 14], was grown in mixed culture with the WT strain (Fig.3). Neither of these mutants in pure cultures shows a significant difference in growth rate as compared to the WT strain either in constant light or in light/dark cycles [11, 12]. When the short period mutants, B22a or C22a, were grown in mixed cultures with the WT strain in LD 11:11 cycles (11 hours of light followed by 11 hours of darkness), the short period mutant (either B22a or C22a) out-competed the WT strain (Fig.3B, top panel). Similarly, both of the long period mutants, A30a and C28a were able to defeat the WT strain when these strains were co-cultured in LD 15:15 cycles (15 hours of light followed by 15 hours of darkness) (Fig. 3B, bottom panel). Conversely, the WT strain was the predominant strain in mixed cultures with short period mutants when grown in LD15:15 (Fig.3B, bottom panel) or when grown in mixed cultures with long period mutants in LD11:11 cycles (Fig. 3B, upper panel). Our analyses suggested that all of these mutants entrained to the LD cycles, but they entrained with different phase relationships relative to WT that were based on the difference between their FRP and the period of the LD cycle [12]. It appears to be clear from these results that the cyanobacterial strain whose circadian clock was optimally entrained to the environmental cycle was more fit than the strains whose clock was entrained to the LD cycles in non-optimal phase relationships.
Figure 3. Competition of the WT strain with period-altered mutants under LD 11:11 and LD 15:15 cycles [12].
A, circadian phenotypes of the WT strain and period-altered mutants used in these competition experiments. The short period mutants (FRP ~ 22 h) include the kaiB mutant B22a and the kaiC mutant C22a, and the long period mutants (FRP ~ 30 h) include the kaiA mutant A30a and the kaiC mutant C28a. All strains have a luciferase construct that reports the clock-regulated promoter activity of the psbA1 gene by time-dependent luminescence intensity. B, competitions between the WT strain and the period-altered mutants under LD 11:11 cycles (upper) or LD 15:15 cycles (lower). Data are plotted as the fraction of the mutant strain in the mixed culture versus the estimated number of generations. Symbols for each strain are identified under the abscissa. Figure modified from [3, 12].
Furthermore, this fitness advantage is not dependent on which clock gene is mutated, indicating that the difference in reproductive fitness is due to the clock phenotype itself, rather than to a mutation in a particular clock gene. One of the most persuasive features of these competition results is that mutants are able to outcompete WT when the period of the LD cycle dovetailed better with the mutants’ FRP than with WT’s FRP. When the period mutants were competed against the WT strain in constant light, the proportions of the WT and clock mutant in the mixed culture remained relatively constant, providing additional evidence that the adaptive value of the circadian clock is extrinsic rather than intrinsic. In many other cases of tests of adaptive significance, mutant strains are uniformly out-competed by WT strains. But these studies of the cyanobacterial clock provide an example where mutants can out-compete the WT strain if their particular properties (e.g., FRP) resonance optimally with an imposed environmental condition (e.g., the period of the environmental light/dark cycle).
Taken together, the competition experiments between cyanobacteria with a normally functioning circadian clock and strains carrying mutations in clock genes have demonstrated (i) that the circadian clock enhances the reproductive fitness of cyanobacteria in cyclic environments but not in non-cyclic environments, and (ii) that this enhancement is the greatest when the period of the internal clock optimally matches the period of the external cycle [11, 12].
Potential Mechanisms of Clock-mediated Fitness Enhancement
While competition experiments have clearly demonstrated a clock-mediated fitness enhancement in cyanobacteria, the cellular mechanism remains unknown. Cyanobacterial strains with different clock properties all showed a similar growth rate when cultured alone; however, the reproductive fitness was negatively affected in mixed cultures in a way that is dependent on the light/dark cycles [11, 12]. To explain these observations, three models have been proposed: the “limiting resource model,” the “diffusible inhibitor model” and the “cell-to-cell communication model” [3].
The Limiting Resource Model
The limiting resource model proposes that the circadian system enables individual cyanobacterial cells to optimally utilize some limiting environmental resource by phasing their metabolism to the environmental cycle [3]. For instance, transcription of genes that encode components of the photosynthetic machinery in cyanobacteria is up-regulated during the day and down-regulated at night [16], and this rhythmic gene expression may facilitate cyanobacterial cells to perform photosynthesis more efficiently and to consume less energy at night by limiting unnecessary transcription and translation. In contrast, cyanobacterial cells without a functioning circadian clock or with a non-optimally entrained clock may be less efficient metabolically, thereby having an inherent disadvantage when competing for a limited resource with cells that have a clock that is favorably entrained to the environmental cycle.
Our published results that the growth rates of the various pure cultures (WT and mutants) were experimentally indistinguishable led us to believe that the Limiting Resource Model was incorrect [11, 12], but a more recent modeling paper from Hellweger has forced us to re-evaluate the experimental evidence for and against this model [17] (see below).
To show that the limiting resource model could be hypothetically true, Hellweger’s mathematical modeling of the competition experiments attributed small differences in the growth rates between the cyanobacterial strains as a key determining factor in predicting the outcome of the competition [17]. This model simulated the growth of the WT strain and period-altered mutants in both pure and mixed cultures and was able to successfully reproduce the experimental outcomes [17]. After examining parameters in the simulation, it was found that a circadian clock will have a higher amplitude when its FRP matches the period of the LD cycle, and this higher amplitude leads to greater expression of photosynthesis genes and ultimately to higher growth rates. The difference in predicted gene expression and growth rates was therefore suggested by this model to be the mechanism of the clock-mediated fitness enhancement. Although our experimental measurements of overall growth rates had detected no differences in pure cultures of the strains used in competition [11, 12], Hellweger’s model suggested that small differences may exist between the strains which are difficult to detect in the batch cultures used for the competition experiments; however, these hypothetically small differences in growth rates between the strains might be detectable when cyanobacteria are cultured using chemostats [17]. Using a chemostat culturing method, cells can be grown in a physiological steady state where they grow at a constant rate, and all culture parameters remain constant [18]. Previously, competition experiments between the WT strain and the long period mutant, C28a, conducted in chemostats produced results that are quite similar to the competition results using the batch culture method [12], but a re-evaluation of this approach is warranted based upon the modeling study of Hellweger [17].
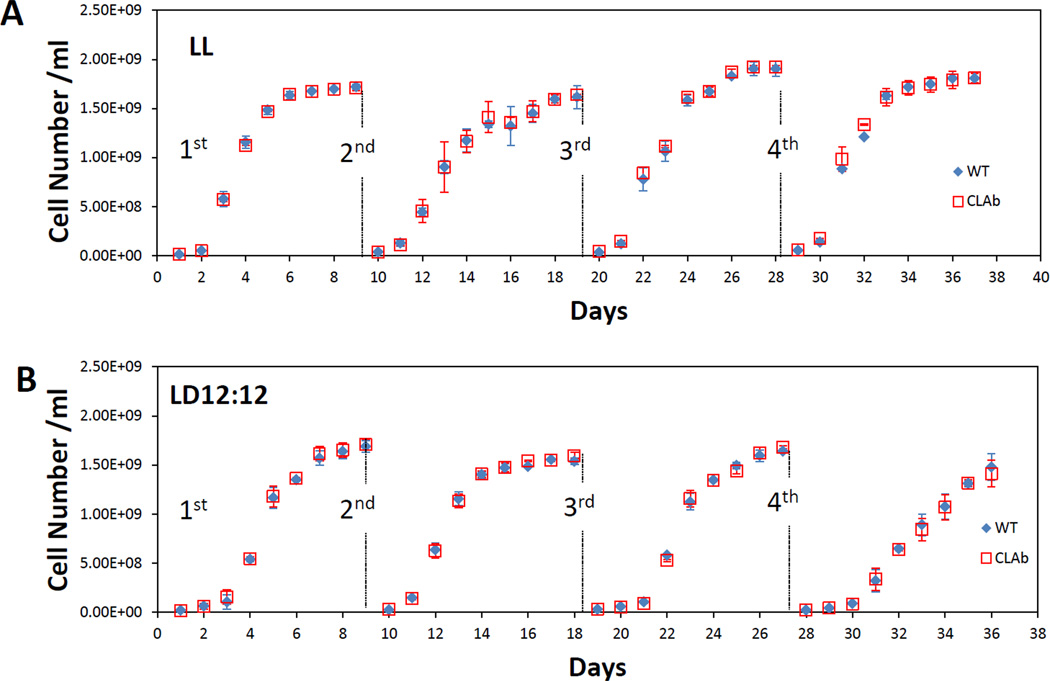
Another way to reconcile the Limiting Resource Model with our experimental data is to consider the impact of transfers/dilutions in the competition assay. In our previous studies, we did not detect any significant differences in the growth rates between strains with different circadian phenotypes during exponential growth [11, 12]. We therefore considered the possibility that there could be small differences in the latency of these strains to start growing at the time of each transfer to new culture medium. If so, the initial growth rates might be different among the strains and these differences might accumulate over successive transfers. Therefore, to test the Limiting Resource Model experimentally, we focused on testing a hypothesis that small differences in initial growth rates may be cumulative over repeated dilutions of cell cultures and therefore are responsible for the result of the competition studies (Dr. Tetsuya Mori, personal communication). By analogy to a race, it is as if one runner gets started “off the blocks” from the starting line a little earlier than the other runners. If one strain has the innate ability to adapt to the new medium more quickly than the other such that it enters the exponential phase of growth in a shorter period of time after each transfer to new medium, small differences in this period of adaptation between strains with different circadian phenotypes could give rise to large differences in the composition of the mixed cultures after several generations [3]. To test this hypothesis, we examined whether there were small differences in initial growth rates over several serial dilutions/transfers of pure cultures of the WT strain and of the arhythmic mutant, CLAb. In previous studies, the WT strain and clock mutants did not show any significant difference in their overall growth rates in pure cultures [11, 12], however, the difference might have been too subtle to be observed with the sampling techniques that were used. If there were small differences in the ability of cells to adapt to new medium, we would expect that this difference could be enhanced by a series of dilutions to new medium over many generations. To experimentally test this version of the Limiting Resource Model, stationary phase cultures of the WT strain and CLAb cells were diluted one thousand fold every 8 days, and the growth in constant light or in a light/dark cycle of each of the two strains was monitored by measuring the optical density. As shown in Fig.4, during a series of four dilutions, no obvious differences were observed by eye in the growth curves of the WT and arhythmic strains in pure cultures in either constant light or in LD 12:12 cycles. However, some small differences were detected when the initial growth rates were calculated (Table 1; the initial growth rates were calculated as the doubling time in the first 24 hours after dilution). In constant light, even though the initial doubling time of CLAb was significantly greater (i.e., slower) than that of the WT strain after the second dilution, no significant difference was observed after the third dilution, and after the fourth dilution, the direction of the difference was the opposite to that in the second dilution. Therefore, the end result was no difference. In LD 12:12 cycles, the CLAb strain grew significantly faster than the WT strain in the lag phase after the first transfer to fresh medium, whereas in the subsequent dilutions, CLAb showed slower initial growth rates than the WT strain (and the difference was not significant after the fourth dilution). Although these results indicate that the WT strain and the CLAb strain may have different abilities to adapt to new medium, the different initial growth rates in pure cultures were not amplified in a straightforward way by subsequent dilutions (Fig.4). Therefore, whether the outcome of these competition experiments is caused by small differences in the initial growth rates between strains remains unclear, and indicates that a better experimental design is needed to more rigorously test this model. Previous work utilizing chemostats to culture cyanbacterial cells in competition yielded results that were substantially the same as those obtained when competition is conducted in batch cultures [12]. This culture method may allow us to more accurately determine whether there are differences between cyanobacterial strains in their ability to adapt to the introduction of new medium.
Figure 4. Growth curves of the WT strain and the clock mutant (CLAb) in pure cultures that were serially diluted four times.
A, pure cultures of the WT strain (blue diamond) and CLAb (red squares) were set up under LL conditions. B, pure cultures of the WT strain and CLAb were set up under LD conditions. After the cells reached the stationary phase, they were diluted 1:1000 into fresh BG-11 medium. When the diluted cultures reached the stationary phase, they were diluted again. The cultures were serially diluted four times. Cell density was measured as OD750 value.
Table 1.
| Initial doubling time (DT) in constant light conditions | ||||
|---|---|---|---|---|
| Dilutions | 1st | 2nd | 3rd | 4th |
| WT DT (h) | 12.61 (1.24)* | 13.89 (0.28)* | 13.74 (2.07)* | 19.05 (2.06)* |
| CLAb DT (h) | 14.4 0(1.77)* | 16.38 (0.25)* | 13.90 (0.18)* | 15.72 (0.12)* |
| p-value** | 0.15 | 0.01*** | 0.88 | 0.02*** |
| Initial doubling time (DT) in LD 12:12 cycles | ||||
| Dilutions | 1st | 2nd | 3rd | 4th |
| WT DT (h) | 15.19 (1.12)* | 9.35 (1.14)* | 25.22 (4.95)* | 27.45 (6.49)* |
| CLAb DT (h) | 10.85 (1.58)* | 10.54 (0.33)* | 38.43 (1.86)* | 33.06 (3.64)* |
| p-value** | 0.01*** | 0.09 | 0.01*** | 0.18 |
Numbers in ( ) represent standard deviation. n=4
T-test, n=4
P<0.05, significantly different
The Limiting Resource Model is supported by mathematical modeling that predicts there are differences in the physiological states between WT and clock mutants when grown in pure cultures versus mixed cultures. However, the clock mutants used in the competition experiments have not yet been found to differ significantly from the WT strain in physiological properties that have been measured experimentally [3]. Further research is needed to address these questions.
The Diffusible Inhibitor Model
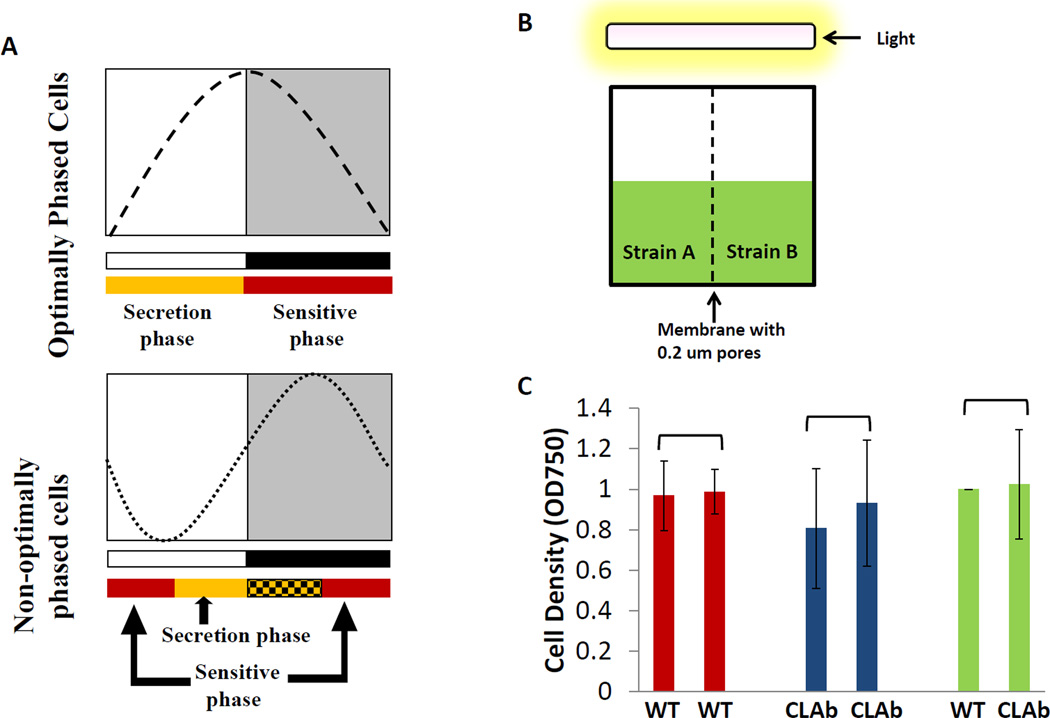
The Diffusible Inhibitor Model proposes that cyanobacterial cells rhythmically secrete a diffusible molecule that acts to inhibit the growth of other cells (i.e., cells of the same strain/species or cells of another strain/species) in the same environment [3]. If cyanobacteria are sensitive to their own inhibitory molecule, it would be advantageous for them to possess some mechanism to be insensitive to or to inactivate this inhibitor. For example, cyanobacterial cells could avoid inhibition by regulating both the timing of secretion of the inhibitor and their own sensitivity to the inhibitor. This model has two underlying assumptions: 1) the secretion of the inhibitor is light-dependent and is therefore limited to the light phase and the subjective day in constant conditions (clock and light dependent); 2) cyanobacterial cells are only sensitive to the inhibitor during the dark phase and in the subjective night (clock dependent) as depicted in Fig. 5A [3]. This hypothetical phenomenon could allow the cyanobacterial cells to produce an inhibitor that retards the growth of competitors (inter- or intra-species competitors) without inhibiting themselves. The subjective day of the cyanobacterial strain whose clock is optimally entrained to a cycling environment will coincide with the light phase, but strains that are entrained to a non-optimal phase relationship would have their subjective day in a different temporal regime (e.g., one hypothetical non-optimal phase relationship could be the subjective day starting in the middle of the light phase and ending in the middle of dark phase). In this scenario, the secretion of the inhibitor from these poorly entrained cells occurs during only part of the light phase and the secretion phase that overlaps with the dark phase does not result in active secretion (because the secretion is light-dependent, see underlying assumption # 1 above). However, the sensitive phase spans part of the subjective night starting in the middle of the night and ending during the middle of the day (Fig.5A) [3]. Using this line of reasoning, in a mixed culture of an optimally entrained strain with a non-optimally entrained strain, one would expect the growth of the poorly entrained cells to be inhibited because their sensitive phase overlaps with the secretion phase of the optimally entrained strain. Therefore, the growth of the poorly entrained cells would be inhibited by the secretions from the optimally entrained cells (but not vice versa). Similarly, if an arhythmic strain is grown in mixed culture with the WT cells in a LD 12:12 cycle, arhythmic cells might be sensitive to the inhibitor all the time due to the lack of a functioning clock. There have been reports suggesting that cyanobacteria secrete secondary metabolites that are toxic to other species [19, 20] (and even to themselves). For example, a secreted secondary metabolite of the cyanobacterium Scytonema hofmanni, cyanobacterin, inhibits the growth of several other cyanobacterial species [19]. A proteomic study demonstrates that some proteins in S. elongatus are secreted into medium, but the function of these secreted proteins remains unknown [21]. Therefore, it is possible that the competition between the WT and clock mutants could be mediated by secreted metabolites or proteins.
Figure 5. Test of the Diffusible Inhibitor Model.
A, Depiction of the Diffusible Inhibitor Model. Entrained phases of two strains are modeled in terms of their entrained phase relationship to an LD 12:12 cycle. For the optimally-phased cells, the subjective day overlaps with the daytime (white box), and the subjective night phase overlaps with the nighttime (black box). Therefore, their secretion phase (yellow box) coincides with the daytime, and the sensitive phase (red box) coincides with the nighttime. For non-optimally-phased cells, their subjective day starts from middle of the daytime, and their subjective night starts from the middle of the night and ends in the middle of the daytime. Therefore, the secretion phase (yellow box) of the non-optimal-phased cells is only from the middle of the day to the end of the day, while the secretion-competent phase that overlaps with the dark phase (yellow-black box) does not result in secretion because the secretion is postulated to be light-dependent. The sensitive phase (red box) of the non-optimally-phased cells starts in the middle of the night and ends in the middle of the day. B, the semi-co-culture apparatus used to test the existence of a diffusible inhibitor. Two chambers (left and right) were separated by a membrane with 0.2 µm pores. Cells of different strains could be cultured separately in these two chambers, but their media passes freely through this membrane such that the putative inhibitor could diffuse to the other side. The cultures were illuminated by white fluorescent light from the top, and the light intensity was 50 uE*m−2*s−1. C, the WT strain and CLAb were semi-co-cultured in this apparatus under LD 12:12 cycles. Cell densities (OD750) were measured on the fifth day. Bars which share the same color represent cultures in the same apparatus. Panel A modified from [3, 12].
In contrast to Hellweger’s mathematical modeling that best supports the Limiting Resource Model, an earlier modeling of the competition experiments provided support for the Diffusible Inhibitor Model [22, 23]. By postulating the existence of a diffusible inhibitor, Roussel et al. successfully reproduced all the experimental observations, whereas the theoretical model based on differences in resource exploitation did not [22]. Gonze et al. proposed and tested a more sophisticated model that supported the Diffusible Inhibitor Model and predicted that the outcome of the competition depends on the initial proportions of cells and on the FRPs of the different cyanobacterial strains [23]. However, this prediction was not supported by experimental testing–in contrast to the model predictions, competition experiments between the WT and CLAb strains found that CLAb was defeated by the WT strain even though the starting proportion of CLAb was as high as 90% of the whole culture [11].
As a different strategy to test the Diffusible Inhibitor Model experimentally, we designed a growth chamber in which two cyanobacterial strains are separated by a permeable membrane that allows the exchange of medium and small molecules, but prevents the two strains from directly contacting each other (Fig.5B). Thus, if a diffusible inhibitor is secreted by one strain, it could pass through the membrane and affect the growth of the strain on the other side of the membrane. As discussed earlier, the WT strain rapidly becomes the predominant strain in mixed cultures with the arhythmic strain CLAb in LD 12:12 cycles [11]; therefore, we conducted a series of experiments by using these two strains separated by a membrane. The pore size of the membrane was 0.2 µm that allows most molecules and small proteins to pass through but not the cyanobacterial cells themselves. If the Diffusible Inhibitor Model is correct, we would expect that the arhythmic strain would show a significantly slower growth rate; however, CLAb displayed the same growth rate as WT in LD 12:12 cycles (Fig.5C). One possible explanation for this result is that the putative diffusing molecule is too large to pass through the pores of the membrane; therefore, membranes with varying pore sizes could be tested (a pore size of 0.2 µm was used in the experiments whose results are depicted in Fig. 5C). Alternatively, the inhibitor could be a cell-surface molecule rather than a diffusible factor. Contact-dependent inhibition (CDI) has been reported in E.coli [24, 25] and interestingly, a few potential homologs of genes involved in CDI in E.coli have been identified in S.elongatus. Knock-out mutants of these potential homologs could be constructed in order to address whether a contact-dependent inhibition mechanism is involved in the competitions between cyanobacterial strains.
The Cell-to-Cell Communication Model
The Cell-to-Cell Communication Model, which is a combination of the two hypotheses discussed above, postulates that circadian clocks regulate some pathways involved in cell to cell communication in cyanobacteria such that individual cells can cooperate as a group in order to adapt to the environment and/or optimally utilize a limiting resource [3]. This hypothesis postulates that when the circadian clock is disrupted or is not optimally entrained to the environment, cells do not effectively communicate due to the absence of proper circadian regulation. Under these conditions, cells with mutations in the circadian clock compete as individuals with WT cells that can act as a group, and thus are at a competitive disadvantage in utilizing a limited resource.
Quorum sensing is recognized as a mechanism by which many bacterial species communicate and cooperate [26]. Bacterial species that engage in quorum sensing secrete signaling molecules called autoinducers (AI), and the detection of the AI allows cells to switch between two distinct patterns of gene expression, depending upon cell density. When the cell density is high and AI reaches a threshold concentration, individual cells cooperate with others such that the entire population turns on a gene expression mode that triggers biological activity, for example the formation of a biofilm or virulence factor production [26].
To date, there are only a few cyanobacterial species in which quorum-sensing has been reported [27, 28]. One of the AIs, N-octanoyl homoserine lactone (C8-AHL), was found in Gloeothece sp. PCC6909 cultures, and 43 genes were expressed differently in response to C8-AHL treatment. It was suggested that this quorum sensing may mediate the formation of a biofilm [28]. Another cyanobacterium, Trichodesmium consortia, was reported to respond to quorum-sensing molecules called acylated homoserine lactones (AHLs) that are produced by epibionts attached to its surface, and colonies of Trichodesmium that were treated by AHLs doubled their activity of alkaline phosphatases, which are enzymes used by epibionts in the acquisition of phosphate from dissolved-organic phosphorus molecules [28]. Homologs of luxO and luxU, genes that encode components of the quorum sensing pathway, are present in the cyanobacterial species Synechocystis sp. PCC 6803 [29]. When quorum sensing genes from Vibrio harveyi [26] were used to search the genome sequence of S. elongatus, potential homologs were identified; the homologs identified included the AI receptor cqsS [30], and cqsA (an enzyme involved in AI synthesis [30]), as well as the signal transduction pathway component, luxO [26] (Table 2). In addition, a homolog of aphA, the gene that encodes the transcription factor that is a master regulator of the quorum sensing pathway and is active at low cell density (LCD) in V. harveyi [31], is also present in the S. elongatus genome. Taken together, the data from this bioinformatics approach suggest that S. elongatus may be capable of quorum sensing with clock-enhanced reproductive fitness mediated by this form of cell-to-cell communication. This possibility could be tested by knocking out these potential homologs.
Table 2.
| QS gene in V. harveyi [26] |
Function [26] | Potential Homologs in S.elongatus | E-value |
|---|---|---|---|
| LuxN | Receptor | periplasmic sensor hybrid histidine kinase | 4e-22 |
| LuxPQ | Receptor | N/A | N/A |
| CqsS | Receptor | periplasmic sensor hybrid histidine kinase | 8e-22 |
| LuxM | AI* synthesis | N/A | N/A |
| LuxS | AI* synthesis | N/A | N/A |
| CqsA | AI* synthesis | 8-amino-7-oxononanoate synthase | 6e-23 |
| LuxO | components of the AI signaling pathway | nitrogen assimilation regulatory protein | 2e-14 |
| LuxU | components of the AI signaling pathway | N/A | N/A |
| LuxR | Transcriptional Regulator at HCD | N/A | N/A |
| AphA | Transcriptional Regulator at LCD | histone deacetylase/AcuC/AphA family protein-like | 5e-67 |
AI: autoinducer
Future Directions
Competition experiments have demonstrated the adaptive value of the circadian clock in cyanobacteria. It is clear that the circadian clock enhances the fitness of cyanobacterial cells in light/dark cycles, and that this enhancement in reproductive fitness is greatest when the circadian clock resonates with the environmental cycle. In contrast, the circadian clock in cyanobacteria provides little or no reproductive advantage under constant conditions, indicating that the adaptive value conferred by circadian clock is an extrinsic rather than intrinsic property [11, 12].
To date, the underlying mechanism by which the circadian clock enhances reproductive fitness remains elusive. Although several models have been proposed and tested, each has some evidence that supports it and none can be excluded at this time. Each of the models discussed here will be examined further using a complementary approach. These include alternative culture methods aimed at accurately detecting small (but significant) differences in growth rates between wild-type and clock mutant strains. These small differences in growth rate could explain the competition outcomes. In addition, a genetic approach guided by bioinformatics and mathematical modeling could also be used to identify genes involved in the pathway and define the mechanism of fitness enhancement.
Acknowledgements
We thank Dr. David McCauley and Dr. Doug Taylor for helpful suggestions and statistical advice concerning the population biology experiments. We are grateful for the support and assistance of current and former members of the Johnson Laboratory. Grant support for these experiments came from the USA National Institutes of Health, specifically the National Institute of General Medical Sciences (GM067152 and GM088595 to CHJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson CH, Elliott J, Foster R, Honma K, Kronauer R. Fundamental Properties of Circadian Rhythms. In: Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological timekeeping. Sunderland: Sinauer; 2004. pp. 67–105. [Google Scholar]
- 2.Edmunds LN., Jr Chronobiology at the cellular and molecular levels: models and mechanisms for circadian timekeeping. Am. J. Anat. 1983;168:389–431. doi: 10.1002/aja.1001680404. [DOI] [PubMed] [Google Scholar]
- 3.Woelfle MA, Johnson CH. The adaptive value of the circadian clock system in cyanobacteria. In: Ditty JL, Mackey SR, Johnson CH, editors. Bacterial circadian programs. New York: Springer-Verlag LLC; 2009. pp. 207–224. [Google Scholar]
- 4.DeCoursey PJ, Walker JK, Smith SA. A circadian pacemaker in free-living chipmunks: essential for survival. J. Comp. Physiol. A. 2000;186:169–180. doi: 10.1007/s003590050017. [DOI] [PubMed] [Google Scholar]
- 5.Klarsfeld A, Rouyer F. Effects of circadian mutations and LD periodicity on the life span of Drosophila melanogaster. J. Biol. Rhythms. 1998;13:471–478. doi: 10.1177/074873098129000309. [DOI] [PubMed] [Google Scholar]
- 6.Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM. Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 2002;99:2134–2139. doi: 10.1073/pnas.032426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 8.Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U S A. 2012;109:4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futuyma DJ. Evolutionary Biology. Sinauer Associates. (third ed.) 1998 [Google Scholar]
- 11.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 12.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. U S A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 14.Kondo T, Tsinoremas NF, Golden SS, Johnson CH, Kutsuna S, Ishiura M. Circadian clock mutants of cyanobacteria. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 15.Johnson CH. Testing the adaptive value of circadian systems. Methods Enzymol. 2005;393:818–837. doi: 10.1016/S0076-6879(05)93043-7. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, et al. Circadian orchestration of gene expression in cyanobacteria. Genes. Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 17.Hellweger FL. Resonating circadian clocks enhance fitness in cyanobacteria in silico. Ecol. Model. 2010;221:1620–1629. [Google Scholar]
- 18.Harder W, Kuenen JG. A review: Microbial selection in continuous culture. J. Appl. Bacteriol. 1977;43:1–24. doi: 10.1111/j.1365-2672.1977.tb00717.x. [DOI] [PubMed] [Google Scholar]
- 19.Gleason FK, Paulson JL. Site of action of the natural algicide, cyanobacterin, in the blue-green alga Synechococcus sp. Arch. Microbiol. 1984:273–277. [Google Scholar]
- 20.Zaccaro MC, Kato A, Zulpa G, Storni MM, Steyerthal N, Lobasso K, et al. Bioactivity of Scytonema hofmanni (Cyanobacteria) in Lilium alexandrae in vitro propagation. Electron. J. Biotechn. 2006;9:211–214. [Google Scholar]
- 21.Koksharova OA, Klint J, Rasmussen U. The protein map of Synechococcus sp. PCC 7942 - the first overlook. Quant. Biol. 2005 arXiv:q-bio/0510013. [PubMed] [Google Scholar]
- 22.Roussel MR, Gonze D, Goldbeter A. Modeling the differential fitness of cyanobacterial strains whose circadian oscillators have different free-running periods: comparing the mutual inhibition and substrate depletion hypotheses. J. theor. Biol. 2000;205:321–340. doi: 10.1006/jtbi.2000.2072. [DOI] [PubMed] [Google Scholar]
- 23.Gonze D, Roussel MR, Goldbeter A. A model for the enhancement of fitness in cyanobacteria based on resonance of a circadian oscillator with the external light-dark cycle. J. theor. Biol. 2002;214:577–597. doi: 10.1006/jtbi.2001.2476. [DOI] [PubMed] [Google Scholar]
- 24.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 26.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharif DI, Gallon J, Smith CJ, Dudley E. Quorum sensing in Cyanobacteria: N-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium Gloeothece PCC6909. Isme. J. 2008;2:1171–1182. doi: 10.1038/ismej.2008.68. [DOI] [PubMed] [Google Scholar]
- 28.Van Mooy BA, Hmelo LR, Sofen LE, Campagna SR, May AL, Dyhrman ST, et al. Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 2012;6:422–429. doi: 10.1038/ismej.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Daniel R, Wagner-Dobler I, Zeng AP. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 2004;4:36–47. doi: 10.1186/1471-2148-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011;79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in Vibrios. Genes. Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]